Abstract
Similar to chloroplast loci, mitochondrial markers are frequently used for genotyping, phylogenetic studies, and population genetics, as they are easily amplified due to their multiple copies per cell. In a recent study, it was revealed that the chloroplast offers little variation for this purpose in central European populations of beech. Thus, it was the aim of this study to elucidate, if mitochondrial sequences might offer an alternative, or whether they are similarly conserved in central Europe. For this purpose, a circular mitochondrial genome sequence from the more than 300-year-old beech reference individual Bhaga from the German National Park Kellerwald-Edersee was assembled using long and short reads and compared to an individual from the Jamy Nature Reserve in Poland and a recently published mitochondrial genome from eastern Germany. The mitochondrial genome of Bhaga was 504,730 bp, while the mitochondrial genomes of the other two individuals were 15 bases shorter, due to seven indel locations, with four having more bases in Bhaga and three locations having one base less in Bhaga. In addition, 19 SNP locations were found, none of which were inside genes. In these SNP locations, 17 bases were different in Bhaga, as compared to the other two genomes, while 2 SNP locations had the same base in Bhaga and the Polish individual. While these figures are slightly higher than for the chloroplast genome, the comparison confirms the low degree of genetic divergence in organelle DNA of beech in central Europe, suggesting the colonisation from a common gene pool after the Weichsel Glaciation. The mitochondrial genome might have limited use for population studies in central Europe, but once mitochondrial genomes from glacial refugia become available, it might be suitable to pinpoint the origin of migration for the re-colonising beech population.
1. Introduction
1.1. Importance of European Beech and Its Population Structure
European beech is the climax tree species in most regions of Germany and adjacent areas [1]. Due to its valuable hardwood, it is also widely planted. Climate change has impacted some beech stands in drier regions of Germany, especially during the past few years, which were characterised by record heat and drought events [2,3], and might eventually lead to a shift to oak forests in those areas [4]. However, at the northern edge of its distribution, beech is also gaining ground, replacing oaks in north-eastern habitats [5]. The ecological importance and dominance of beech in central Europe and its peculiar post-glacial migration pattern, with an arrival in northern Europe well after the post-glacial optimum [6], have triggered several population genetics studies. Most of these relied on nuclear markers [7,8,9,10,11]. However, nuclear markers are inherited both maternally and paternally, obscuring local migration patterns due to pollen influx from more distant populations.
1.2. Maternally Inherited Markers Are Important for Tracking Migration Routes
As seeds of beech are rarely dispersed over long distances, markers derived from maternally inherited organelle genomes of the chloroplast and mitochondria are likely to resolve fine-scale migrations better than nuclear markers [12]. Some studies have considered chloroplast markers for tracing migration [13] and beech population genetics [14,15,16,17,18]. However, recently it was shown that chloroplast genomes vary hardly in central Europe, providing limited resolution for investigation [18]. Mitochondrial genomes are also maternally inherited [19] but have larger genomes in plants [20], thus, offering potentially more loci for analysis, as also mutation rates in mitochondria are higher than in chloroplasts [21]. Recently, a mitochondrial genome was reported for European Beech [22] for the same individual from whom a chloroplast sequence had been generated before.
Thus, it was the aim of this study to provide additional complete mitochondrial sequences from the two other individuals from which chloroplast sequences had been obtained previously [23]. These were the more than 300-year-old reference individual Bhaga from the National Park Kellerwald-Edersee in Germany [24], which corresponds to the centre of the current distribution range, and a younger individual, Jamy, sampled from the Jamy Nature Reserve in Poland, corresponding to the north-eastern area of the distribution range. The sequences from all three individuals were then compared to each other for detecting variable positions in the mitochondrial genomes, as these positions might be suitable for future population-scale migration studies focusing on maternally inherited mutations.
2. Materials and Methods
2.1. Data Acquisition and Initial Draft Assembly of the Mitochondrial Genome of Bhaga
DNA was obtained from the European beech reference individual Bhaga from the National Park Kellerwald-Edersee (51°10′09″ N 8°57′47″ E) in a previous study [24] and sequenced as outlined therein.
The mitochondrial genome was assembled using both short (Illumina) and long (PacBio) reads obtained previously [24]. The PacBio reads were double corrected by first performing a correction with Illumina reads using proovread [25], which was followed by self-correction using Canu [26]. Double-corrected PacBio reads were aligned with five different plant mitochondrial genomes using BLAST, as described previously [23]. All PacBio reads with a match over more than 50% of their length to any of the targets were used for the construction of an assembly using Canu. The SSpaceLong scaffolder [27] was used to link contigs using all initially used PacBio reads. Subsequently, reads matching to mitochondria with at least 10% of their length were added. The subsequent reassembly using Canu resulted in a circular contig and of 531,393 bp without ambiguous nucleotide positions. This contig was considered to be the initial mitochondrial genome assembly and was subjected to further refinements, as detailed below.
2.2. Manual Curation of the Assembly
Illumina and PacBio reads were aligned over the draft assembly to check for coverage continuity, and a stretch was found with half of the average coverage but otherwise complete identity, revealing a misassembly. Consequently, the duplication was removed, and the continuity of the resulting mitochondrial genome was verified using read mapping, which now revealed an even coverage without breaks. Pilon [28] was subsequently used for refinement of the final circular mitochondrial genome assembly with the raw data initially used.
2.3. Assembly of the Mitochondrial Genome of Jamy
The mitochondrial genome of the individual Jamy originating from the Jamy Nature Reserve, Poland (18°56′6.07″ E, 53°35′9.67″ N) was assembled on the basis of 18,306,393 (paired end, 150 bp) Illumina reads, using the de novo assembler NOVOPlasty v 4.2 [29] with AY453092.1 as seed sequence [30] and the Bhaga mitochondrial genome as reference for guiding the assembler in repeat regions.
2.4. Functional Annotation and Circular Representation
The sequence of the mitochondrial genome was annotated as described previously for the chloroplast genome of Bhaga [23], using GeSeq ChloroBox [31] but with the sequence MT446430.1 [22] as reference for CDS + rRNA prediction and manual corrections performed using Unipro UGENE [32] assisted by bwa-mem [33]. Duplicate annotations were removed from the GenBank file manually, and the resulting file was used to obtain a graphical representation using OGDraw of the GeSeq Chlorobox package. The mitochondrial genomes of Bhaga and Jamy were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MW771358 and MW582695, respectively.
3. Results
3.1. General Features of the Mitochondrial Genome Assemblies
The circular genome of the mitochondrion of the Fagus sylvatica reference individual, Bhaga, was 504,730 bp long (Figure 1), with an average coverage of 1145×, while that of Jamy was 504,715 bp long, with an average 160× coverage. The GC content was 45.8%, in both cases. In both Bhaga and Jamy 40.65 kb of the mitochondrial genome consisted of repeat elements. In total, 49 protein-coding genes, 17 tRNAs, and 5 rRNAs were annotated in the mitochondrial genomes of Bhaga and Jamy, while 35 protein-coding genes, 20 tRNA genes, and 3 rRNA genes had been reported in the recently published mitochondrial genome of Fagus sylvatica [22]. However, when the previously reported mitochondrial genome [22] was re-annotated using the approach described above, we found the same number of genes as in Bhaga and Jamy.
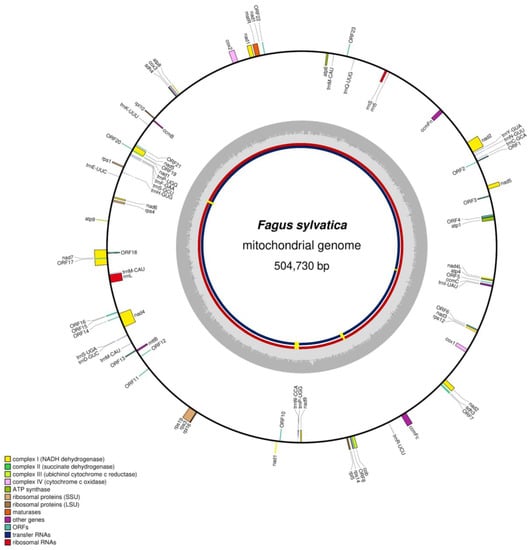
Figure 1.
Circular representation of the mitochondrial genome of Fagus sylvatica (Bhaga) with gene locations on both strands and an indication of SNP and indel locations indicated in yellow on the inner red and blue circles, representing Jamy and the individual sequenced previously [22].
3.2. Comparison of Mitochondrial Genome Assemblies
Very few variable locations in the mitochondrial genome were found in the comparison of the mitochondrial genomes of Bhaga, Jamy, and a genome from eastern Germany recently published (GenBank accession MT446430) [22]. None of the variable positions were present within genes (Table 1). Interestingly, the mitochondrial genome of Jamy from north-central Poland and the previously published genome from eastern Germany were highly similar, differing only in two SNPs and no indel positions. In both of these SNPs, the alternate base in Jamy was identical to the one called in Bhaga. Bhaga differed from Jamy and the previously published mitochondrial genome in 17 and 19 SNPs, respectively. In addition, Bhaga differed in seven indel positions from Jamy and the individual from eastern Germany. The longest of these indels was a stretch of 7 bp present only in Bhaga (Table 1).

Table 1.
Variable positions in the mitochondrial genomes of the three individuals investigated in this study. Indel positions are shaded in grey. SNPs private to Bhaga are shaded in blue, SNPs private to the mitochondrial genome reported in Mader et al. [22] are shaded in green.
4. Discussion
4.1. Why Variable Regions in Maternally Inherited Genomes Are Important to Identify in Beech
European Beech has received much research attention over the past decades since it has developed into one of the most important forestry trees in central Europe [34]. While European Beech is able to cope with significant climatic and edaphic differences, e.g., from the semi-arid Gargano area in southern Italy [35] to the perhumid areas of eastern Sweden [36], its migration by seed dispersal is slow [37], despite occasional distribution by seed predators [38], and probably a limiting factor in its distribution range expansion [39]. As a consequence, it was the last dominant broadleaf tree to colonise northern Europe after the Weichsel glaciation had ended, arriving in Sweden when Mycenaean Greece flourished [40]. This also means that genotypes fitting to a specific climatic niche in their future migration might not hold pace with predicted future warming, leading to a reduction in its distribution area [41] and probably ending its dominance in large parts of its current distribution range. Thus, for assessing the risk of decline of beech stands throughout the 21st century, it is important to understand its past migration and the contribution of pollen flow and seed dispersal in shaping current beech populations.
To disentangle these processes, maternally inherited markers that travel solely by seed dispersal and markers not bound to the female line need to be considered. While the nuclear genomic diversity can disperse via pollen, organelle DNA is, apart from rare events [42], bound to the female gametophyte and thus can be used to trace migration via seed dispersal. However, chloroplast genomes have only a very low diversity in the central to the northern current range of European Beech [18] and thus likely insufficient resolution for tracing fine-scale population dynamics. Thus, the current study was focused on the much larger mitochondrial genome, which is also maternally inherited, to identify potential additional markers for tracing migration by seed dispersal, including anthropogenic dispersal in the Holocene.
4.2. Utility of the Genomic Diversity of Organelle Genomes in Beech for Tracing Migration
The mitochondrial genome of the reference individual Bhaga from the National Park Kellerwald-Edersee in central Germany and the individual Jamy from the Jamy Nature Reserve in northern Poland were assembled and compared with the mitochondrial genome previously published for an individual from eastern Germany [22]. The same three individuals had previously been used for assessing chloroplast genome diversity in northern central Europe [23]. In line with the low diversity found among the three chloroplast genomes [18,23], also the mitochondrial genome showed little variation when comparing the three individuals. Apart from seven indel positions, 19 SNP positions were found, but none was located within a gene. This is noteworthy since it suggests a higher degree of conservation of the gene space than of the non-coding mitochondrial genome content. Interestingly, the mitochondrial genome previously reported from Brandenburg in eastern Germany [22] and that of Jamy from northern Poland are highly similar and distinguished only by two SNPs, suggesting a common ancestry for these populations. However, they differ from Bhaga in central Germany by 19 and 17 SNPs, respectively (Table 1). Whether this is best explained by Bhaga being a relic genotype in an extreme habitat or by migration history needs to be clarified in future studies. Although the number of SNP positions found in this study is not high, it might be sufficient to resolve the routes of migration to central Europe.
Based on population genetics studies conducted so far, including those based on organelle DNA (e.g., [17]), it can be expected that a much higher diversity of mitochondrial genomes will exist in Pleistocene refugia of Fagus sylvatica, since the diversity in organelle genomes is reflected by the diversity in nuclear markers in general (e.g., [43]). It thus seems likely that mitochondrial genomes will be useful in pinpointing the populations of European Beech that migrated to the previously glaciated areas of Europe. In addition, such studies might also help to estimate potential future migration and identify areas in which assisted migration of genotypes [44] could be used as a strategy to save beech forests for future generations, if identified limitations, such as potential maladaptations, e.g. with respect to prevailing pathogens, can be resolved [45,46].
Author Contributions
Conceptualization, M.T.; formal analysis, B.M., M.T., B.U. and J.B.; resources, M.T., S.P., J.B. and J.M.; data curation, B.M. and B.U.; writing—original draft preparation, M.T. and B.M.; writing—review and editing, J.B., B.U., J.M., M.T. and B.M.; visualization, B.M. and M.T.; supervision, M.T. and J.B.; project administration, M.T. and J.B.; funding acquisition, M.T. and J.B. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the National Science Center, Poland (2012/04/A/NZ9/00500), the Polish Ministry of Science and Higher Education under the program “Regional Initiative of Excellence” in 2019–2022 (Grant No. 008/RID/2018/19), and the German Science Foundation (Grant No. Thi1362/18-1). Additional support by the LOEWE initiative in the framework of the Centre for Translational Biodiversity Genomics (TBG) is gratefully acknowledged.
Data Availability Statement
The mitochondrial genome sequences have been deposited in GenBank under the accession numbers MW771358 for Bhaga and MW582695 for Jamy.
Acknowledgments
The authors gratefully acknowledge the permission of the office of the National Park Kellerwald-Edersse to sample the reference individual Bhaga.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Giesecke, T.; Hickler, T.; Kunkel, T.; Sykes, M.T.; Bradshaw, R.H.W. ORIGINAL ARTICLE: Towards an understanding of the Holocene distribution of Fagus sylvatica L. J. Biogeogr. 2006, 34, 118–131. [Google Scholar] [CrossRef]
- Zscheischler, J.; Fischer, E.M. The record-breaking compound hot and dry 2018 growing season in Germany. Weather Clim. Extrem. 2020, 29, 100270. [Google Scholar] [CrossRef]
- Kron, W.; Löw, P.; Kundzewicz, Z.W. Changes in risk of extreme weather events in Europe. Environ. Sci. Policy 2019, 100, 74–83. [Google Scholar] [CrossRef]
- Albert, R.E.I.F.; Xystrakis, F.; Gaertner, S.; Sayer, U. Floristic change at the drought limit of European beech (Fagus sylvatica L.) to downy oak (Quercus pubescens) forest in the temperate climate of central Europe. Not. Bot. Horti Agrobot. Cluj Napoca 2017, 45, 646–654. [Google Scholar]
- Sandurska, E.; Ulaszewski, B.; Burczyk, J. Genetic Insights into Ecological Succession from Oak- (Quercus robur L.) to Beech- (Fagus sylvatica L.) Dominated Forest Stands. Acta Biol. Crac. Ser. Bot. 2017, 59, 23–33. [Google Scholar] [CrossRef]
- Magri, D. Patterns of post-glacial spread and the extent of glacial refugia of European beech (Fagus sylvatica). J. Biogeogr. 2008, 35, 450–463. [Google Scholar] [CrossRef]
- Comps, B.; Thiébaut, B.; Paule, L.; Merzeau, D.; Letouzey, J. Allozymic variability in beechwoods (Fagus sylvatica L.) over central Europe: Spatial differentiation among and within populations. Heredity 1990, 65, 407–417. [Google Scholar] [CrossRef]
- Leonardi, S.; Menozzi, P. Genetic variability of Fagus sylvatica L. in Italy: The role of postglacial recolonization. Heredity 1995, 75, 35–44. [Google Scholar] [CrossRef][Green Version]
- Sjölund, M.J.; González-Díaz, P.; Moreno-Villena, J.J.; Jump, A.S. Understanding the legacy of widespread population translocations on the post-glacial genetic structure of the European beech, Fagus sylvatica L. J. Biogeogr. 2017, 44, 2475–2487. [Google Scholar] [CrossRef]
- Cuervo-Alarcon, L.; Arend, M.; Müller, M.; Sperisen, C.; Finkeldey, R.; Krutovsky, K.V. Genetic variation and signatures of natural selection in populations of European beech (Fagus sylvatica L.) along precipitation gradients. Tree Genet. Genomes 2018, 14, 84. [Google Scholar] [CrossRef]
- Müller, M.; Lopez, P.A.; Papageorgiou, A.C.; Tsiripidis, I.; Gailing, O. Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd. Diversity 2019, 11, 90. [Google Scholar] [CrossRef]
- Xia, H.; Wang, B.; Zhao, W.; Pan, J.; Mao, J.F.; Wang, X.R. Combining mitochondrial and nuclear genome analyses to dissect the effects of colonization, environment, and geography on population structure in Pinus tabuliformis. Evol. Appl. 2018, 11, 1931–1945. [Google Scholar] [CrossRef] [PubMed]
- Magri, D.; Vendramin, G.G.; Comps, B.; Dupanloup, I.; Geburek, T.; Gömöry, D.; Latałowa, M.; Litt, T.; Paule, L.; Roure, J.M.; et al. A new scenario for the Quaternary history of European beech populations: Palaeobotanical evidence and genetic consequences. New Phytol. 2006, 171, 199–221. [Google Scholar] [CrossRef]
- Demesure, B.; Comps, B.; Petit, R.J. Chloroplast DNA Phylogeography of the Common Beech (Fagus sylvatica L.) in Europe. Evolution 1996, 50, 2515–2520. [Google Scholar] [CrossRef]
- Sebastiani, F.; Carnevale, S.; Vendramin, G.G. A new set of mono- and dinucleotide chloroplast microsatellites in Fagaceae. Mol. Ecol. Notes 2004, 4, 259–261. [Google Scholar] [CrossRef]
- Vettori, C.; Vendramin, G.G.; Anzidei, M.; Pastorelli, R.; Paffetti, D.; Giannini, R. Geographic distribution of chloroplast variation in Italian populations of beech (Fagus sylvatica L.). Theor. Appl. Genet. 2004, 109, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hatziskakis, S.; Papageorgiou, A.C.; Gailing, O.; Finkeldey, R. High chloroplast haplotype diversity in Greek populations of beech (Fagus sylvatica L.). Plant Biol. 2009, 11, 425–433. [Google Scholar] [CrossRef]
- Meger, J.; Ulaszewski, B.; Vendramin, G.G.; Burczyk, J. Using reduced representation libraries sequencing methods to identify cpDNA polymorphisms in European beech (Fagus sylvatica L). Tree Genet. Genomes 2019, 15, 7. [Google Scholar] [CrossRef]
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef]
- Smith, D.R. Mutation Rates in Plastid Genomes: They Are Lower than You Might Think. Genome Biol. Evol. 2015, 7, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Mader, M.; Schroeder, H.; Schott, T.; Schöning-Stierand, K.; Montalvão, A.P.L.; Liesebach, H.; Liesebach, M.; Fussi, B.; Kersten, B. Mitochondrial Genome of Fagus sylvatica L. as a Source for Taxonomic Marker Development in the Fagales. Plants 2020, 9, 1274. [Google Scholar] [CrossRef]
- Mishra, B.; Ulaszewski, B.; Ploch, S.; Burczyk, J.; Thines, M. A Circular Chloroplast Genome of Fagus sylvatica Reveals High Conservation between Two Individuals from Germany and One Individual from Poland and an Alternate Direction of the Small Single-Copy Region. Forests 2021, 12, 180. [Google Scholar] [CrossRef]
- Mishra, B.; Gupta, D.K.; Pfenninger, M.; Hickler, T.; Langer, E.; Nam, B.; Paule, J.; Sharma, R.; Ulaszewski, B.; Warmbier, J.; et al. A reference genome of the European beech (Fagus sylvatica L.). GigaScience 2018, 7, 063. [Google Scholar] [CrossRef] [PubMed]
- Hackl, T.; Hedrich, R.; Schultz, J.; Förster, F. proovread: Large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 2014, 30, 3004–3011. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. SSPACE-LongRead: Scaffolding bacterial draft genomes using long read sequence information. BMC Bioinform. 2014, 15, 211. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Barkman, T.J.; Lim, S.-H.; Salleh, K.M.; Nais, J. Mitochondrial DNA sequences reveal the photosynthetic relatives of Rafflesia, the world’s largest flower. Proc. Natl. Acad. Sci. USA 2004, 101, 787–792. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Brunet, J.; Felton, A.; Lindbladh, M. From wooded pasture to timber production—Changes in a European beech (Fagus sylvatica) forest landscape between 1840 and 2010. Scand. J. For. Res. 2012, 27, 245–254. [Google Scholar] [CrossRef]
- Dittmar, C.; Zech, W.; Elling, W. Growth variations of Common beech (Fagus sylvatica L.) under different climatic and environmental conditions in Europe—A dendroecological study. For. Ecol. Manag. 2003, 173, 63–78. [Google Scholar] [CrossRef]
- Diekmann, M.; Eilertsen, O.; Fremstad, E.; Lawesson, J.E.; Aude, E. Beech forest communities in the Nordic countries—A multivariate analysis. Plant Ecol. 1999, 140, 203–220. [Google Scholar] [CrossRef]
- Wang, K.; Hattemer, H.H. Dispersal of seed and effective pollen in small stands of European beech (Fagus sylvatica L.). In Genetic Response of Forest Systems to Changing Environmental Conditions; Springer: Dordrecht, The Netherlands, 2001; pp. 259–269. [Google Scholar]
- Zwolak, R.; Bogdziewicz, M.; Wróbel, A.; Crone, E.E. Advantages of masting in European beech: Timing of granivore satiation and benefits of seed caching support the predator dispersal hypothesis. Oecologia 2016, 180, 749–758. [Google Scholar] [CrossRef]
- Saltré, F.; Saint-Amant, R.; Gritti, E.S.; Brewer, S.; Gaucherel, C.; Davis, B.A.S.; Chuine, I. Climate or migration: What limited European beech post-glacial colonization? Glob. Ecol. Biogeogr. 2013, 22, 1217–1227. [Google Scholar] [CrossRef]
- Björkman, L. The establishment of Fagus sylvatica at the stand-scale in southern Sweden. Holocene 1999, 9, 237–245. [Google Scholar] [CrossRef]
- Saltré, F.; Duputié, A.; Gaucherel, C.; Chuine, I. How climate, migration ability and habitat fragmentation affect the projected future distribution of European beech. Glob. Change Biol. 2015, 21, 897–910. [Google Scholar] [CrossRef]
- McCauley, D.E. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 2013, 200, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Muñoz-Fuentes, V.; Buch, A.-K.; Çelik, A.; Dutbayev, A.; Gabrielyan, I.; Glynou, K.; Kachour, L.; Khaliq, I.; Kitner, M.; et al. Out of Transcaucasia: Origin of Western and Central Palearctic populations of Microthlaspi perfoliatum. Flora Morphol. Distrib. Funct. Ecol. Plants 2019, 253, 127–141. [Google Scholar] [CrossRef]
- Vitt, P.; Havens, K.; Kramer, A.T.; Sollenberger, D.; Yates, E. Assisted migration of plants: Changes in latitudes, changes in attitudes. Biol. Conserv. 2010, 143, 18–27. [Google Scholar] [CrossRef]
- Bucharova, A. Assisted migration within species range ignores biotic interactions and lacks evidence. Restor. Ecol. 2016, 25, 14–18. [Google Scholar] [CrossRef]
- Park, A.; Talbot, C. Information Underload: Ecological Complexity, Incomplete Knowledge, and Data Deficits Create Challenges for the Assisted Migration of Forest Trees. Bioscience 2018, 68, 251–263. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).