Tree Shape Variability in a Mixed Oak Forest Using Terrestrial Laser Technology: Implications for Mating System Analysis
Abstract
1. Introduction
2. Materials and Methods
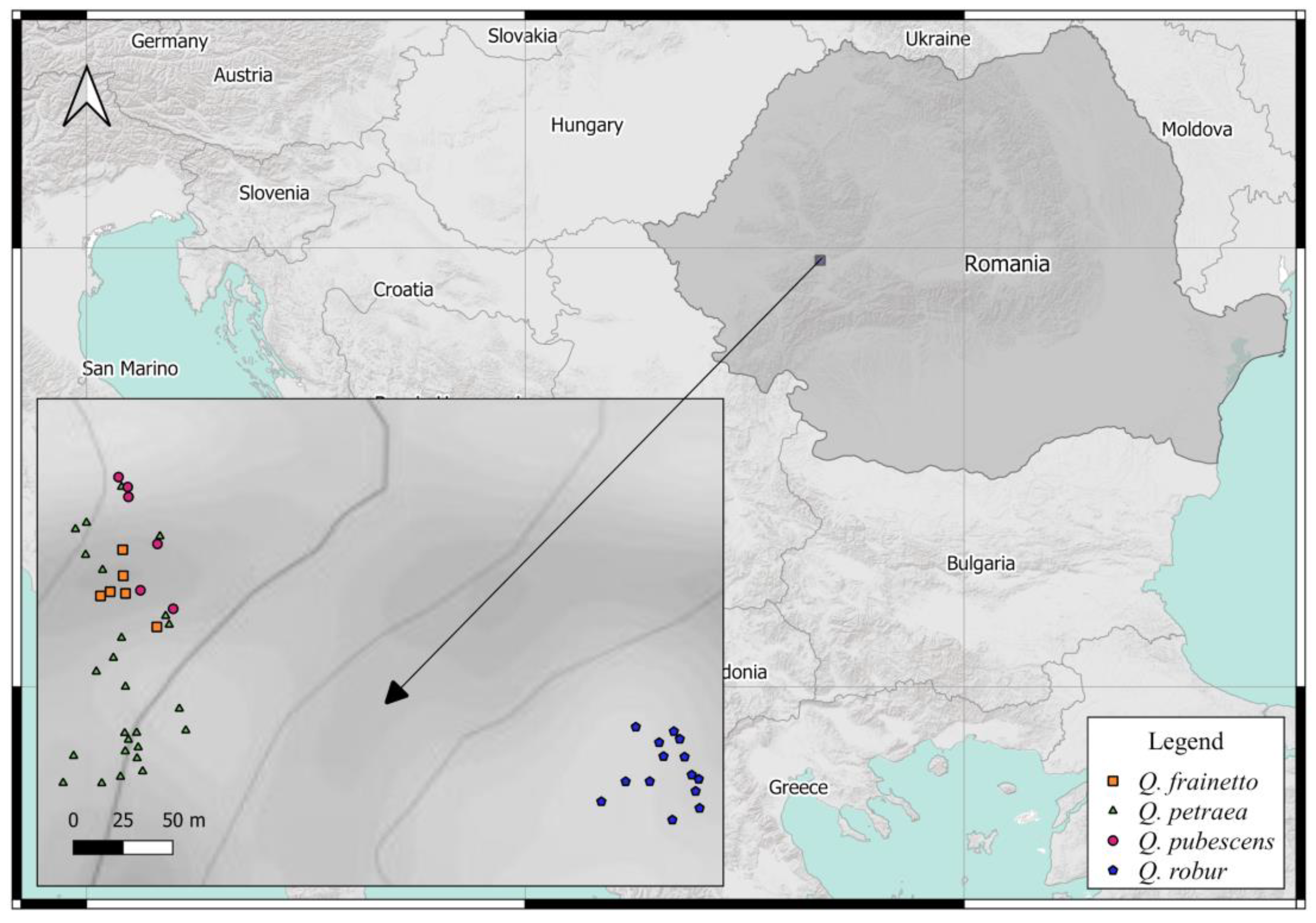
2.1. Study Site
2.2. Materials
2.3. Method
2.3.1. Field Work
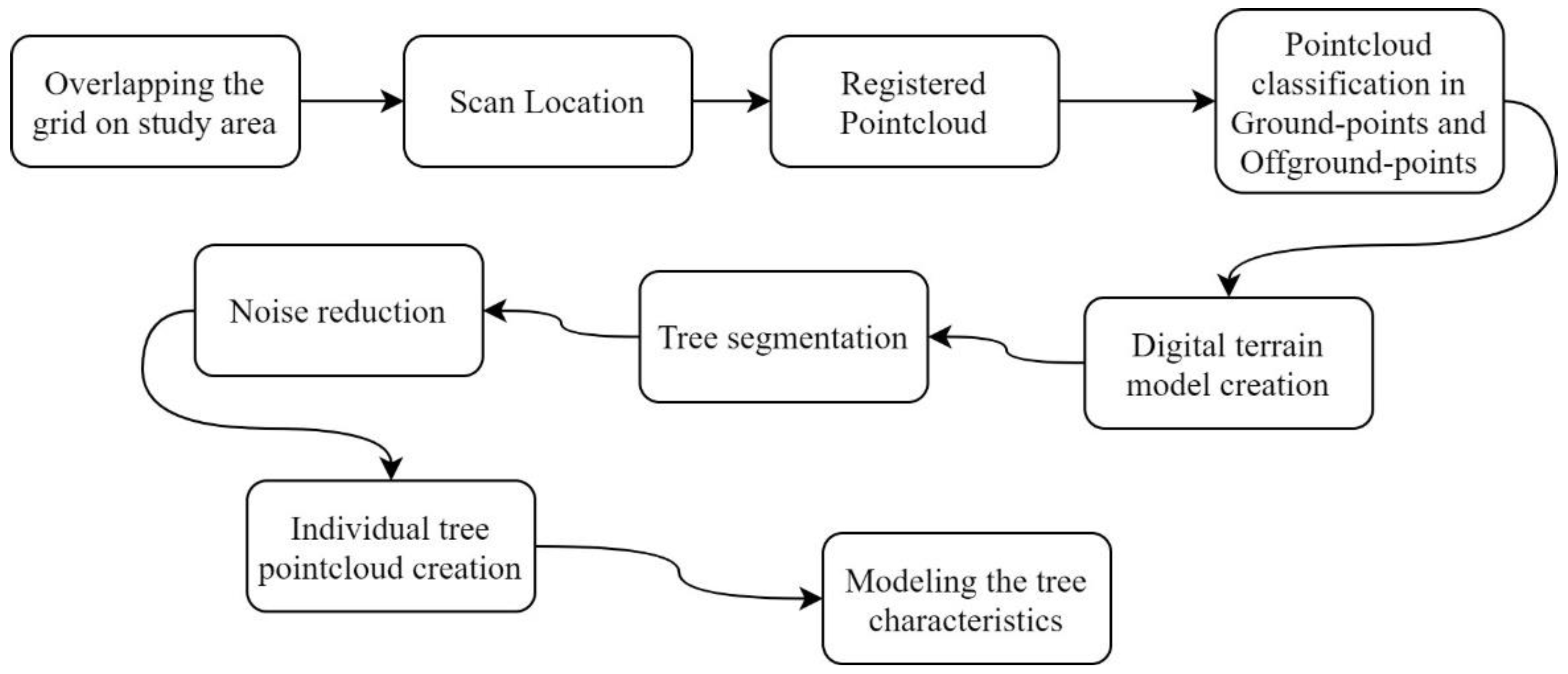
2.3.2. Data Processing and Analysis
2.3.3. Genetic Data Analysis
3. Results
3.1. Scan Registration Accuracy and Point Cloud Statistics
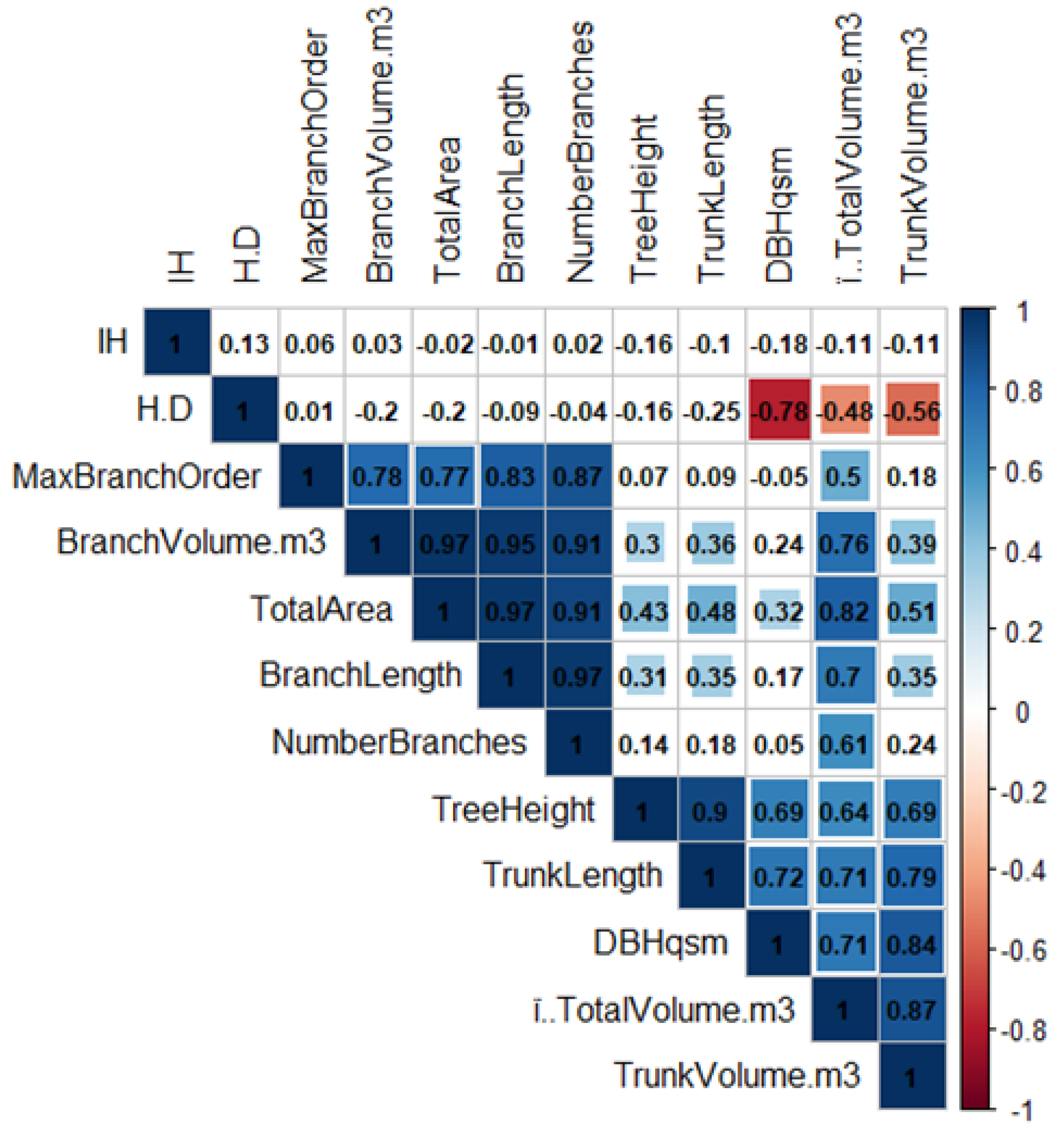
3.2. Tree Shape Variability
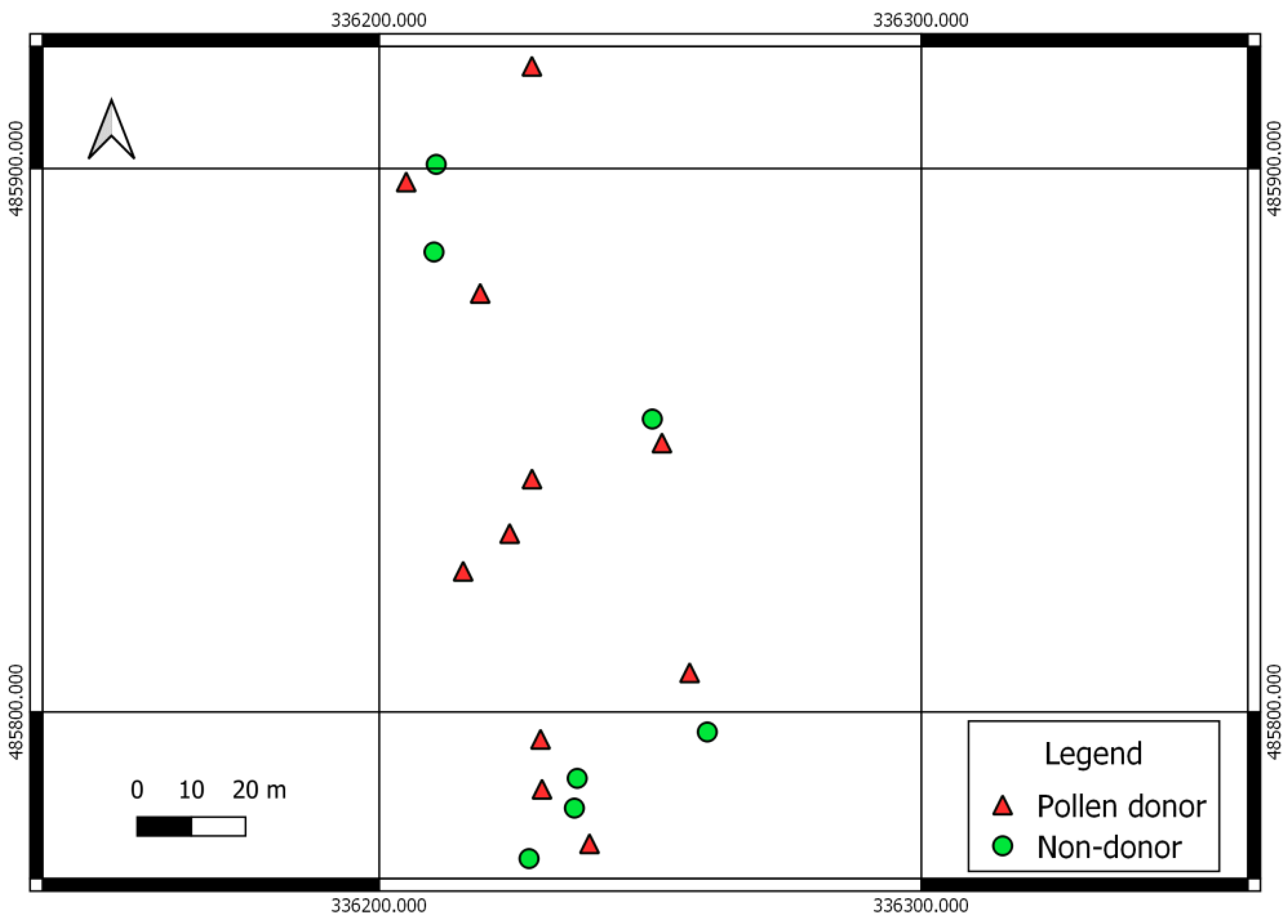
3.3. Tree Shape and Male Fecundity
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| p-Value | Total Volume | Trunk Volume | Branch Volume | Tree Height | Trunk Length | Branch Length | Number Branches | Max Branch Order | Total Area | DBH qsm | IH | H.D |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Volume | NA | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.449 | 0.000 |
| Trunk Volume | 0.000 | NA | 0.005 | 0.000 | 0.000 | 0.011 | 0.086 | 0.218 | 0.000 | 0.000 | 0.449 | 0.000 |
| Branch Volume | 0.000 | 0.005 | NA | 0.031 | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.088 | 0.822 | 0.151 |
| Tree Height | 0.000 | 0.000 | 0.031 | NA | 0.000 | 0.027 | 0.312 | 0.617 | 0.002 | 0.000 | 0.260 | 0.265 |
| Trunk Length | 0.000 | 0.000 | 0.009 | 0.000 | NA | 0.012 | 0.197 | 0.522 | 0.000 | 0.000 | 0.507 | 0.079 |
| Branch Length | 0.000 | 0.011 | 0.000 | 0.027 | 0.012 | NA | 0.000 | 0.000 | 0.000 | 0.227 | 0.937 | 0.511 |
| No. Branches | 0.000 | 0.086 | 0.000 | 0.312 | 0.197 | 0.000 | NA | 0.000 | 0.000 | 0.753 | 0.879 | 0.767 |
| Max Branch Ord. | 0.000 | 0.218 | 0.000 | 0.617 | 0.522 | 0.000 | 0.000 | NA | 0.000 | 0.704 | 0.677 | 0.950 |
| Total Area | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.000 | 0.000 | 0.000 | NA | 0.022 | 0.869 | 0.152 |
| DBHqsm | 0.000 | 0.000 | 0.088 | 0.000 | 0.000 | 0.227 | 0.753 | 0.704 | 0.022 | NA | 0.194 | 0.000 |
| IH | 0.449 | 0.449 | 0.822 | 0.260 | 0.507 | 0.937 | 0.879 | 0.677 | 0.869 | 0.194 | NA | 0.358 |
| H.D | 0.000 | 0.000 | 0.151 | 0.265 | 0.079 | 0.511 | 0.767 | 0.950 | 0.152 | 0.000 | 0.358 | NA |
Appendix B
| Q. robur vs. Q. pubescens | Q. robur vs. Q. petraea | Q. robur vs. Q. frainetto | |||||||
| U | Z | p-Level | U | Z | p-Level | U | Z | p-Level | |
| Total Volume | 27 | 1.237 | 0.21602 | 154 | −0.615 | 0.53000 | 38 | −0.412 | 0.68005 |
| Trunk Volume | 33 | 0.742 | 0.45790 | 98.5 | −2.240 | 0.02511 | 34 | −0.660 | 0.50936 |
| Branch Volume | 24 | 1.485 | 0.13765 | 113 | 1.815 | 0.06950 | 37 | −0.412 | 0.68005 |
| Tree Height | 10 | 2.639 | 0.00831 | 152 | 0.673 | 0.50070 | 21 | 1.732 | 0.08327 |
| Trunk Length | 10 | 2.639 | 0.00831 | 165 | 0.293 | 0.76960 | 22 | 1.650 | 0.09903 |
| Branch Length | 22 | 1.650 | 0.09903 | 100 | 2.196 | 0.02811 | 34 | −0.660 | 0.50936 |
| Number Branches | 36 | 0.495 | 0.62069 | 115 | 1.757 | 0.07898 | 26 | −1.320 | 0.18695 |
| Max Branch Order | 30.5 | −0.949 | 0.34287 | 140.5 | 1.010 | 0.31247 | 36 | −0.495 | 0.62069 |
| Total Area | 19 | 1.897 | 0.05783 | 121 | 1.581 | 0.11389 | 36 | −0.495 | 0.62069 |
| DBH | 25 | 1.402 | 0.16088 | 122 | −1.552 | 0.12074 | 40 | 0.165 | 0.86898 |
| IH | 22 | −1.650 | 0.09903 | 152.5 | 0.659 | 0.51007 | 34.5 | 0.619 | 0.53619 |
| Q. pubescensvs. Q. petraea | Q. pubescensvs. Q. frainetto | Q. petraeavs. Q. frainetto | |||||||
| U | Z | p-Level | U | Z | p-Level | U | Z | p-Level | |
| Total Volume | 33 | −2.100 | 0.03573 | 11 | −1.121 | 0.26233 | 75 | 0.000 | 1.00000 |
| Trunk Volume | 25 | −2.500 | 0.01242 | 13 | −0.801 | 0.42334 | 61 | 0.700 | 0.48393 |
| Branch Volume | 72 | 0.150 | 0.88077 | 10 | −1.281 | 0.20019 | 41 | −1.700 | 0.08913 |
| Tree Height | 26 | −2.450 | 0.01429 | 8 | −1.601 | 0.10932 | 49.5 | 1.275 | 0.20231 |
| Trunk Length | 29 | −2.300 | 0.02145 | 8.5 | −1.521 | 0.12754 | 55 | 1.000 | 0.31726 |
| Branch Length | 66 | 0.450 | 0.65271 | 7 | −1.761 | 0.07817 | 37 | −1.900 | 0.05744 |
| Number Branches | 56 | 0.950 | 0.34211 | 6 | −1.922 | 0.05467 | 33 | −2.100 | 0.03573 |
| Max Branch Order | 47.5 | 1.375 | 0.16913 | 16 | 0.320 | 0.74442 | 54 | −1.050 | 0.28821 |
| Total Area | 65 | −0.500 | 0.61708 | 8 | −1.601 | 0.10932 | 42 | −1.650 | 0.09894 |
| DBH | 22.5 | −2.650 | 0.08665 | 13 | −0.801 | 0.42334 | 46 | 1.450 | 0.14706 |
| IH | 23 | 2.600 | 0.09323 | 5 | 2.082 | 0.03216 | 73 | 0.100 | 0.91878 |
| Trunk and Crown Characteristics | Kruskal-Wallis Test: H |
|---|---|
| Total Volume m3 | (3, N = 51) = 3.955066 p =0.2664 |
| Trunk Volume m3 | (3, N = 51) = 8.505381 p =0.0366 |
| Branch Volume m3 | (1, N = 20) = 2.204082 p =0.1376 |
| Tree Height | (1, N = 20) = 6.965986 p =0.0083 |
| Trunk Length | (1, N = 20) = 6.965986 p =0.0083 |
| Branch Length | (1, N = 20) = 2.723136 p =0.0989 |
| Number of Branches | (1, N = 20) = 0.244898 p =0.6207 |
| Max Branch Order | (1, N = 20) = 0.942164 p =0.3317 |
| Total Area | (1, N = 20) = 3.598639 p = 0.0578 |
| DBH qsm | (1, N = 20) = 1.965986 p = 0.1609 |
| IH (individual heterozygosity) | (1, N = 20) = 2.838469 p = 0.0920 |
References
- Pretzsch, H. Canopy space filling and tree crown morphology in mixed-species stands compared with monocultures. For. Ecol. Manag. 2014, 327, 251–264. [Google Scholar] [CrossRef]
- Chen, H.Y.H.; Klinka, K.; Kayahara, G.J. Effects of light on growth, crown architecture, and specific leaf area for naturally established Pinus contorta var. latifolia and Pseudotsuga menziesii var. glauca saplings. Can. J. For. Res. 1996, 26, 1149–1157. [Google Scholar] [CrossRef]
- Forrester, D.I.; Benneter, A.; Bouriaud, O.; Bauhus, J. Diversity and competition influence tree allometric relationships—Developing functions for mixed-species forests. J. Ecol. 2017, 105, 761–774. [Google Scholar] [CrossRef]
- Jensen, E.C.; Long, J.N. Crown structure of a codominant Douglas-fir (Pseudotsuga menziesii). Can. J. For. Res. 1983, 13, 264–269. [Google Scholar] [CrossRef]
- Lau, A.; Bentley, L.P.; Martius, C.; Shenkin, A.; Bartholomeus, H.; Raumonen, P.; Malhi, Y.; Jackson, T.; Herold, M. Quantifying branch architecture of tropical trees using terrestrial LiDAR and 3D modelling. Trees Struct. Funct. 2018, 32, 1219–1231. [Google Scholar] [CrossRef]
- Van der Zande, D.; Hoet, W.; Jonckheere, I.; van Aardt, J.; Coppin, P. Influence of measurement set-up of ground-based LiDAR for derivation of tree structure. Agric. For. Meteorol. 2006, 141, 147–160. [Google Scholar] [CrossRef]
- Murray, J.; Fennell, J.T.; Blackburn, G.A.; Whyatt, J.D.; Li, B. The novel use of proximal photogrammetry and terrestrial LiDAR to quantify the structural complexity of orchard trees. Precis. Agric. 2020, 21, 473–483. [Google Scholar] [CrossRef]
- Garms, C.G.; Simpson, C.H.; Parrish, C.E.; Wing, M.G.; Strimbu, B.M. Assessing lean and positional error of individual mature douglas-fir (Pseudotsuga menziesii) trees using active and passive sensors. Can. J. For. Res. 2020, 50, 1228–1243. [Google Scholar] [CrossRef]
- Zhou, T.; Popescu, S.; Lawing, A.; Eriksson, M.; Strimbu, B.; Bürkner, P. Bayesian and Classical Machine Learning Methods: A Comparison for Tree Species Classification with LiDAR Waveform Signatures. Remote Sens. 2017, 10, 39. [Google Scholar] [CrossRef]
- Holopainen, M.; Vastaranta, M.; Kankare, V.; Räty, M.; Vaaja, M.; Liang, X.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Viitala, R.; et al. Biomass estimation of individual trees using stem and crown diameter TLS measurements. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 3812, 91–95. [Google Scholar] [CrossRef]
- Pascu, I.S.; Dobre, A.C.; Badea, O.; Tănase, M.A. Estimating forest stand structure attributes from terrestrial laser scans. Sci. Total Environ. 2019, 691, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Apostol, B.; Chivulescu, S.; Ciceu, A.; Petrila, M.; Pascu, I.S.; Apostol, E.N.; Leca, S.; Lorent, A.; Tanase, M.; Badea, O. Data collection methods for forest inventory: A comparison between an integrated conventional equipment and terrestrial laser scanning. Ann. For. Res. 2018, 61, 189–202. [Google Scholar] [CrossRef]
- Nita, M.D.; Clinciu, I.; Popa, B. Evaluation of stream bed dynamics from Vidas torrential valley using terrestrial measurements and GIS techniques. Environ. Eng. Manag. J. 2016, 15, 1387–1395. [Google Scholar] [CrossRef]
- Tang, S.; Dong, P.; Buckles, B.P. Three-dimensional surface reconstruction of tree canopy from lidar point clouds using a region-based level set method. Int. J. Remote Sens. 2013, 34, 1373–1385. [Google Scholar] [CrossRef]
- Seidel, D. A holistic approach to determine tree structural complexity based on laser scanning data and fractal analysis. Ecol. Evol. 2018, 8, 128–134. [Google Scholar] [CrossRef]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef]
- Hackenberg, J.; Morhart, C.; Sheppard, J.; Spiecker, H.; Disney, M. Highly Accurate Tree Models Derived from Terrestrial Laser Scan Data: A Method Description. Forests 2014, 5, 1069–1105. [Google Scholar] [CrossRef]
- Krůček, M.; Král, K.; Cushman, K.; Missarov, A.; Kellner, J.R. Supervised Segmentation of Ultra-High-Density Drone Lidar for Large-Area Mapping of Individual Trees. Remote Sens. 2020, 12, 3260. [Google Scholar] [CrossRef]
- White, T.L.; Adams, W.T.; Neale, D.B. Forest Genetics; CABI Publishing: Wallingford, UK, 2007; ISBN 1845932854. [Google Scholar]
- Müller-Starck, G. Genetic differences between “tolerant” and “sensitive” beeches (Fagus sylvatica L.) in an environmentally stressed adult forest stand. Silvae Genet. 1985, 34, 241–247. [Google Scholar]
- Burkardt, K.; Pettenkofer, T.; Ammer, C.; Gailing, O.; Leinemann, L.; Seidel, D.; Vor, T. Influence of heterozygosity and competition on morphological tree characteristics of Quercus rubra L.: A new single-tree based approach. New For. 2020, 1–17. [Google Scholar] [CrossRef]
- Dow, B.D.; Ashley, M. V High levels of gene flow in bur oak revealed by paternity analysis using microsatellites. J. Hered. 1998, 89, 62–70. [Google Scholar] [CrossRef]
- Chybicki, I.J.; Burczyk, J. Seeing the forest through the trees: Comprehensive inference on individual mating patterns in a mixed stand of Quercus robur and Q. petraea. Ann. Bot. 2013, 112, 561–574. [Google Scholar] [CrossRef]
- Barrett, S.; Kohn, J. Genetic and evolutionary consequences of small population size in plants: Implications for conservation. In Genetics and Conservation of Rare Plants; Oxford University Press: New York, NY, USA, 1991; pp. 3–30. [Google Scholar]
- Franceschinelli, E.V.; Bawa, K.S. The effect of ecological factors on the mating system of a South American shrub species (Helicteres brevispira). Heredity 2000, 84, 116–123. [Google Scholar] [CrossRef]
- Farris, M.A.; Mitton, J.B. Population density, outcrossing rate, and heterozygote superiority in ponderosa pine. Evolution 1984, 38, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J. Density-dependent pollinator foraging, flowering phenology, and temporal pollen dispersal patterns in Linanthus bicolor. Evolution 1983, 37, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Ennos, R.A.; Clegg, M.T. Effect of population substructuring on estimates of outcrossing rate in plant populations. Heredity 1982, 48, 283–292. [Google Scholar] [CrossRef]
- Menitsky, Y.L. Oaks of Asia; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Johnson, P.S.; Shifley, S.R.; Rogers, R. The Ecology and Silviculture of Oaks; CABI Publishing: Wallingford, UK, 2002. [Google Scholar]
- Nixon, K.C. Infrageneric classification of Quercus (Fagaceae) and typification of sectional names. Ann. Des. Sci. For. 1993, 50, 25–34. [Google Scholar] [CrossRef]
- Curtu, A.L.; Gailing, O.; Finkeldey, R. Patterns of contemporary hybridization inferred from paternity analysis in a four-oak-species forest. BMC Evol. Biol. 2009, 9, 284. [Google Scholar] [CrossRef]
- Curtu, A.L.; Craciunesc, I.; Enescu, C.M.; Vidalis, A.; Sofletea, N. Fine-scale spatial genetic structure in a multi-oak-species (Quercus spp.) forest. iForest Biogeosci. For. 2015, 8, 324–332. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- StatSoft STATISTICA for Windows, Version 8.0; Software-System for Data Analysis; StatSoft, Inc.: Tulsa, OK, USA, 2008.
- Li, Y.; Su, Y.; Zhao, X.; Yang, M.; Hu, T.; Zhang, J.; Liu, J.; Liu, M.; Guo, Q. Retrieval of tree branch architecture attributes from terrestrial laser scan data using a Laplacian algorithm. Agric. For. Meteorol. 2020, 284. [Google Scholar] [CrossRef]
- Ferrara, R.; Virdis, S.G.P.; Ventura, A.; Ghisu, T.; Duce, P.; Pellizzaro, G. An automated approach for wood-leaf separation from terrestrial LIDAR point clouds using the density based clustering algorithm DBSCAN. Agric. For. Meteorol. 2018, 262, 434–444. [Google Scholar] [CrossRef]
- Coșofreț, C.; Barnoaiea, I.; Scriban, R.E.; Dănilă, I.C.; Duduman, M.L.; Bouriaud, O. Utilizarea scanerului laser terestru în măsurătorile forestiere: Cerințe metodologice și precauții necesare la aplicarea în practică. Bucov. For. 2018, 18, 137–153. [Google Scholar] [CrossRef]
- Savolainen, O.; Hedrick, P. Heterozygosity and fitness: No association in Scots pine. Genetics 1995, 140, 755–766. [Google Scholar] [CrossRef]
- Bergmann, F.; Ruetz, W. Isozyme genetic variation and heterozygosity in random tree samples and selected orchard clones from the same Norway spruce populations. For. Ecol. Manag. 1991, 46, 39–47. [Google Scholar] [CrossRef]
- Asuka, Y.; Tani, N.; Tsumura, Y.; Tomaru, N. Development and characterization of microsatellite markers for Fagus crenata Blume. Mol. Ecol. Notes 2004, 4, 101–103. [Google Scholar] [CrossRef]
- Șofletea, N.; Curtu, L. Dendrologie; Editura Universitatii Transilvania: Brasov, Romania, 2007; ISBN 9789736358852. [Google Scholar]
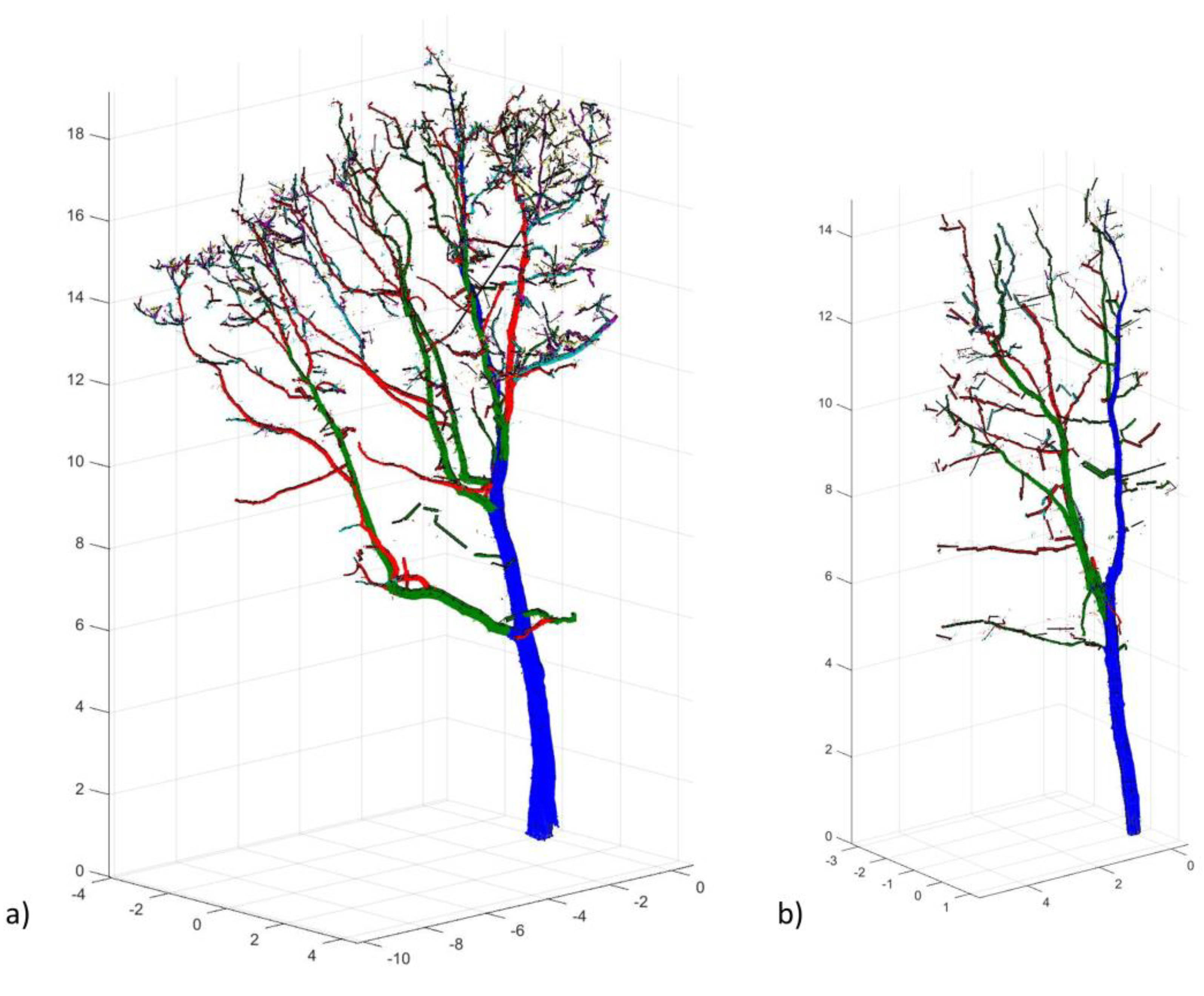
| Tree Characteristics | Description |
|---|---|
| Total Volume | total volume of the tree (sum of all cylinder volumes) in liters |
| Trunk Volume | volume of the stem in liters |
| Branch Volume | volume of all the branches in liters |
| Tree Height | height of the tree in meters vertical distance between the base of the tree and the tip of the highest branch on the tree |
| Trunk Length | length of the stem in meters between the base of the tree and the tip of the highest branch of the tree |
| Branch Length | total length of all the branches in meters |
| Number Branches | number of branches |
| Max Branch Order | maximum branching order |
| Total Area | total surface area of the tree in sq.m. (sum of all cylinder surface area) |
| DBHqsm | DBH in m, the diameter of the cylinder in the QSM at the right height |
| Value | Trunk | Crown | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total Vol | Trunk Vol | Tree Hgt. | Trunk Len | DBH | Branch Vol | Branch Len | No. Branch | Max Branch Order | Total Area | |
| m3 | m3 | m | m | cm | m3 | m | - | - | m2 | |
| Quercus spp. (51 individuals) | ||||||||||
| Mean | 2.003 | 1.304 | 20.015 | 21.133 | 45.86 | 0.699 | 489.9 | 615.314 | 6 | 62.45 |
| SD | 1.323 | 0.898 | 3.736 | 4.7 | 14.98 | 0.669 | 464.1 | 655.356 | 2.175 | 46.74 |
| Q. petraea (25 individuals) | ||||||||||
| Mean | 2.189 | 1.646 | 20.746 | 22.168 | 51.41 | 0.542 | 360.2 | 459.800 | 5.400 | 51.39 |
| SD | 1.395 | 1.010 | 4.078 | 4.603 | 13.76 | 0.588 | 397.0 | 561.001 | 2.598 | 39.59 |
| Q. robur (14 individuals) | ||||||||||
| Mean | 1.856 | 0.953 | 20.742 | 21.742 | 42.73 | 0.903 | 631.823 | 687.571 | 6.300 | 78.64 |
| SD | 1.049 | 0.587 | 3.215 | 4.946 | 14.10 | 0.642 | 412.894 | 547.247 | 1.499 | 47.62 |
| Q. frainetto (6 individuals) | ||||||||||
| Mean | 2.428 | 1.264 | 18.885 | 19.872 | 42.18 | 1.163 | 899.067 | 1279.333 | 6.667 | 95.33 |
| SD | 1.837 | 0.869 | 2.274 | 2.920 | 17.71 | 1.015 | 734.870 | 1108.775 | 1.751 | 68.20 |
| Q. pubescens (6 individuals) | ||||||||||
| Mean | 1.148 | 0.739 | 16.402 | 16.655 | 33.74 | 0.410 | 290.683 | 430.667 | 7.000 | 37.85 |
| SD | 0.782 | 0.458 | 2.526 | 3.831 | 20.30 | 0.332 | 159.239 | 265.82 | 1.414 | 20.30 |
| Trunk | Crown | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | Total Vol. | Trunk Vol. | Tree Hgt. | Trunk Len. | DBH | Branch Vol. | Branch Len. | No Branches | Max Branch Order | Total Area |
| m3 | m3 | m | m | cm | m3 | m | - | - | m2 | |
| Pollen Donors | ||||||||||
| Mean | 2.703 | 1.846 | 20.409 | 22.682 | 53.026 | 0.857 | 554.679 | 723.3 | 6.5 | 72.100 |
| SD | 1.157 | 0.889 | 3.922 | 4.271 | 10.532 | 0.596 | 421.900 | 607.0 | 2.7 | 38.790 |
| Non-Donors | ||||||||||
| Mean | 1.276 | 1.039 | 19.633 | 20.353 | 39.553 | 0.238 | 179.912 | 243.4 | 5 | 30.272 |
| SD | 0.522 | 0.531 | 4.129 | 4.040 | 9.888 | 0.918 | 195.907 | 304.9 | 2.3 | 14.015 |
| p-value | 0.010 | 0.042 | 0.683 | 0.556 | 0.021 | 0.021 | 0.052 | 0.077 | 0.249 | 0.027 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomșa, V.R.; Curtu, A.L.; Niță, M.D. Tree Shape Variability in a Mixed Oak Forest Using Terrestrial Laser Technology: Implications for Mating System Analysis. Forests 2021, 12, 253. https://doi.org/10.3390/f12020253
Tomșa VR, Curtu AL, Niță MD. Tree Shape Variability in a Mixed Oak Forest Using Terrestrial Laser Technology: Implications for Mating System Analysis. Forests. 2021; 12(2):253. https://doi.org/10.3390/f12020253
Chicago/Turabian StyleTomșa, Vlăduț Remus, Alexandru Lucian Curtu, and Mihai Daniel Niță. 2021. "Tree Shape Variability in a Mixed Oak Forest Using Terrestrial Laser Technology: Implications for Mating System Analysis" Forests 12, no. 2: 253. https://doi.org/10.3390/f12020253
APA StyleTomșa, V. R., Curtu, A. L., & Niță, M. D. (2021). Tree Shape Variability in a Mixed Oak Forest Using Terrestrial Laser Technology: Implications for Mating System Analysis. Forests, 12(2), 253. https://doi.org/10.3390/f12020253








