Filamentous Fungi and Yeasts Associated with Mites Phoretic on Ips typographus in Eastern Finland
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Collection of Samples
2.2. DNA Extraction, PCR and Sequencing
2.3. Sequence Analyses and Fungal Identification
3. Results
3.1. Collection of Beetles and Mites
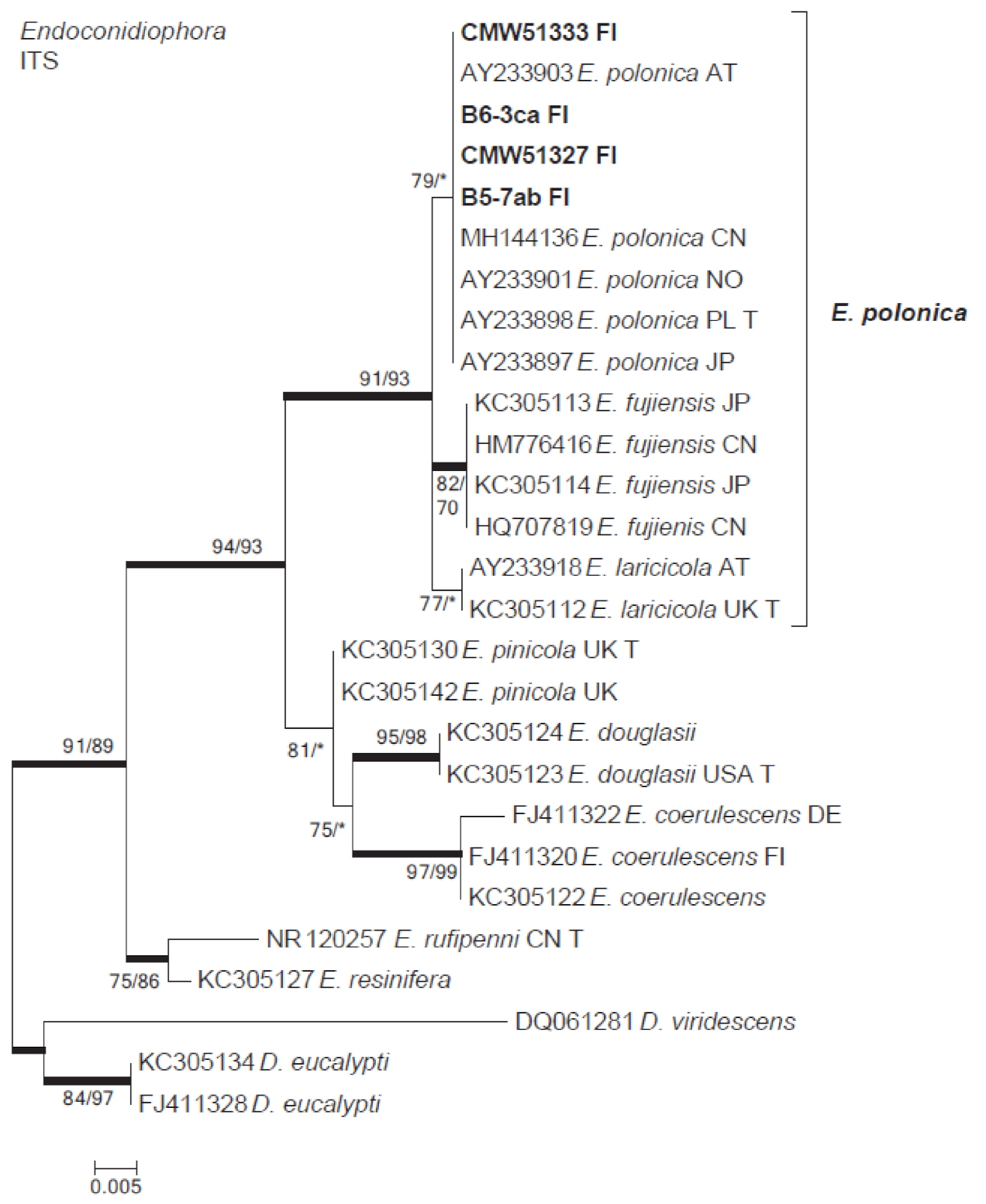
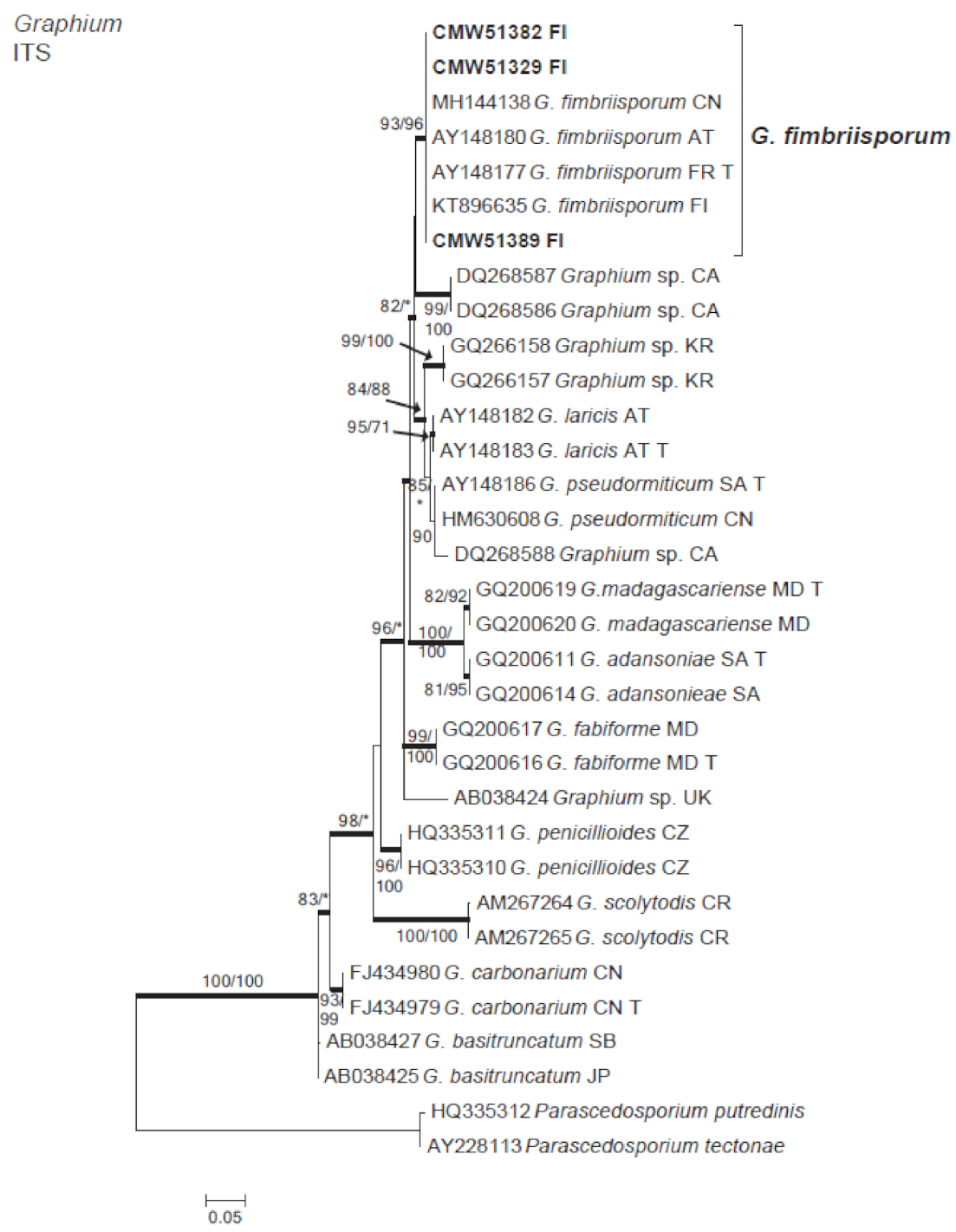
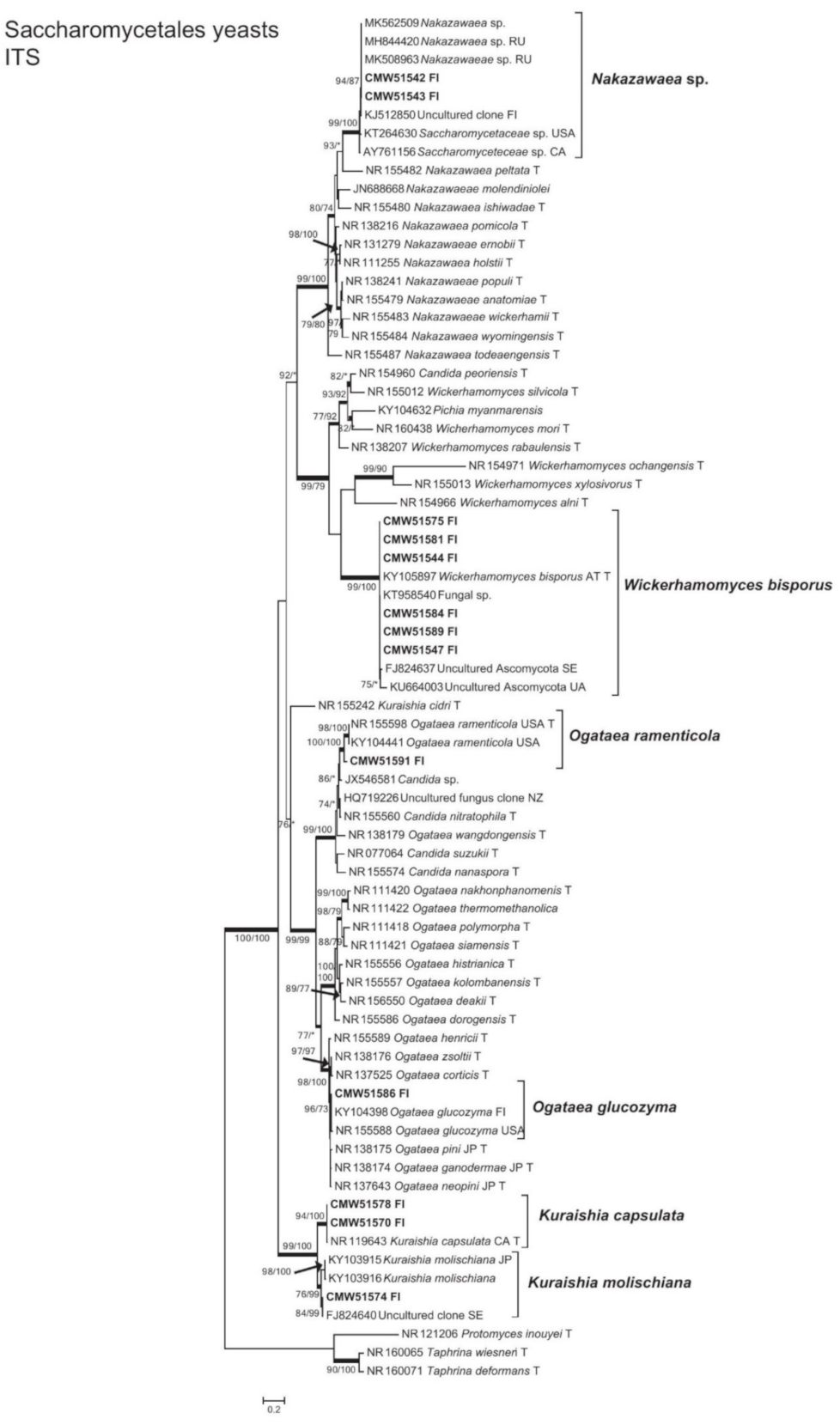
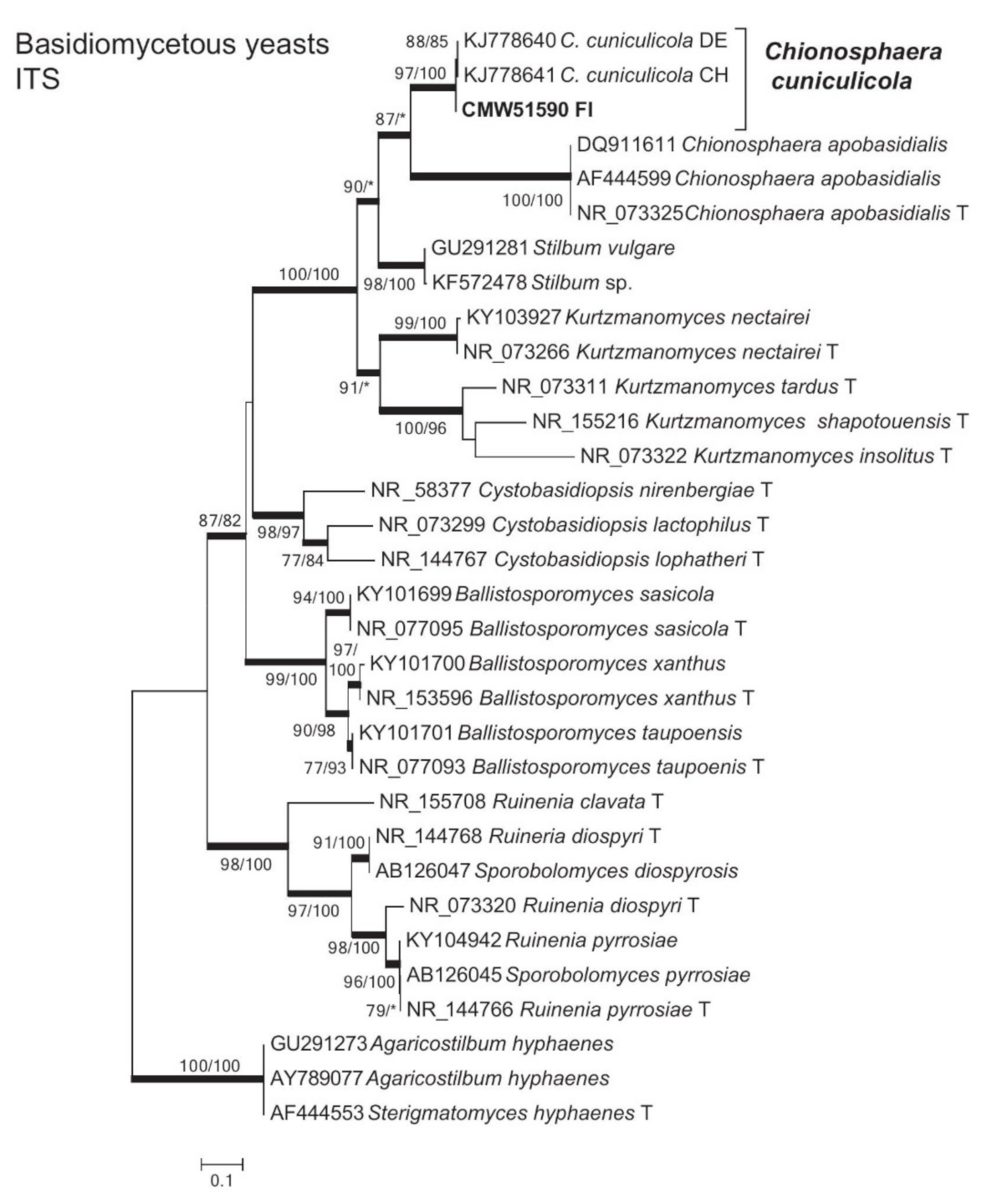
3.2. Isolation and Identification of Fungi
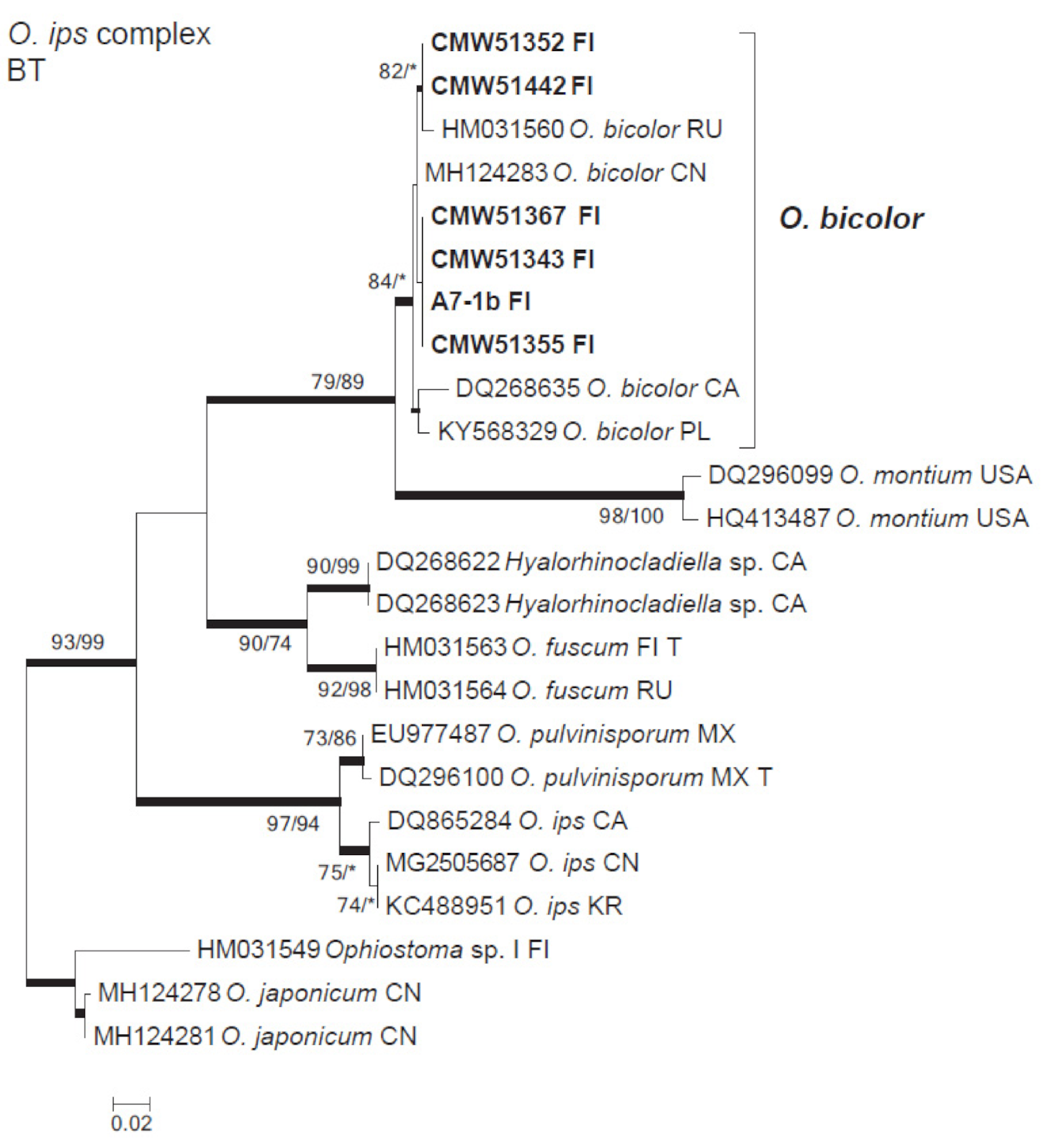
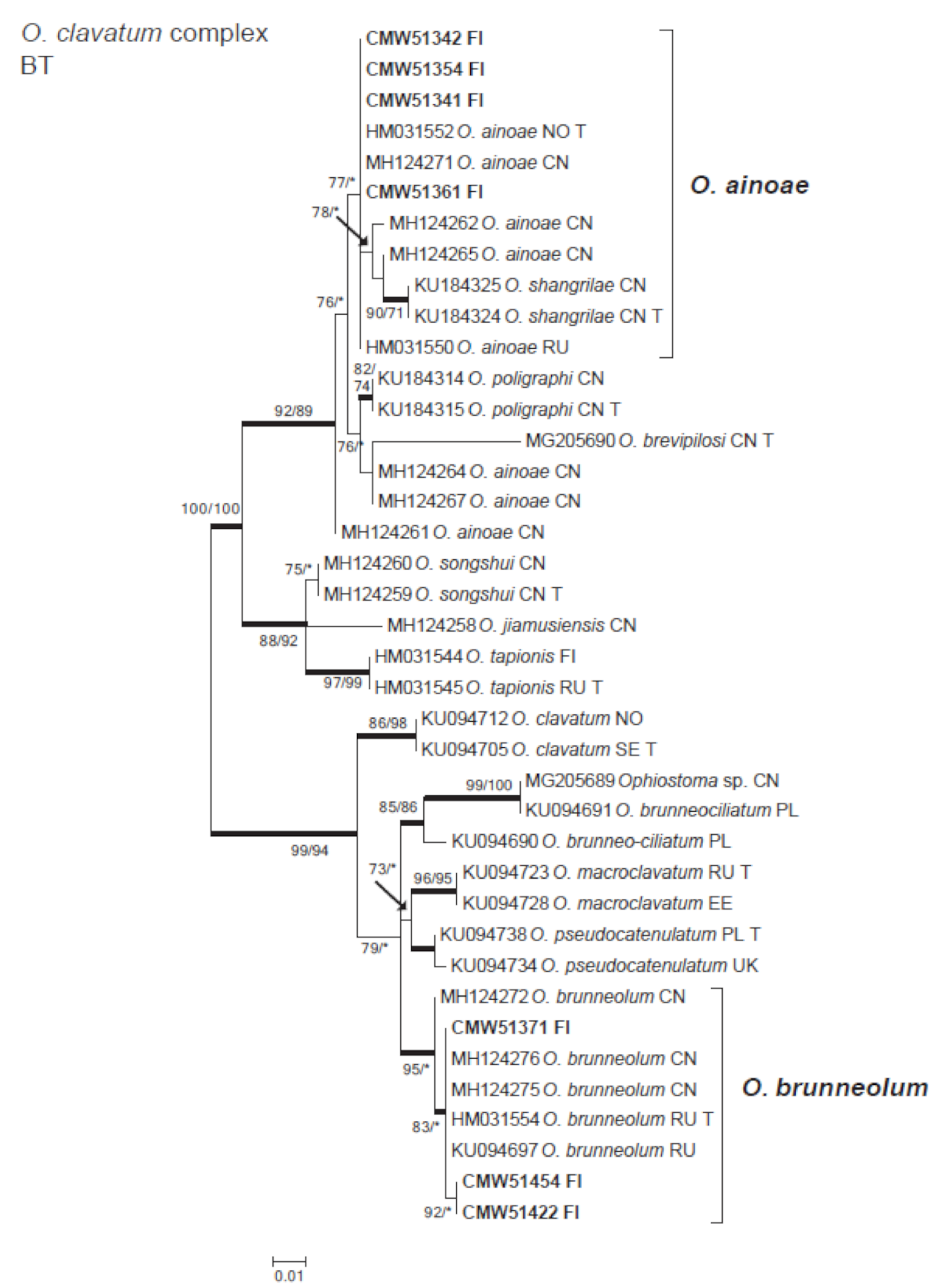
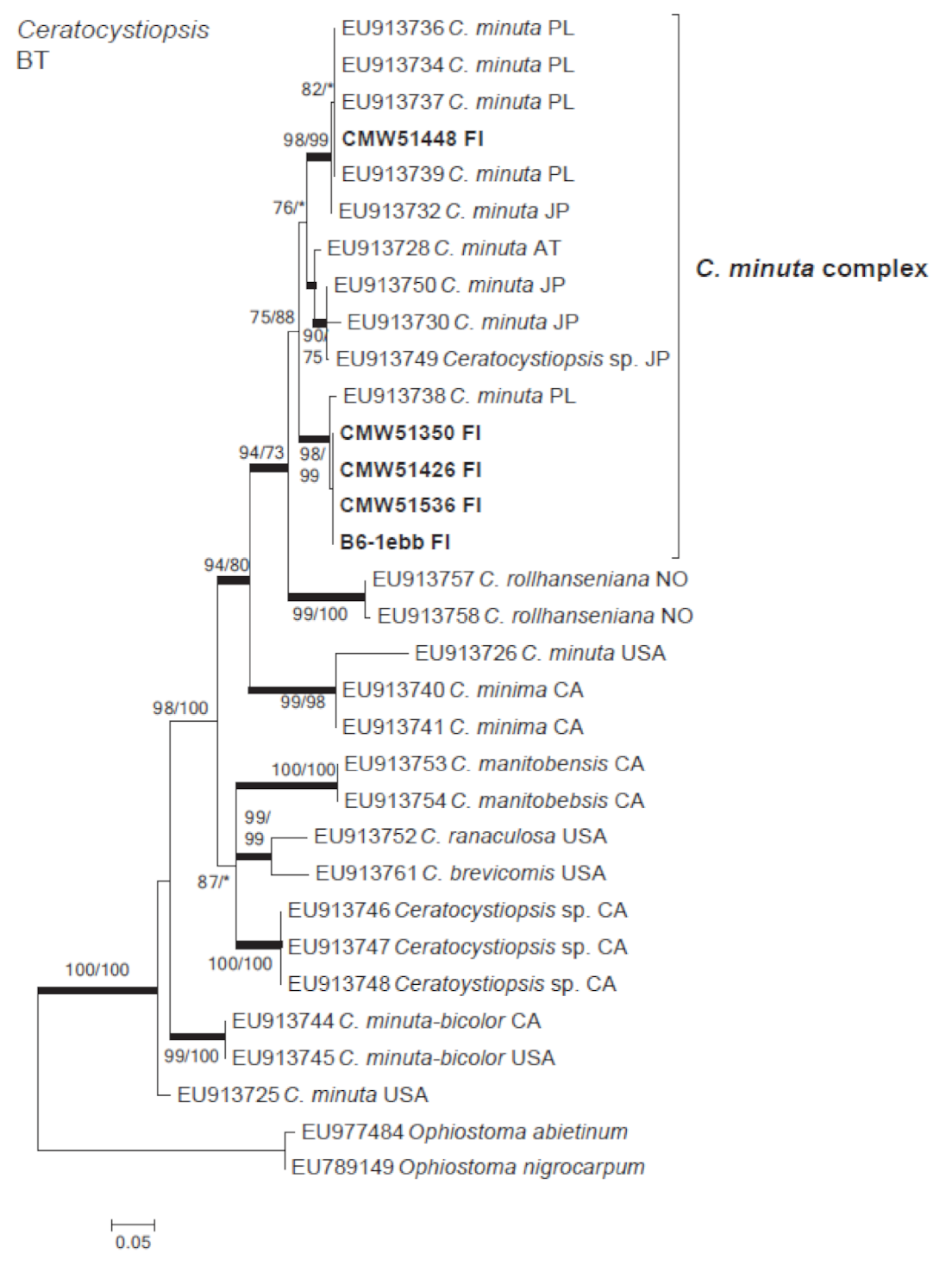
3.3. Ophiostomatales
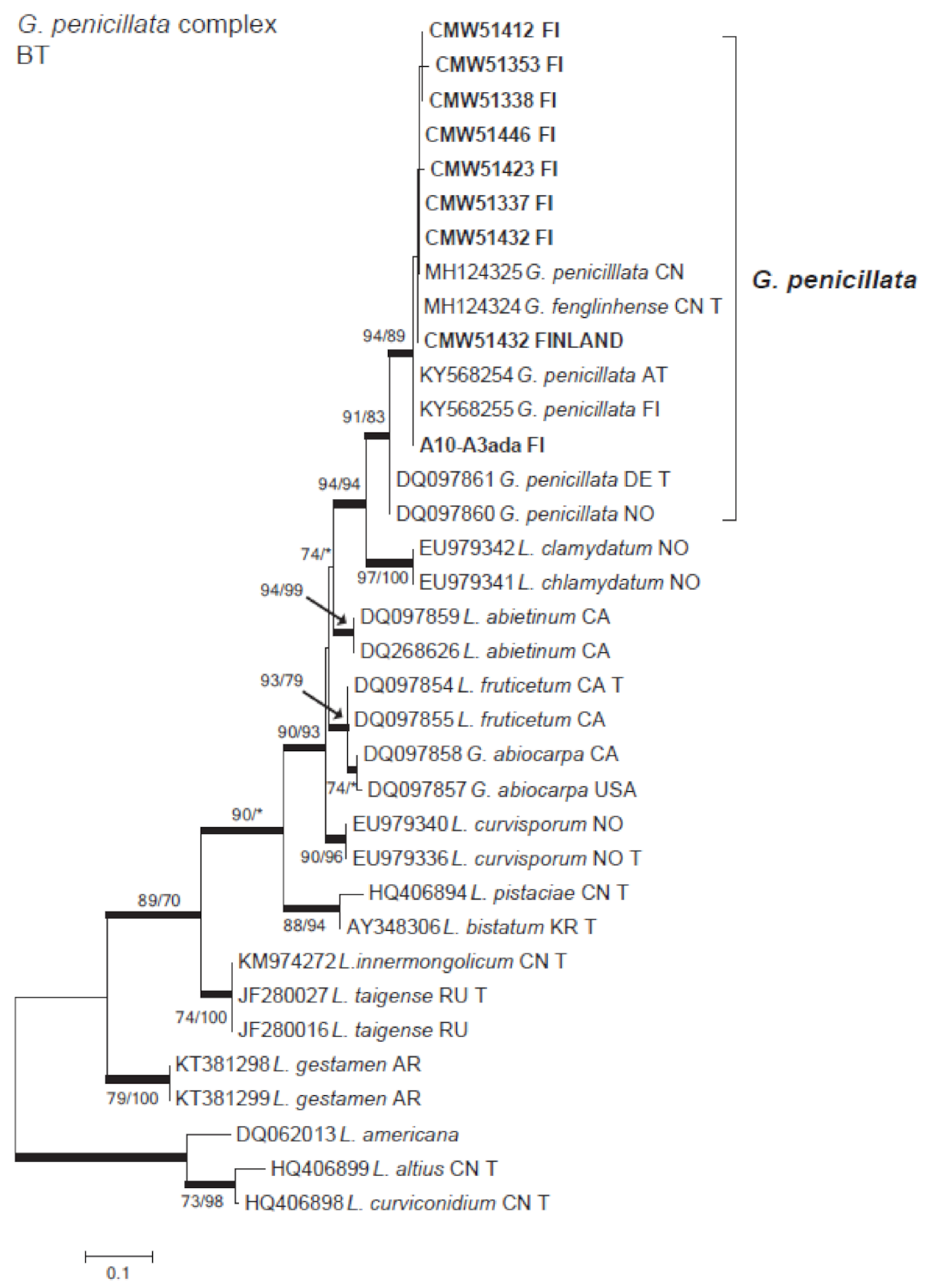
3.4. Microascales
3.5. Yeasts
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neuvonen, S.; Viiri, H. Changing climate and outbreaks of forest pest insects in a cold northern country, Finland. In The Interconnected Arctic—UArctic Congress 2016; Latola, K., Savela, H., Eds.; Springer Polar Sciences: Cham, Switzerland, 2017. [Google Scholar]
- Økland, B.; Netherer, S.; Marini, L. The Eurasian spruce bark beetle: The role of climate. In Climate Change and Insect Pests; Björkman, C., Niemelä, P., Eds.; CAB International: Wallingford, UK, 2015; pp. 202–219. [Google Scholar]
- Chang, R.; Duong, T.; Taerum, S.; Wingfield, M.; Zhou, X.; Yin, M.; De Beer, Z. Ophiostomatoid fungi associated with the spruce bark beetle Ips typographus, including 11 new species from China. Persoonia Mol. Phylogeny Evol. Fungi 2019, 42, 50–74. [Google Scholar] [CrossRef] [PubMed]
- Grucmanová, S.; Holuša, J. Nematodes associated with bark beetles, with focus on the genus Ips (Coleoptera: Scolytinae) in Central Europe. Acta Zool. Bulg. 2013, 65, 547–554. [Google Scholar]
- Penttinen, R.; Viiri, H.; Moser, J. The mites (Acari) associated with bark beetles in the Koli national park in Finland. Acarologia 2013, 53, 3–15. [Google Scholar] [CrossRef]
- Kandasamy, D.; Gershenzon, J.; Andersson, M.N.; Hammerbacher, A. Volatile Organic Compounds Influence the Interaction of the Eurasian Spruce Bark Beetle (Ips Typographus) with Its Fungal Symbionts. ISME J. 2019, 13, 1788–1800. [Google Scholar] [CrossRef]
- Seifert, K.A.; De Beer, Z.W.; Wingfield, M.J. (Eds.) The Ophiostomatoid Fungi: Expanding Frontiers; CBS: Utrecht, The Netherlands, 2013. [Google Scholar]
- Jankowiak, R. Fungi associated with Ips typographus on Picea abies in southern Poland and their succession into the phloem and sapwood of beetle infested trees and logs. For. Pathol. 2005, 35, 37–55. [Google Scholar] [CrossRef]
- Kirisits, T. Fungal associates of European bark beetles with special emphasis on the ophiostomatoid fungi. In Bark and Wood Boring Insects in Living Trees in Europe, a Synthesis; Lieutier, F., Day, K.R., Battisti, A., Gregoire, J.-C., Evans, H.F., Eds.; Springer: Amsterdam, The Netherlands, 2004; pp. 181–236. [Google Scholar]
- Linnakoski, R.; Mahilainen, S.; Harrington, A.; Vanhanen, H.; Eriksson, M.; Mehtätalo, L.; Pappinen, A.; Wingfield, M.J. Seasonal succession of fungi associated with Ips typographus beetles and their phoretic mites in an outbreak region of Finland. PLoS ONE 2016, 11, e0155622. [Google Scholar] [CrossRef]
- Viiri, H.; Lieutier, F. Ophiostomatoid fungi associated with the spruce bark beetle, Ips typographus, in three areas in France. Ann. For. Sci. 2004, 61, 215–219. [Google Scholar] [CrossRef][Green Version]
- Jankowiak, R.; Strzałka, B.; Bilański, P.; Kacprzyk, M.; Lukášová, K.; Linnakoski, R.; Matwiejczuk, S.; Misztela, M.; Rossa, R. Diversity of Ophiostomatales species associated with conifer-infesting beetles in Western Carpathians. Eur. J. For. Res. 2017, 136, 939–956. [Google Scholar] [CrossRef]
- Linnakoski, R.; De Beer, Z.W.; Ahtiainen, J.; Sidorov, E.; Niemelä, P.; Pappinen, A.; Wingfield, M.J. Ophiostoma spp. associated with pine- and spruce-infesting bark beetles in Finland and Russia. Persoonia 2010, 25, 72–93. [Google Scholar] [CrossRef]
- Linnakoski, R.; Jankowiak, R.; Villari, C.; Kirisits, T.; Solheim, H.; De Beer, Z.W.; Wingfield, M.J. The Ophiostoma clavatum species complex: A newly defined group in the Ophiostomatales including three novel taxa. Antonie van Leeuwenhoek 2016, 109, 987–1018. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, P.H.; Vega, F.E. Ecology and evolution of insect-fungus mutualisms. Annu. Rev. Entomol. 2020, 65. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Duong, T.A.; Taerum, S.J.; Wingfield, M.J.; Zhou, X.; de Beer, Z.W. Ophiostomatoid fungi associated with conifer-infesting beetles and their phoretic mites in Yunnan, China. MycoKeys 2017, 28, 19–64. [Google Scholar] [CrossRef]
- Chang, R.; Duong, T.A.; Taerum, S.J.; Wingfield, M.J.; Zhou, X.; De Beer, Z.W. Ophiostomatoid fungi associated with mites phoretic on bark beetles in Qinghai, China. IMA Fung. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Vissa, S.; Hofstetter, R.W. The role of mites in bark and ambrosia beetle-fungal interactions. In Insect Physiology and Ecology; Shields, V., Ed.; InTech: Rijeka, Croatia, 2017; pp. 135–156. [Google Scholar]
- Moser, J.C. Use of sporothecae by phoretic Tarsonemus mites to transport ascospores of coniferous bluestain fungi. Trans. Br. Mycol. Soc. 1985, 84, 750–753. [Google Scholar] [CrossRef]
- Lombardero, M.J.; Klepzig, K.D.; Moser, J.C.; Ayres, M.P. Biology, demography and community interactions of Tarsonemus (Acarina: Tarsonemidae) mites phoretic on Dendroctonus frontalis (Coleoptera: Scolytidae). Agric. For. Entomol. 2000, 2, 193–202. [Google Scholar] [CrossRef]
- Hofstetter, R.; Moser, J. The role of mites in insect-fungus associations. Annu. Rev. Entomol. 2014, 59, 537–557. [Google Scholar] [CrossRef]
- Siemaszko, W. Zespoly grzbow towarzyszacych kornikom polskim. Planta Pol. 1939, 7, 1–54. [Google Scholar]
- Davis, T.S. The ecology of yeasts in the bark beetle holobiont: A century of research revisited. Microb. Ecol. 2014, 69, 723–732. [Google Scholar] [CrossRef]
- Chakraborty, A.; Modlinger, R.; Ashraf, M.Z.; Synek, J.; Schlyter, F.; Roy, A. Core mycobiome and their ecological relevance in the gut of five Ips bark beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Kurtzman, C.D.; Fell, J.W.; Boekhout, T. (Eds.) The Yeasts: A Taxonomic Study; Elsevier: New York, NY, USA, 2011. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhiza and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–321. [Google Scholar]
- O’Donnel, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-S.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 3, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 9, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, N. Further analysts of the data by akaike’ s information criterion and the finite corrections. Commun. Stat. Theory Methos 1978, 1, 13–26. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, T.D. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP* 4.0; Phylogenetic analysis using parsimony (* and other methods); Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2; Program distributed by the author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rambaut, A. Figtree, a Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 20 January 2021).
- Gwiazdowicz, D.J.; Kamczyc, J.; Teodorowicz, E.; Błoszyk, J. Mite communities (Acari, Mesostigmata) associated with Ips typographus (Coleoptera, Scolytidae) in managed and natural Norway spruce stands in Central Europe. Cent. Eur. J. Biol. 2012, 7, 910–916. [Google Scholar] [CrossRef]
- Manu, M.; Poliză, D.; Onete, M. Comparative analysis of the phoretic mites communities (Acari: Mesostigmata) associated with Ips typographus from natural and planted Norway spruce stands—Romania. Rom. Biotech. Lett. 2018, 23. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Garnas, J.R.; Hajek, A.; Hurley, B.P.; de Beer, Z.W.; Taerum, S.J. Novel and Co-Evolved Associations between Insects and Microorganisms as Drivers of Forest Pestilence. Biol Invasions 2016, 18, 1045–1056. [Google Scholar] [CrossRef]
- Moser, J.C.; Perry, T.J.; Solheim, H. Ascospores hyperphoretic on mites associated with Ips Typographus. Mycol. Res. 1989, 93, 513–517. [Google Scholar] [CrossRef]
- Moser, J.C.; Perry, T.J.; Furuta, K. Phoretic mites and their hyperphoretic fungi associated with flying Ips typographus japonicus Niijima (Col., Scolytidae) in Japan. J. Appl. Entomol. 1997, 121, 425–428. [Google Scholar] [CrossRef]
- Solheim, H. Species of Ophiostomataceae isolated from Picea abies infested by the bark beetle Ips Typographus. Nord. J. Bot. 1986, 6, 199–207. [Google Scholar] [CrossRef]
- Savonmäki, S. Tärkeimmät Kaarnakuoriaisten Mäntyyn ja Kuuseen Levittämät Sinistäjäsienilajit. Master’s Thesis, Department of Plant Pathology, University of Helsinki, Helsinki, Finland, 1990. [Google Scholar]
- Ahtiainen, J. The Blue-Stain Fungi Associated with the Spruce Bark Beetle (Ips typographus L.) in Lake Vodla Area in Eastern Russia. Master’s Thesis, University of Joensuu, Joensuu, Finland, 2008. [Google Scholar]
- Plattner, A.; Kim, J.-J.; Reid, J.; Hausner, G.; Lim, Y.W.; Yamaoka, Y.; Breuil, C. Resolving taxonomic and phylogenetic incongruence within species Ceratocystiopsis minuta. Mycologia 2009, 101, 878–887. [Google Scholar] [CrossRef]
- Persson, Y.; Vasaitis, R.; Långström, B.; Öhrn, P.; Ihrmark, K.; Stenlid, J. Fungi vectored by the bark beetle Ips typographus following hibernation under the bark of standing trees and in the forest litter. Microb. Ecol. 2009, 58, 651–659. [Google Scholar] [CrossRef]
- Giordano, L.; Garbelotto, M.; Nicolotti, G.; Gonthier, P. Characterization of fungal communities associated with the bark beetle Ips typographus varies depending on detection method, location, and beetle population levels. Mycol. Prog. 2012, 12, 127–140. [Google Scholar] [CrossRef]
- Hui, F.-L.; Chen, L.; Chu, X.-Y.; Niu, Q.-H.; Ke, T. Wicherhamomyces mori sp. nov., an anamorphic yeast species found in the guts of wood-boring insect larvae. Int. J. Syst. Evol. Microbiol. 2013, 63, 1174–1178. [Google Scholar] [CrossRef]
- Ninomiya, S.; Mikata, K.; Kajimura, H.; Kawasaki, H. Two novel ascomycetous yeast species, Wickerhamomyces scolytoplatypi sp. nov. and Cyberlindnerea xylebori sp. nov., isolated from ambrosia beetle galleries. Int. J. Syst. Evol. Microbiol. 2013, 63, 2706–2711. [Google Scholar] [CrossRef]
- Leufvén, A.; Nehls, L. Quantification of different yeasts associated with the bark beetle, Ips typographus, during its attack on a spruce tree. Microb. Ecol. 1986, 12, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Leufvén, A.; Bergström, G.; Falsen, E. Interconversion of verbenols and verbenone by identified yeasts isolated from the spruce bark beetle Ips typographus. J. Chem. Ecol. 1984, 10, 1349–1361. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, R.; Begerow, D.; Oberwinkler, F. A new Chionosphaera species associated with conifer inhabiting bark beetles*. Mycol. Res. 2001, 105, 1403–1408. [Google Scholar] [CrossRef]
- Lou, Q.-Z.; Lu, M.; Sun, J.-. H Yeast diversity associated with invasive Dendroctonus valens killing Pinus tabuliformis in China using culturing and molecular methods. Microb. Ecol. 2014, 68, 397–415. [Google Scholar] [CrossRef] [PubMed]
| Species | Isolate no | Mite No. | GenBank Acc. No. | ||
|---|---|---|---|---|---|
| Personal 1 | CMW 2 | ITS | β-Tubulin | ||
| Ceratocystiopsis minuta (Siemaszko) H.P. Upadhyay & W.B. Kendr. | A10-1aba | 51,536 | A7-1d | MW345787 | |
| A7-1d | 51,426 | A7-1 | MW345786 | ||
| B3-1aab | B3-1 | ||||
| B3-6balla | 51,448 | B3-6 | MW345784 | ||
| B6-1ebb | 51,350 | B6-1 | MW345785 | ||
| Chionosphaera cuniculicola R. Kirschner, Begerow & Oberw. | B1-2aaaa | 51,590 | B1-1 | MW256646 | |
| Endoconidiophora polonica | B3-7ab | 51,333 | B3-7 | MW256648 | |
| B5-7ab | B5-7 | MW256650 | |||
| B6-2aba | 51,327 | B6-2 | MW256649 | ||
| B6-2ac | 51,463 | B6-2 | |||
| B6-3ca | B6-3 | MW256647 | |||
| Fontanospora fusiramosa Marvanová, Peter J. Fisher & Descals | B1-1b2 | 51,453 | B1-1 | ||
| Graphium fimbriisporum (M. Morelet) K. Jacobs, Kirisits & M.J. Wingf. | A4-2aab | A4-2 | |||
| A6-1ad | 51,382 | A6-1 | MW256653 | ||
| A7-2aca | 51,389 | A7-2 | MW256652 | ||
| B6-2ad | 51,329 | B6-2 | MW256651 | ||
| B6-3cb | B6-3 | ||||
| Grosmannia penicillata | A6-2b | 51,417 | A6-2 | ||
| A6-2cbb | 51,432 | A6-2 | MW345794 | ||
| A6-3adb | A6-3 | ||||
| A6-3c | 51,353 | A6-3 | MW345789 | ||
| A9-1cba | 51,412 | A9-1 | MW345788 | ||
| B5-1b | 51,337 | B5-1 | MW345793 | ||
| B6-1c | 51,338 | B6-1 | MW345790 | ||
| B6-1ca | B6-1 | ||||
| B6-2b | 51,435 | B6-2 | |||
| B6-2c | B6-2 | ||||
| B6-2eca | 51,446 | B6-2 | MW345791 | ||
| B5-1dbb | 51,423 | B5-1 | MW345792 | ||
| A10-1b | 51,376 | A10-1 | MW345795 | ||
| A10-3ada | A10-1 | MW345796 | |||
| A6-3da | 51,470 | A6-3 | |||
| Kuraishia capsulata (Wick.) Y. Yamada, K. Maeda & Mikata | A4-2aaaa | 51,570 | A4-2 | MW256635 | |
| G83-1aa | 51,578 | G83 | MW256634 | ||
| Kuraishia molischiana Dlauchy, G. Péter, Tornai-Leh. & Kurtzman | B2-3aa | 51,574 | B2-3 | MW256633 | |
| Nakazawaea sp. | B8-1aaa | B8-1 | |||
| B8-1abb | 51,543 | B8-1 | MW256638 | ||
| B2-3b | 51,542 | B2-3 | MW256639 | ||
| Ogataea glucozyma (Wick.) Y. Yamada, K. Maeda & Mikata | F30-1caa | 51,585 | F30 | MW256636 | |
| Ogataea ramenticola (Kurtzman) Kurtzman & Robnett | B3-1aaaa | 51,591 | B3-1 | MW256637 | |
| Ophiostoma ainoae H. Solheim | A4-2aab | 51,342 | A4-2 | MW345797 | |
| A4-2b-1 | 51,464 | A4-2 | |||
| A7-2db | 51,361 | A7-2 | MW345800 | ||
| A9-1cbbb | 51,354 | A9-1 | MW345798 | ||
| B6-2ea | 51,459 | B6-2 | |||
| B6-2ebb | B6-2 | ||||
| B6-2ecb | 51,341 | B6-2 | MW345799 | ||
| Ophiostoma bicolor | A10-2b | 51,355 | A10-1 | MW345809 | |
| A7-2ab | 51,367 | A7-2 | MW345806 | ||
| A7-2b | 51,364 | A7-2 | |||
| A7-2da | 51,462 | A7-2 | |||
| B1-1d | 51,442 | B1-1 | MW345805 | ||
| B3-1aba | B3-1 | ||||
| B3-1b | 51,352 | B3-1 | MW345804 | ||
| B3-1d | 51,425 | B3-1 | |||
| B3-7ad | 51,343 | B3-7 | MW345807 | ||
| A10-1dac | 51,456 | A10-1 | |||
| A10-1db | 51,427 | A10-1 | |||
| B3-7ac | B3-7 | ||||
| A7-1b | A7-1 | MW345808 | |||
| Ophiostoma brunneolum Linnak., Z.W. de Beer & M.J. Wingf. | B1-3ab | B1-3 | |||
| B1-3gab | 51,328 | B1-3 | |||
| B1-3gbb | 51,371 | B1-3 | MW345801 | ||
| B3-7cb | B3-7 | ||||
| B8-1ac | 51,454 | B8-1 | MW345802 | ||
| B8-2ba | 51,422 | B8-2 | MW345803 | ||
| B8-2bb | 51,346 | B8-2 | |||
| B8-cb | 51,375 | B8-2 | |||
| Wickerhamomyces bisporus (O. Beck) Kurtzman, Robnett & Basehoar-Powers | A6-1aaba | 51,547 | A6-1 | MW256640 | |
| A6-3aaa | 51,544 | A6-3 | MW256641 | ||
| A7-2aaa | 51,575 | A7-2 | MW256644 | ||
| B1-1a | B1-1 | ||||
| B1-3aaa | 51,584 | B1-1 | MW256642 | ||
| B2-1aaa | 51,581 | B2-1 | MW256645 | ||
| B2-2aaa | 51,589 | B2-2 | MW256643 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linnakoski, R.; Lasarov, I.; Veteli, P.; Tikkanen, O.-P.; Viiri, H.; Jyske, T.; Kasanen, R.; Duong, T.A.; Wingfield, M.J. Filamentous Fungi and Yeasts Associated with Mites Phoretic on Ips typographus in Eastern Finland. Forests 2021, 12, 743. https://doi.org/10.3390/f12060743
Linnakoski R, Lasarov I, Veteli P, Tikkanen O-P, Viiri H, Jyske T, Kasanen R, Duong TA, Wingfield MJ. Filamentous Fungi and Yeasts Associated with Mites Phoretic on Ips typographus in Eastern Finland. Forests. 2021; 12(6):743. https://doi.org/10.3390/f12060743
Chicago/Turabian StyleLinnakoski, Riikka, Ilmeini Lasarov, Pyry Veteli, Olli-Pekka Tikkanen, Heli Viiri, Tuula Jyske, Risto Kasanen, Tuan A. Duong, and Michael J. Wingfield. 2021. "Filamentous Fungi and Yeasts Associated with Mites Phoretic on Ips typographus in Eastern Finland" Forests 12, no. 6: 743. https://doi.org/10.3390/f12060743
APA StyleLinnakoski, R., Lasarov, I., Veteli, P., Tikkanen, O.-P., Viiri, H., Jyske, T., Kasanen, R., Duong, T. A., & Wingfield, M. J. (2021). Filamentous Fungi and Yeasts Associated with Mites Phoretic on Ips typographus in Eastern Finland. Forests, 12(6), 743. https://doi.org/10.3390/f12060743







