Comparative Plastome Analyses and Phylogenetic Applications of the Acer Section Platanoidea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Chloroplast Genome Sequencing, Assembling and Annotation
2.3. Divergence Hotspot Identification
2.4. Phylogenomic Reconstruction
3. Results and Discussion
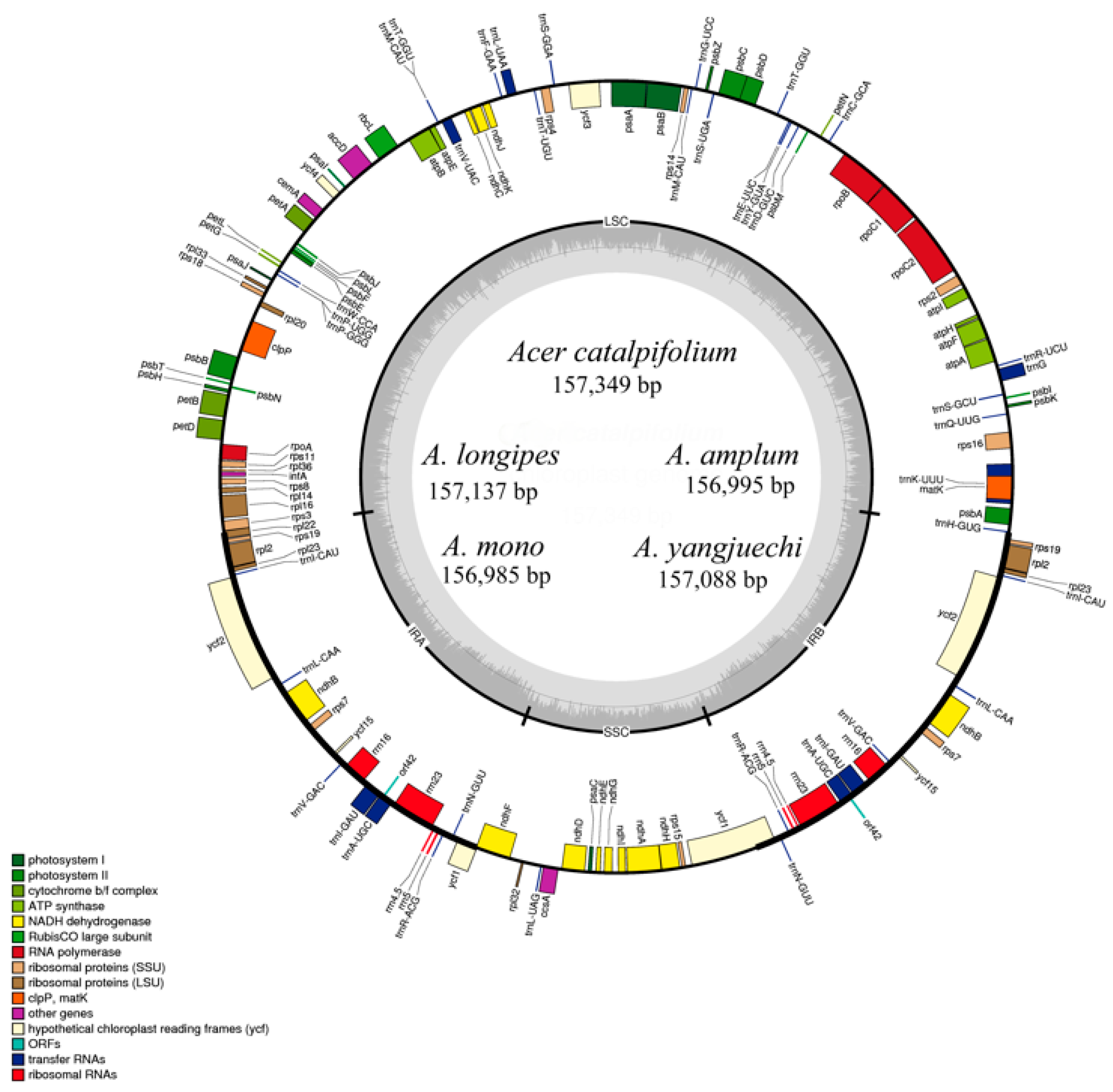
3.1. Chloroplast Genome Organisation of the Acer sect. Platanoidea
3.2. Comparative Analysis of the Genomic Structure
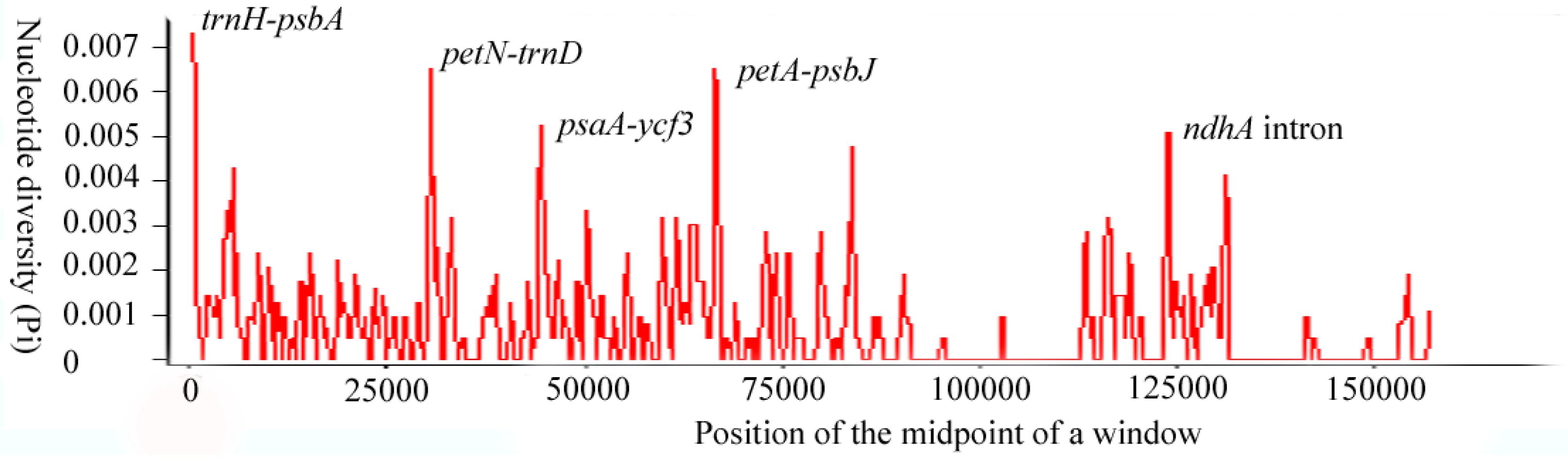
3.3. Divergence Hotspot of the Acer sect. Platanoidea Species
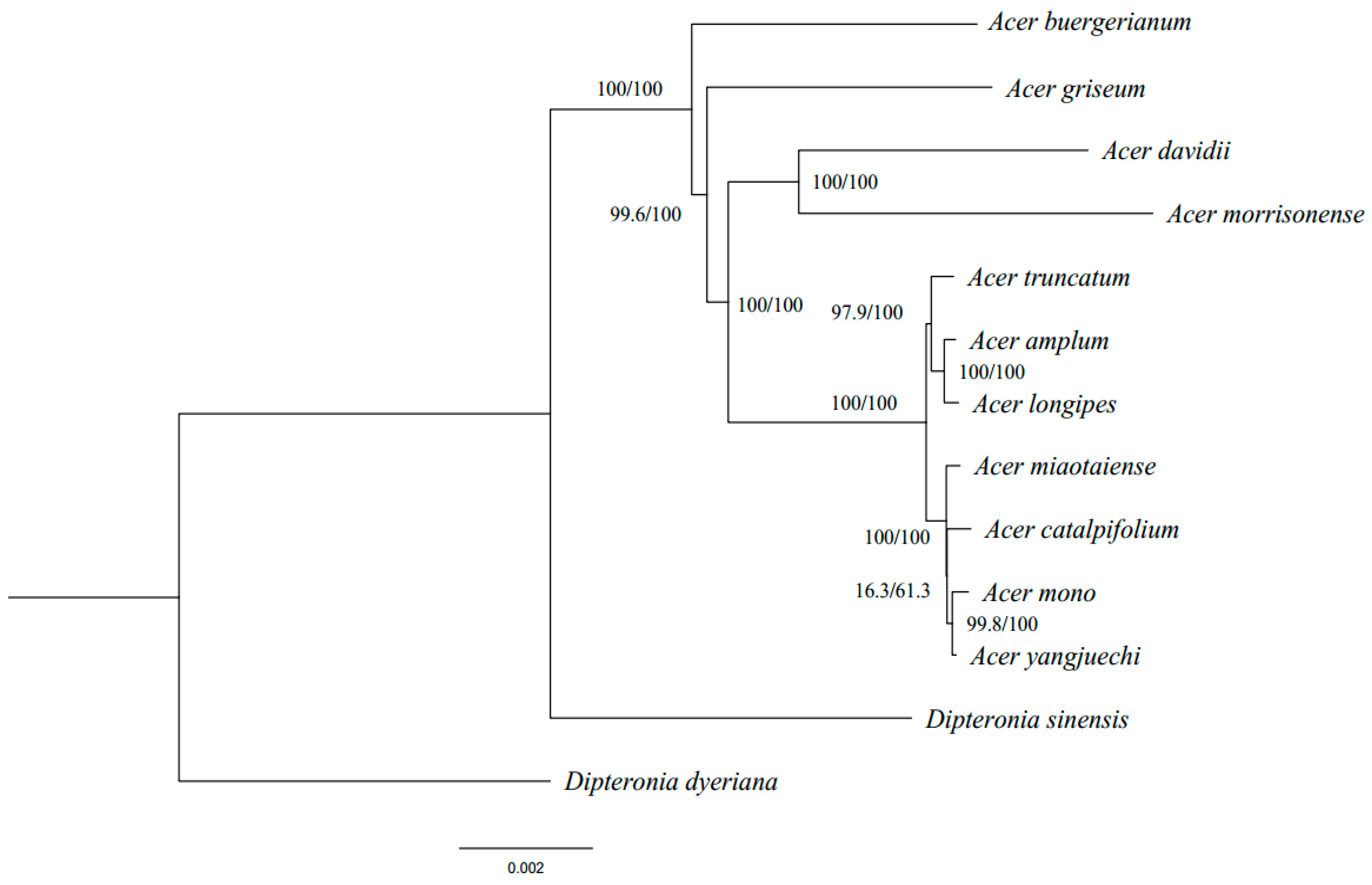
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Neuhaus, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef] [PubMed]
- Armbrust, E.V. Uniparental inheritance of chloroplast genomes. Adv. Photosynth. Respir. 1998, 7, 93–113. [Google Scholar]
- Choi, K.S.; Chung, M.G.; Park, S.J. The complete chloroplast genome sequences of three veroniceae species (plantaginaceae): Comparative analysis and highly divergent regions. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Lv, J.; Li, J.; Du, F.K.; Yin, K. The complete chloroplast genome of the dove tree Davidia involucrata (Nyssaceae), a relict species endemic to China. Conserv. Genet. Resour. 2016, 8, 1–4. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.K.; Singh, V.P.; Gupta, D.K.; Singh, N.K.; Sharma, T.R. Genomic resources in horticultural crops: Status, utility and challenges. Biotechnol. Adv. 2011, 29, 199–209. [Google Scholar] [CrossRef]
- Xiong, J.S.; Ding, J.; Li, Y. Genome-editing technologies and their potential application in horticultural crop breeding. Hortic. Res. 2015, 2. [Google Scholar] [CrossRef]
- Cai, J.; Ma, P.F.; Li, H.T.; Li, D.Z. Complete plastid genome sequencing of fourtiliaspecies (malvaceae): A comparative analysis and phylogenetic implications. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Ruhsam, M.; Rai, H.S.; Mathews, S.; Ross, T.G.; Graham, S.W.; Raubeson, L.A.; Mei, W.; Thomas, P.I.; Gardner, M.F.; Ennos, R.A.; et al. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Resour. 2015, 15, 1067–1078. [Google Scholar] [CrossRef]
- Yi, X.; Gao, L.; Wang, B.; Su, Y.J.; Wang, T. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol. Evol. 2013, 5, 688–698. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Brozynska, M.; Furtado, A.; Waters, D.L.; Henry, R.J. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.Z.; Chen, Y.S.; de Jong, P.C.; Oterdoom, H.J. Aceraceae. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science press: Beijing, China, 2008; Volume 11, pp. 515–553. [Google Scholar]
- Van Gelderen, D.M.; De Jong, P.C.; Oterdoom, H.J. Maples of the World; Timber Press: Portland, OR, USA, 1994; ISBN 0881920002. [Google Scholar]
- Renner, S.S.; Grimm, G.W.; Schneeweiss, G.M.; Stuessy, T.F.; Ricklefs, R.E. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: Implications for north American/Asian disjunctions. Syst. Biol. 2008, 57, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Rosado, A.; Vera-Vélez, R.; Cota-Sánchez, J.H. Floral morphology and reproductive biology in selected maple ( Acer, L.) species (Sapindaceae). Braz. J. Bot. 2018, 41, 1–14. [Google Scholar] [CrossRef]
- Guo, X.; Wang, H.; Bao, L.; Wang, T.; Bai, W.; Ye, J.; Ge, J. Evolutionary history of a widespread tree species Acer mono in East Asia. Ecol. Evol. 2014, 4, 4332–4345. [Google Scholar] [PubMed]
- Grimm, G.W.; Denk, T. The Colchic region as refuge for relict tree lineages: Cryptic speciation in field maples. Turk. J. Bot. 2014, 38, 1050–1066. [Google Scholar] [CrossRef]
- Kumar, S.; Hahn, F.M.; McMahan, C.M.; Cornish, K.; Whalen, M.C. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009, 9, 131. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; Chen, H.; Wang, Y. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conserv. Genet. Resour. 2016, 8, 1–3. [Google Scholar] [CrossRef]
- Wang, R.; Fan, J.; Chang, P.; Zhu, L.; Zhao, M.; Li, L. Genome survey sequencing of acer truncatum bunge to identify genomic information, simple sequence repeat (ssr) markers and complete chloroplast genome. Forests 2019, 10, 87. [Google Scholar] [CrossRef]
- Doyle, J.J. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Dai, M.; Thompson, R.C.; Maher, C.; Contrerasgalindo, R.; Kaplan, M.H.; Markovitz, D.M.; Omenn, G.; Meng, F. NGSQC: Cross-platform quality analysis pipeline for deep sequencing data. BMC Genom. 2010, 11. [Google Scholar] [CrossRef]
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–A baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41, e129. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yang, J.; He, Y.L.; He, Y.; Niu, C.; Gong, L.L.; Li, Z.H. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conserv. Genet. Resour. 2016, 8, 1–3. [Google Scholar] [CrossRef]
- Li, Z.-H.; Xie, Y.-S.; Zhou, T.; Jia, Y.; He, Y.-L.; Yang, J. The complete chloroplast genome sequence of Acer morrisonense (Aceraceae). Mitochondrial DNA Part A 2015, 28, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Guindon, S.; Lethiec, F.; Duroux, P.; Gascuel, O. PHYML Online–A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005, 33, 557–559. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Guo, H.; Liul, J.; Luo, L.; Wei, X.; Zhang, J.; Qi, Y.; Zhang, B.; Liu, H.; Xiao, P. Complete chloroplast genome sequences of Schisandra chinensis: Genome structure, comparative analysis, and phylogenetic relationship of basal angiosperms. Sci. China Life Sci. 2017, 60, 1286–1290. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; Paola, D.D.; Danzi, D.; Vendramin, G.G.; Sonnante, G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other asteraceae. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H. The complete chloroplast genome sequence of the medicinal plant salvia miltiorrhiza. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-F.; Yu, Y.; Deng, Y.-Q.; Li, J.; Liu, H.-Y.; Zhou, S.-D.; He, X.-J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef]
- Han, Y.W.; Duan, D.; Ma, X.F.; Jia, Y.; Liu, Z.L.; Zhao, G.F.; Li, Z.H. Efficient identification of the forest tree species in aceraceae using DNA barcodes. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2. [Google Scholar] [CrossRef]
- Pang, X.; Chang, L.; Shi, L.; Rui, L.; Dong, L.; Li, H.; Cherny, S.S.; Chen, S. Utility of the trnh–Psba intergenic spacer region and its combinations as plant dna barcodes: a meta-analysis. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Lahaye, R.R.Y.; Savolainen, V.; Duthoit, S.; Maurin, O.; Bank, M. A test of psbK-psbI and atpF-atpH as potential plant DNA barcodes using the flora of the Kruger National Park (South Africa) as a model system. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Timme, R.E.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007, 94, 302–312. [Google Scholar] [CrossRef]
- Falchi, A.; Paolini, J.; Desjobert, J.M.; Melis, A.; Costa, J.; Varesi, L. Phylogeography of Cistus creticus L. on Corsica and Sardinia inferred by the TRNL-F and RPL32-TRNL sequences of cpDNA. Mol. Phylogenetics Evol. 2009, 52, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Baillie, J.E.M.; Hilton, C.; Stuart, S.N. IUCN Red List of Threatened Species: A Global Species Assessment; IUCN-The World Conservation Union: Grand, Switzerland, 2004. [Google Scholar]
- Zhang, Y.; Yu, T.; Ma, W.; Tian, C.; Sha, Z.; Li, J. Morphological and physiological response of Acer catalpifolium Rehd. Seedlings to water and light stresses. Glob. Ecol. Conserv. 2019, 19, 10. [Google Scholar] [CrossRef]
- Zhao, J.; Yao, X.; Xi, L.; Yang, J.; Chen, H.; Zhang, J. Characterization of the chloroplast genome sequence of acer miaotaiense: comparative and phylogenetic analyses. Molecules 2018, 23, 1740. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Yue, J.P.; Shoup, S. Phylogenetics of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harv. Pap. Bot. 2006, 11, 101–115. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.; Su, J.; Cong, Y. Extraction of genomic DNA from Acer yangjuechi and system optimization of SRAP-PCR. Jiangsu J. Agric. Sci. 2009, 25, 538–541. [Google Scholar]
- Liu, C.; Tsuda, Y.; Shen, H.; Hu, L.; Saito, Y.; Ide, Y. Genetic structure and hierarchical population divergence history of Acer mono var. mono in South and Northeast China. PLoS ONE 2014, 9. [Google Scholar] [CrossRef][Green Version]

| Name | Total (bp) | LSC (bp) | SSC (bp) | IR (bp) | GC% |
|---|---|---|---|---|---|
| A. catalpifolium | 157,349 | 85,745 | 18,066 | 26,769 | 37.9 |
| A. miaotaiense | 156,595 | 86,327 | 18,068 | 26,100 | 37.9 |
| A. longipes | 157,137 | 85,531 | 18,068 | 26,769 | 37.9 |
| A. amplum | 156,995 | 85,386 | 18,077 | 26,766 | 37.9 |
| A. mono | 156,985 | 85,378 | 18,069 | 26,769 | 37.9 |
| A. yangjuechi | 157,088 | 85,483 | 18,069 | 26,768 | 37.9 |
| A. truncatum | 156,262 | 86,019 | 18,073 | 26,085 | 37.9 |
| Group of Gene | Genes Name | |
|---|---|---|
| 1 | Photostsyem I | psaA, psaB, psaC, psaI, psaJ |
| 2 | Photostsyem II | psbA, psbB, psbC, psbD, psbE, psbF, psbh, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| 3 | Cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN |
| 4 | ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI |
| 5 | NADH dehydrogenase | ndhA*, ndhB*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| 6 | RubisCO large subunit | rbcL |
| 7 | RNA polymerase | ropA, ropB, ropC1*, ropC2 |
| 8 | Ribosomal proteins(SSU) | rps2, rps3, rps4, rps7, rps8, rps11, rps12**, rps14, rps15, rps16*, rps18, rps19 |
| 9 | Ribosomal proteins (LSU) | rpl2*, rpl14, rpl16*, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 |
| 10 | Other gene | clpP **, matK, accD, ccsA, infA, cemA |
| 11 | Proteins of unknown function | ycf1, ycf2, ycf3**, ycf4, ycf15 |
| 12 | ORFs | Orf42 |
| 13 | Transfer RNAs | 31 tRNAs(six contain a single intron) |
| 14 | Ribosomal RNAs | rrn4.5, rrn5, rrn16, rrn23 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Gao, J.; Huang, B.-H.; Dayananda, B.; Ma, W.-B.; Zhang, Y.-Y.; Liao, P.-C.; Li, J.-Q. Comparative Plastome Analyses and Phylogenetic Applications of the Acer Section Platanoidea. Forests 2020, 11, 462. https://doi.org/10.3390/f11040462
Yu T, Gao J, Huang B-H, Dayananda B, Ma W-B, Zhang Y-Y, Liao P-C, Li J-Q. Comparative Plastome Analyses and Phylogenetic Applications of the Acer Section Platanoidea. Forests. 2020; 11(4):462. https://doi.org/10.3390/f11040462
Chicago/Turabian StyleYu, Tao, Jian Gao, Bing-Hong Huang, Buddhi Dayananda, Wen-Bao Ma, Yu-Yang Zhang, Pei-Chun Liao, and Jun-Qing Li. 2020. "Comparative Plastome Analyses and Phylogenetic Applications of the Acer Section Platanoidea" Forests 11, no. 4: 462. https://doi.org/10.3390/f11040462
APA StyleYu, T., Gao, J., Huang, B.-H., Dayananda, B., Ma, W.-B., Zhang, Y.-Y., Liao, P.-C., & Li, J.-Q. (2020). Comparative Plastome Analyses and Phylogenetic Applications of the Acer Section Platanoidea. Forests, 11(4), 462. https://doi.org/10.3390/f11040462






