Comparative Chloroplast Genomics of Litsea Lam. (Lauraceae) and Its Phylogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sampling and DNA Extraction of L. auriculata
2.2. Illumina Paired-End Sequencing, De Novo Assembly, and Annotation of the Chloroplast Genome of L. auriculata
2.3. Comparative Chloroplast Genome Analysis of Litsea
2.4. Mining of cp Microsatellite Markers and Hypervariable Regions of Litsea
2.5. Phylogenetic Analysis
3. Results and Discussion
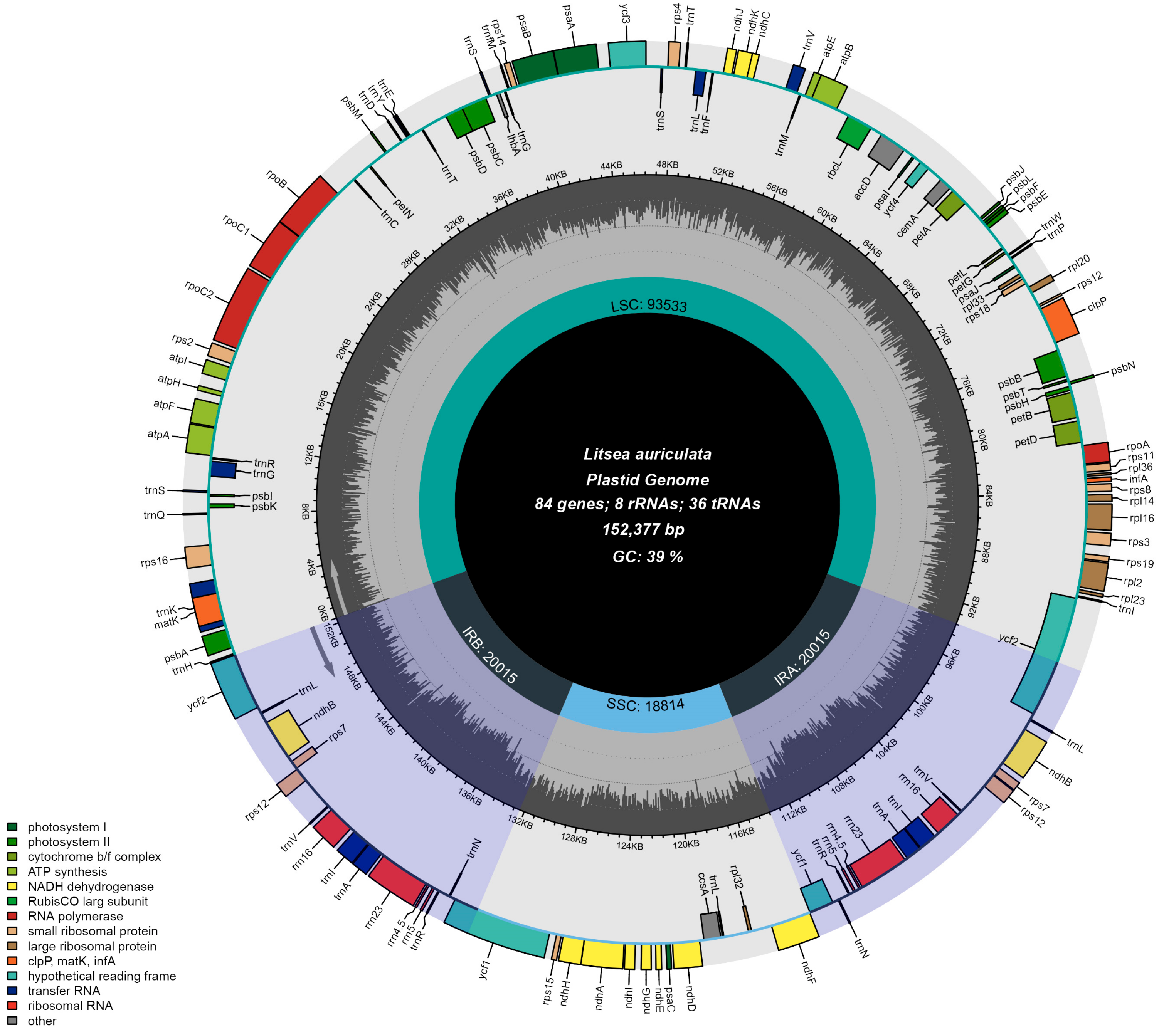
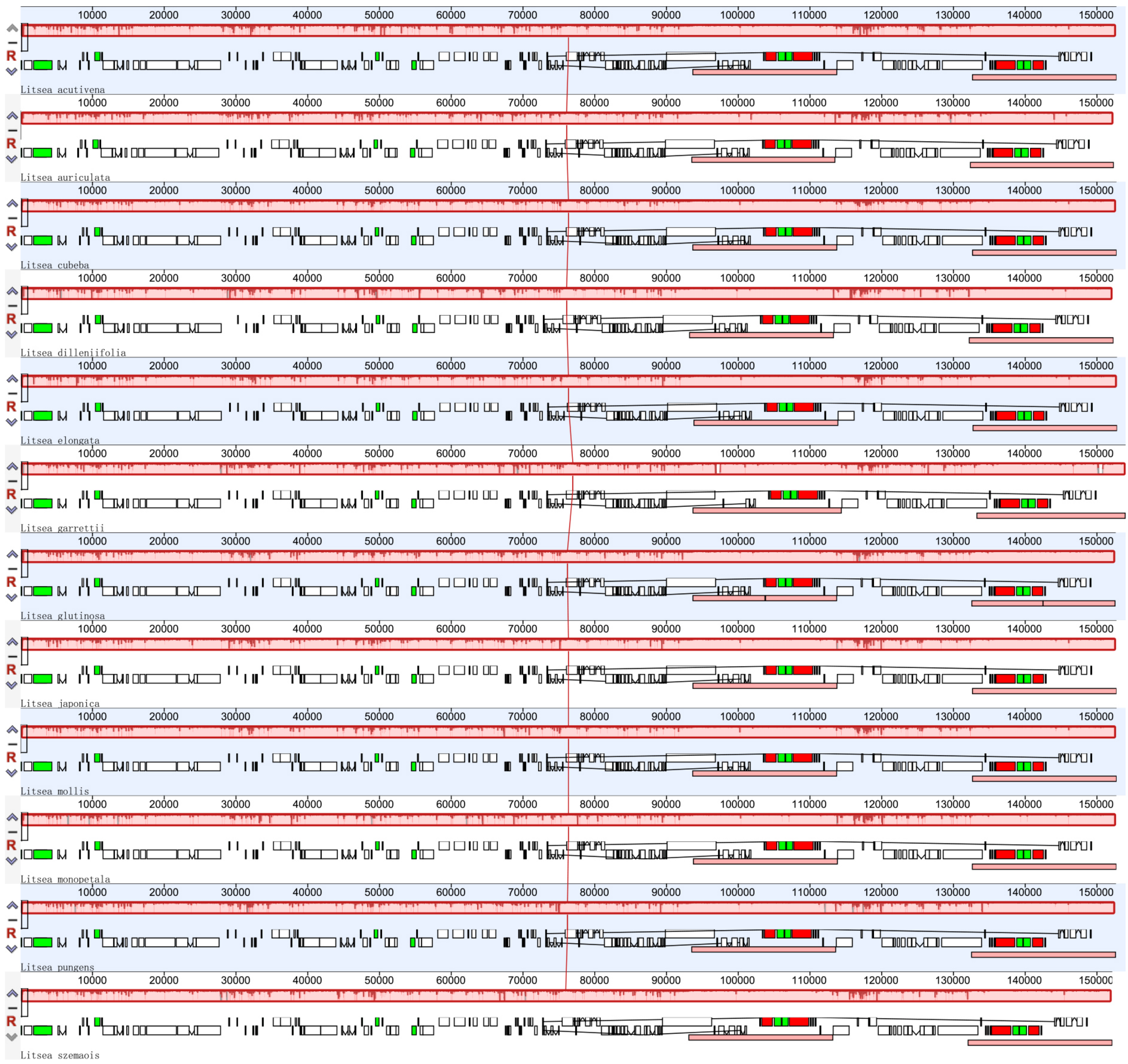
3.1. Conservation of Litsea Chloroplast Genomes
3.2. Enrichment of Chloroplast DNA Genetic Resources of Litsea
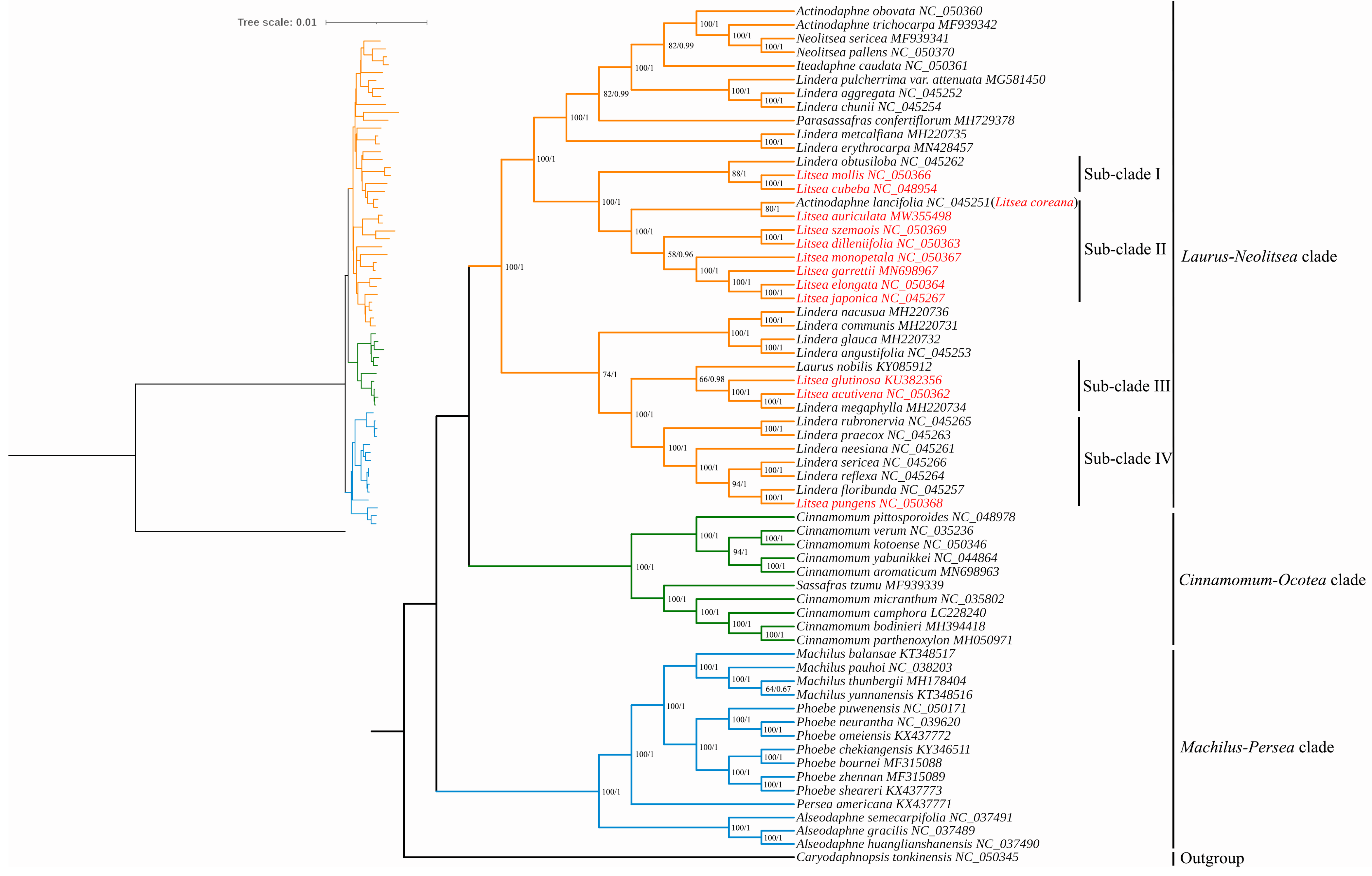
3.3. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chanderbali, A.S.; Werff, H.V.D.; Renner, S.S. Phylogeny and historical biogeography of Lauraceae: Evidence from the chloroplast and nuclear genomes. Ann. Mo. Bot. Gard. 2001, 88, 104–134. [Google Scholar] [CrossRef]
- Renner, S.S. Circumscription and phylogeny of the Laurales: Evidence from molecular and morphological data. Am. J. Bot. 1999, 86, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Christophel, D.C.; Conran, J.G.; Li, H.W. Phylogenetic relationships within the ‘core’ Laureae (Litsea complex, Lauraceae) inferred from sequences of the chloroplast gene matK and nuclear ribosomal DNA ITS regions. Plant Syst. Evol. 2004, 246, 19–34. [Google Scholar] [CrossRef]
- Rohwer, J.G. Toward a phylogenetic classification of the Lauraceae: Evidence from matK sequences. Syst. Bot. 2000, 25, 60–71. [Google Scholar] [CrossRef]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Liu, B.; Yao, X.; Jin, J.; Michael, P.; Yang, J.B.; Corlett, R.T. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids. Genome Biol. Evol. 2017, 9, 2354–2364. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Liu, B.; Corlett, R.T. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Rohwer, J.G.; Rudolph, B. Jumping genera: The phylogenetic positions of Cassytha, Hypodaphnis, and Neocinnamomum (Lauraceae) based on different analyses of trnK intron sequences. Ann. Mo. Bot. Gard. 2005, 92, 153–178. [Google Scholar]
- Chen, Y.C.; Li, Z.; Zhao, Y.X.; Gao, M.; Wang, J.Y.; Liu, K.W.; Wang, X.; Wu, L.W.; Jiao, Y.L.; Xu, Z.L.; et al. The Litsea genome and the evolution of the Laurel family. Nat. Commun. 2020, 11, 16–75. [Google Scholar] [CrossRef]
- Li, H.W. Lauraceae. In Flora of China; Science Press: Beijing, China, 1982; Volume 21, pp. 1–463. [Google Scholar]
- Dai, J.; Sun, B.N.; Xie, S.P.; Lin, Z.C.; Wen, W.W.; Wu, J.Y. Cuticular microstructure of Litsea cf. chunii from the Pliocene of Tengchong, Yunnan province. J. Lanzhou Univ. Nat. Sci. 2010, 46, 22–28. [Google Scholar]
- Huang, L.L.; Sun, J.; Jin, J.H.; Quan, C.; Alexei, A.O. Litseoxylon gen. nov. (Lauraceae): The most ancient fossil angiosperm wood with helical thickenings from southeastern Asia. Rev. Palaeobot. Palynol. 2018, 258, 223–233. [Google Scholar] [CrossRef]
- Li, H.W. The origin and evolution of Litsea genera group (Laureae) in Lauraceae. Acta Bot. Yunnanica 1995, 17, 251–254. [Google Scholar]
- Huang, X.W.; Feng, Y.C.; Huang, Y. Potential cosmetic application of essential oil extracted from Litsea cubeba fruits from China. J. Essent. Oil Res. 2013, 25, 112–119. [Google Scholar] [CrossRef]
- Su, Y.C.; Ho, C.L. Essential oil compositions and antimicrobial activities of various parts of Litsea cubeba from Taiwan. Nat. Prod. Commun. 2016, 11, 515–518. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Meile, J.C.; Lebrun, M.; Caruso, D.; Chu, K.S.; Sarter, S. Litsea cubeba leaf essential oil from Vietnam: Chemical diversity and its impacts on antibacterial activity. Lett. Appl. Microbiol. 2018, 66, 207–214. [Google Scholar] [CrossRef]
- Hasan, H.; Azad, M.S.A.; Islam, M.Z.; Rahman, S.M.; Islam, M.R.; Rahman, S.; Rahmatullah, M. Antihyperglycemic activity of methanolic extract of Litsea monopetala (Roxb.) Pers. leaves. Adv. Nat. Appl. Sci. 2014, 8, 51–55. [Google Scholar]
- Kim, C.S.; Lee, I.S.; Lee, Y.M.; Sohn, E.; Jo, K.; Kim, J.H.; Kim, J.S. Extract of Litsea japonica ameliorates blood–retinal barrier breakdown in db/db mice. Endocrine 2014, 46, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.T.; Wang, X.; Chen, Y.C.; Wang, Y.D.; Song, D.F.; Gu, Q. A natural biopreservative: Antibacterial action and mechanisms of Chinese Litsea mollis Hemsl. extract against Escherichia coli DH5α and Salmonella spp. J. Dairy Sci. 2019, 102, 9663–9673. [Google Scholar] [CrossRef] [PubMed]
- Bhuinya, T.; Singh, P.; Mukherjee, S.K. An account of the species of Litsea Lam. (Lauraceae) endemic to India. Bangladesh J. Plant Taxon. 2010, 17, 183–191. [Google Scholar] [CrossRef]
- Richter, H.G. Anatomie des sekundaren xylems und der rinde der Lauraceae. Sonderbd. Naurwissenschaftlichen Ver. Hambg. 1981, 5, 1–148. [Google Scholar]
- Li, H.W. Parallel evolution in Litsea and Lindera of Lauraceae. Acta Bot. Yunnanica 1985, 7, 129–135. [Google Scholar]
- Raj, B.; Werff, H.V.D. A contribution to the pollen morphology of Neotropical Lauraceae. Ann. Mo. Bot. Gard. 1988, 75, 130–167. [Google Scholar] [CrossRef]
- Werff, H.V.D.; Richter, H.G. Toward an improved classification of Lauraceae. Ann. Mo. Bot. Gard. 1996, 83, 409–418. [Google Scholar] [CrossRef]
- Fijridiyanto, I.A.; Murakami, N. Phylogeny of Litsea and related genera (Laureae–Lauraceae) based on analysis of rpb2 gene sequences. J. Plant Res. 2009, 122, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Kim, C.; Kim, J.H. Insights into phylogenetic relationships and genome evolution of subfamily Commelinoideae (Commelinaceae Mirb.) inferred from complete chloroplast genomes. BMC Genom. 2021, 22, 231. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xie, X.N.; Li, F.; Tian, E.W.; Chao, Z. Chloroplast genomes of two Mediterranean Bupleurum species and the phylogenetic relationship inferred from combined analysis with East Asian species. Planta 2021, 253, 81–84. [Google Scholar] [CrossRef]
- Ping, J.; Feng, P.; Li, J.; Zhang, R.; Su, Y.; Wang, T. Molecular evolution and SSRs analysis based on the chloroplast genome of Callitropsis funebris. Ecol. Evol. 2021, 11, 4786–4802. [Google Scholar] [CrossRef]
- Tian, X.Y.; Ye, J.W.; Song, Y. Plastome sequences help to improve the systematic position of trinerved Lindera species in the family Lauraceae. PeerJ 2019, 7, e7662. [Google Scholar] [CrossRef]
- Zhao, M.L.; Song, Y.; Ni, J.; Yao, X.; Tan, Y.H.; Xu, Z.F. Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae). Sci. Rep. 2018, 8, 8844. [Google Scholar] [CrossRef]
- Geng, Q.F.; Sun, L.; Zhang, P.H.; Wang, Z.S.; Qiu, Y.X.; Liu, H.; Lian, C.L. Understanding population structure and historical demography of Litsea auriculata (Lauraceae), an endangered species in east China. Sci. Rep. 2017, 7, 17343. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Yatsuhashi, S.; Yasuda, T.; Sato, M.; Sakai, E.; Xiao, C.; Murata, H.; Murata, J. A new amide from the leaves and twigs of Litsea auriculata. J. Nat. Med. 2009, 63, 331–334. [Google Scholar] [CrossRef]
- Fu, L.G.; Jin, J.M. China Plant Red Data Book: Rare and Endangered Plants; Science Press: Beijing, China, 1992; Volume 1, pp. 350–351. [Google Scholar]
- Deng, Y.; Pu, F.G. Conservation actualities of rare and endangered plants in Anhui Tianma National Nature Reserve and coping strategies. Anhui For. Sci. Technol. 2015, 41, 35–38. [Google Scholar]
- Cheng, H.Y.; Hu, X.L.; Zhou, M.Y.; Li, M.Q. Seedling techniques by sowing of Litsea auriculata. Anhui For. Sci. Technol. 2004, 4, 33–34. [Google Scholar]
- Wang, F.Z.; Xie, F.; Zhan, M.D.; Yuan, S.H. Preliminary study on Litsea auriculata Chien et Cheng tree twig cottage under full sunshine and automatic spray. J. Henan For. Sci. Technol. 2011, 31, 7–17. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Jian, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.J.; Moore, M.J.; Li, D.Z.; Yi, T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.Y.; Poczai, P.; Hyvonen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef] [PubMed]
- Chris, M.; Michael, B.; Schwartz, J.R.; Alexander, P.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Inna, D. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The codon adaptation index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- John, F.P. Analysis of Codon Usage. Ph.D. Thesis, The University of Nottingham, Nottingham, UK, 1999. [Google Scholar]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.B.; Yin, J.L.; Guo, H.Y.; Zhang, Y.Y.; Xiao, W.; Sun, C.; Wu, J.Y.; Qu, X.B.; Yu, J.; Wang, X.M.; et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front. Plant Sci. 2015, 5, 696. [Google Scholar] [CrossRef]
- Li, P.; Lu, R.S.; Xu, W.Q.; Ohi-Toma, T.; Cai, M.Q.; Qiu, Y.X.; Cameron, K.M.; Fu, C.X. Comparative genomics and phylogenomics of East Asian tulips (Amana, Liliaceae). Front. Plant Sci. 2017, 8, 451. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, F.; Yang, D.G.; Li, W.; Zhou, X.J.; Pei, X.Y.; Liu, Y.G.; He, K.L.; Zhang, W.S.; Ren, Z.Y.; et al. Comparative chloroplast genomics of Gossypium species: Insights into repeat sequence variations and phylogeny. Front. Plant Sci. 2018, 9, 376. [Google Scholar] [CrossRef]
- She, R.; Zhao, P.; Zhou, H.; Yue, M.; Zhang, S. Complete chloroplast genomes of Liliaceae (s.l.) species: Comparative genomic and phylogenetic analyses. Nord. J. Bot. 2020, 38, e02477. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Shi, E.; Yang, Z.P.; Geng, Q.F.; Qiu, Y.X.; Wang, Z.S. Development and application of genomic resources in an endangered palaeoendemic tree, Parrotia subaequalis (Hamamelidaceae) from eastern China. Front. Plant Sci. 2018, 9, 246. [Google Scholar] [CrossRef]
- Khakhlova, O.; Bock, R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006, 46, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Dong, W.; Bing, L.; Chao, X.; Yao, X.; Gao, J.; Corlett, R.T. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front. Plant Sci. 2015, 6, 662. [Google Scholar] [CrossRef]
- Song, Y.; Yao, X.; Tan, Y.; Gan, Y.; Corlett, R.T. Complete chloroplast genome sequence of the avocado: Gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can. J. For. Res. 2016, 46, 1293–1301. [Google Scholar] [CrossRef]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.F.; Li, L.; Werff, H.V.D.; Li, H.W.; Rohwer, J.H.; Crayn, D.M.; Meng, H.H.; Merwe, M.V.D.; Conran, J.H.; Li, J. Origins and evolution of cinnamon and camphor: A phylogenetic and historical biogeographical analysis of the Cinnamomum group (Lauraceae). Mol. Phylogenet. Evol. 2016, 96, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Goodman, D.B.; Church, G.M.; Kosuri, S. Causes and effects of N-terminal codon bias in bacterial genes. Science 2013, 342, 475–479. [Google Scholar] [CrossRef]
- Xia, E.H.; Yao, Q.Y.; Zhang, H.B.; Jiang, J.J.; Zhang, L.P.; Gao, L.Z. CandiSSR: An efficient pipeline used for identifying candidate polymorphic SSRs based on multiple assembled sequences. Front. Plant Sci. 2016, 6, 1171. [Google Scholar] [CrossRef]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; de Pamphilis, C.W.; Leebens-Mack, J. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef]
- Pauwels, M.; Vekemans, X.; Godé, C.; Frérot, H.; Castric, V.; Saumitou-Laprade, P. Nuclear and chloroplast DNA phylogeog-raphy reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytol. 2012, 193, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [PubMed]
- Hinsinger, D.D.; Strijk, J.S. Toward phylogenomics of Lauraceae: The complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene 2017, 9, 71–79. [Google Scholar] [CrossRef]
- Rohwer, J.G.; Lj, J.; Rudolph, B.; Schmidt, S.A.; Werff, H.V.D.; Li, H.W. Is Persea (Lauraceae) monophyletic? Evidence from nuclear ribosomal ITS sequences. Taxon 2009, 58, 1153–1167. [Google Scholar] [CrossRef]
- Li, L.; Li, J.; Rohwer, J.G.; Werff, H.V.D.; Wang, Z.H.; Li, H.W. Molecular phylogenetic analysis of the Persea group (Lauraceae) and its biogeographic implications on the evolution of tropical and subtropical Amphi-Pacifi disjunctions. Am. J. Bot. 2011, 98, 1520–1536. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.Q.; Li, L.; Li, J.W.; Rohwer, J.G.; Li, H.W.; Li, J. Alseodaphnopsis: A new genus of Lauraceae based on molecular and morphological evidence. PLoS ONE 2017, 12, e0186545. [Google Scholar]
- Song, Y.; Yao, X.; Lu, B.; Tan, Y.H.; Corlett, R.T. Complete plastid genome sequences of three tropical Alseodaphne trees in the family Lauraceae. Holzforschung 2018, 72, 337–345. [Google Scholar] [CrossRef]
- Li, J.; Li, H.W. Advances in Lauraceae systematic research on the world scale. Acta Bot. Yunnanica 2004, 26, 1–11. [Google Scholar]
- Friedrich, F. Centralblatt für Sammlung und Veroffentlichung von Einzeldiagnosen neuer Pflanzen. Repert. Spec. Nov. Regni Veg. 1912, 10, 370. [Google Scholar]
- Zhang, Z.; He, Z.W.; Xu, S.H.; Li, X.N.; Guo, W.X.; Yang, Y.C.; Zhong, C.R.; Zhou, R.C.; Shi, S.H. Transcriptome analyses provide insights into the phylogeny and adaptive evolution of the mangrove fern genus. Acrostichum. Sci. Rep. 2016, 6, 35634. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Kelly, L.J.; McAllister, H.A.; Zohren, J.; Buggs, R.J.A. Resolving phylogeny and polyploid parentage using genus-wide genome-wide sequence data from birch trees. Mol. Phylogenet. Evol. 2021, 160, 107126. [Google Scholar] [CrossRef]


| Species | GenBank ID | Whole Sequence Length (bp) | Length of LSC Region (bp) | Length of IR Region (bp) | Length of SSC Region (bp) | Total GC Content (%) | Total Number of Genes | Total Number of CDS Genes | Total Number of tRNA Genes | Total Number of rRNA Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| L. acutivena | NC_050362 | 152,718 | 93,677 | 20,066 | 18,909 | 39.2 | 113 | 79 | 30 | 4 |
| L. auriculata | MW355498 | 152,377 | 93,533 | 20,015 | 18,814 | 39.2 | 113 | 79 | 30 | 4 |
| L. cubeba | NC_048954 | 152,725 | 93,674 | 20,064 | 18,923 | 39.2 | 113 | 79 | 30 | 4 |
| L. dilleniifolia | NC_050363 | 152,298 | 93,218 | 20,094 | 18,892 | 39.2 | 112 | 79 | 29 | 4 |
| L. elongata | NC_050364 | 152,793 | 93,827 | 20,066 | 18,834 | 39.1 | 113 | 79 | 30 | 4 |
| L. garrettii | MN698967 | 154,011 | 93,698 | 20,744 | 18,825 | 39.2 | 113 | 79 | 30 | 4 |
| L. glutinosa | KU382356 | 152,618 | 93,690 | 20,061 | 18,806 | 39.2 | 113 | 79 | 30 | 4 |
| L. japonica | NC_045267 | 152,718 | 93,697 | 20,066 | 18,889 | 39.1 | 113 | 79 | 30 | 4 |
| L. mollis | NC_050366 | 152,736 | 93,655 | 20,063 | 18,936 | 39.2 | 113 | 79 | 30 | 4 |
| L. pungens | NC_050368 | 152,655 | 93,520 | 20,131 | 18,873 | 39.2 | 113 | 79 | 30 | 4 |
| L. szemaois | NC_050369 | 152,132 | 93,119 | 20,090 | 18,833 | 39.2 | 113 | 79 | 30 | 4 |
| L.monopetala | NC_050367 | 152,705 | 93,758 | 20,074 | 18,799 | 39.2 | 113 | 79 | 30 | 4 |
| Groups of Genes | Names of Genes |
|---|---|
| Ribosomal RNAs | rrn4.5 (×2), rrn5 (×2), rrn16 (×2), rrn23 (×2) |
| Transfer RNAs | * trnA-UGC (×2), ^ trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, * trnG-UCC, trnH-GUG, trnI-CAU, * trnI-GAU (×2), * trnK-UUU, trnL-CAA (×2), * trnL-UAA, trnL-UAG, trnfM-CAU, trnM-CAU, trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), * trnV-UAC, trnW-CCA, trnY-GUA |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome | petA, * petB, * petD, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, * atpF, atpH, atpI |
| Rubisco | rbcL |
| NADH dehydrogenease | * ndhA, * ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| ATP-dependent protease subunit P | ** clpP |
| Chloroplast envelop membrane protein | cemA |
| Large units | * rpl2 (×2), rpl14, * rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Small units | rps2, rps3, rps4, rps7 (×2), rps8, rps11, ** rps12, rps14, rps15, * rps16, rps18, rps19 |
| RNA polymerase | rpoA, rpoB, * rpoC1, rpoC2 |
| Translational initiation factor | infA |
| Miscellaneous proteins | matK, accD, ccsA |
| Hypothetical proteins and conserved reading frame | ** ycf3, ycf4, ycf1, ycf2 |
| Pseudogene | ψycf1, ψycf2 |
| Species | SSR Numbers | P1 Loci (N) | P2 Loci (N) | P3 Loci (N) | P4 Loci (N) | P5 Loci (N) | P6 Loci (N) | Pc Loci (N) | LSC | SSC | IR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L. acutivena | 72 | 46 | 9 | 1 | 8 | – | – | 8 | 55 | 13 | 4 |
| L. auriculata | 80 | 55 | 9 | 1 | 5 | – | – | 10 | 62 | 14 | 4 |
| L. cubeba | 71 | 43 | 9 | 1 | 8 | – | 1 | 9 | 55 | 12 | 4 |
| L. dilleniifolia | 71 | 42 | 9 | 2 | 8 | – | – | 10 | 55 | 12 | 4 |
| L. elongata | 74 | 46 | 8 | 2 | 6 | – | – | 12 | 58 | 12 | 4 |
| L. garrettii | 75 | 46 | 9 | 1 | 7 | – | – | 12 | 58 | 13 | 4 |
| L. glutinosa | 72 | 45 | 11 | 1 | 7 | 1 | – | 7 | 57 | 9 | 6 |
| L. japonica | 72 | 44 | 7 | 1 | 6 | – | – | 14 | 56 | 12 | 4 |
| L. mollis | 67 | 41 | 8 | 1 | 7 | – | 1 | 9 | 53 | 10 | 4 |
| L. pungens | 73 | 48 | 9 | 2 | 6 | – | – | 8 | 58 | 11 | 4 |
| L. szemaois | 69 | 43 | 8 | 2 | 7 | – | – | 9 | 54 | 11 | 4 |
| L. monopetala | 80 | 49 | 9 | 1 | 6 | – | 2 | 13 | 58 | 16 | 6 |
| Total/ Percentage | 876 (100%) | 548 (62.56%) | 105 (11.99%) | 16 (1.83%) | 81 (9.25%) | 1 (0.11%) | 4 (0.46%) | 121 (13.81%) | 679 (77.51%) | 145 (16.55%) | 52 (5.94%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Tian, Y.; Tng, D.Y.P.; Zhou, J.; Zhang, Y.; Wang, Z.; Li, P.; Wang, Z. Comparative Chloroplast Genomics of Litsea Lam. (Lauraceae) and Its Phylogenetic Implications. Forests 2021, 12, 744. https://doi.org/10.3390/f12060744
Zhang Y, Tian Y, Tng DYP, Zhou J, Zhang Y, Wang Z, Li P, Wang Z. Comparative Chloroplast Genomics of Litsea Lam. (Lauraceae) and Its Phylogenetic Implications. Forests. 2021; 12(6):744. https://doi.org/10.3390/f12060744
Chicago/Turabian StyleZhang, Yunyan, Yongjing Tian, David Y. P. Tng, Jingbo Zhou, Yuntian Zhang, Zhengwei Wang, Pengfu Li, and Zhongsheng Wang. 2021. "Comparative Chloroplast Genomics of Litsea Lam. (Lauraceae) and Its Phylogenetic Implications" Forests 12, no. 6: 744. https://doi.org/10.3390/f12060744
APA StyleZhang, Y., Tian, Y., Tng, D. Y. P., Zhou, J., Zhang, Y., Wang, Z., Li, P., & Wang, Z. (2021). Comparative Chloroplast Genomics of Litsea Lam. (Lauraceae) and Its Phylogenetic Implications. Forests, 12(6), 744. https://doi.org/10.3390/f12060744






