Impact of Invasive Tree Species on Natural Regeneration Species Composition, Diversity, and Density
Abstract
1. Introduction
2. Materials and Methods
2.1. Species Studied
2.2. Study Area
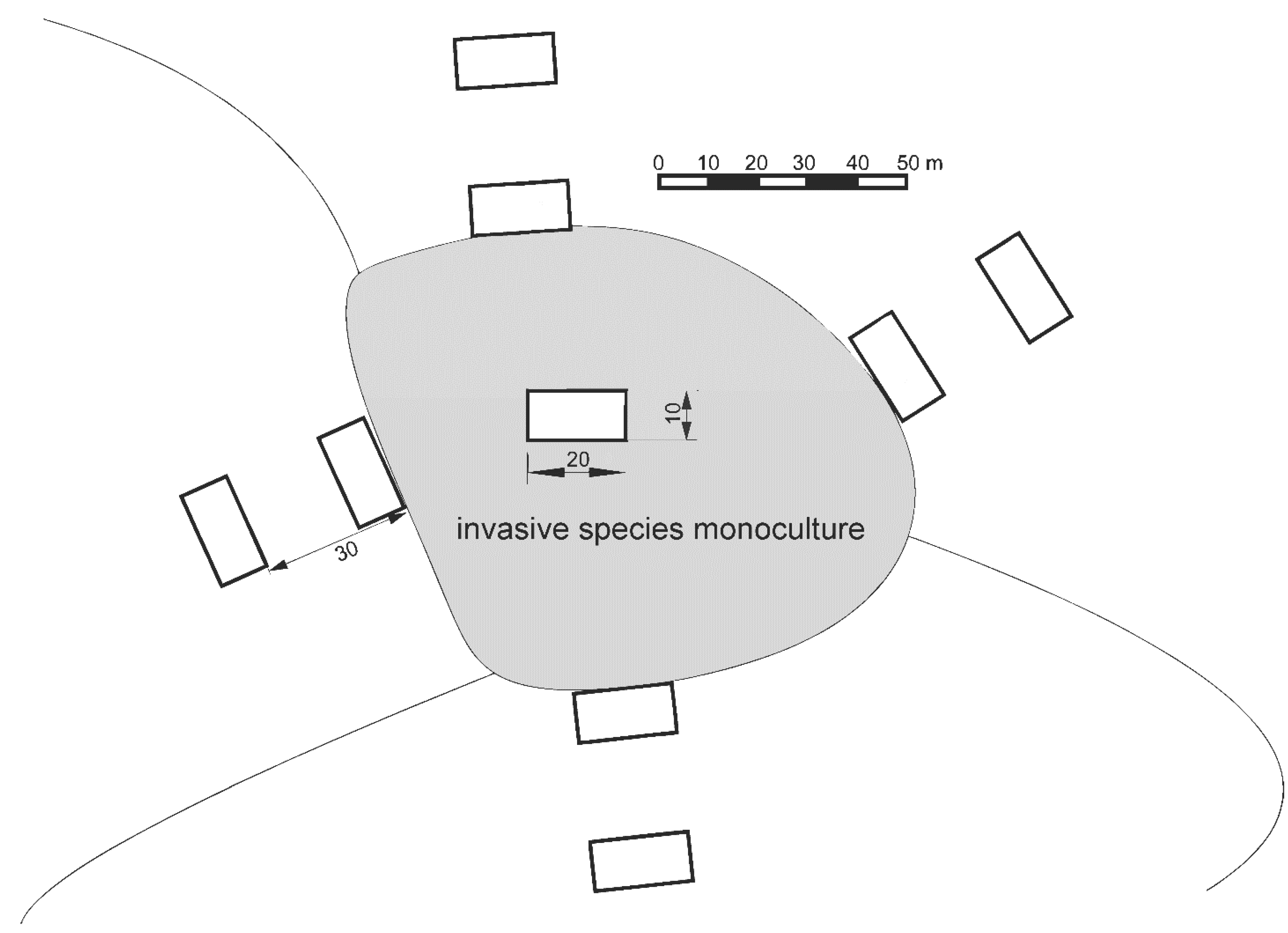
2.3. Study Design
2.4. Data Collection
2.5. Data Analysis
2.5.1. Natural Regeneration Species Composition
2.5.2. Natural Regeneration Species Diversity
2.5.3. Natural Regeneration Density
3. Results
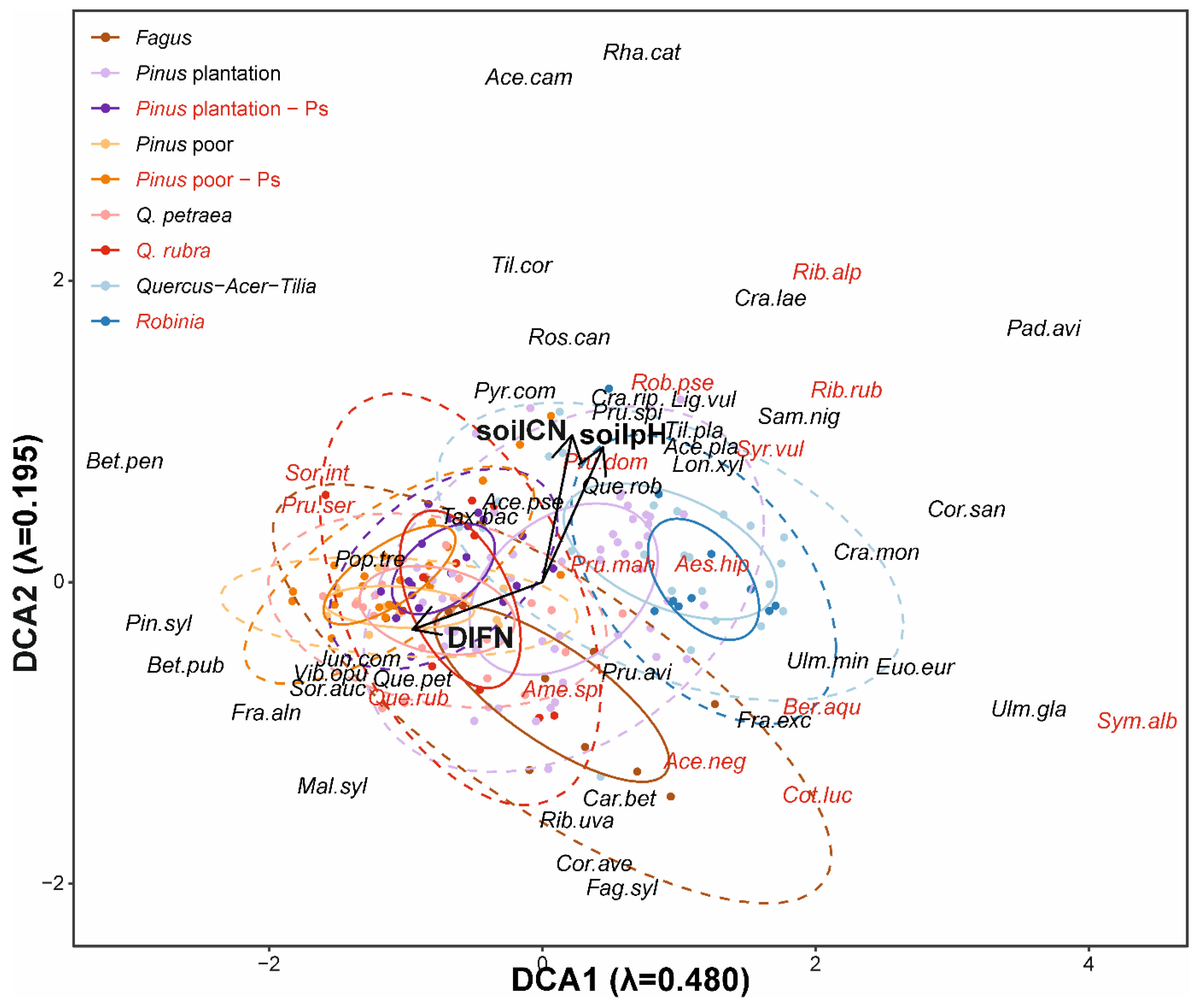
3.1. Impact of Alien Tree Species on Natural Regeneration Species Composition
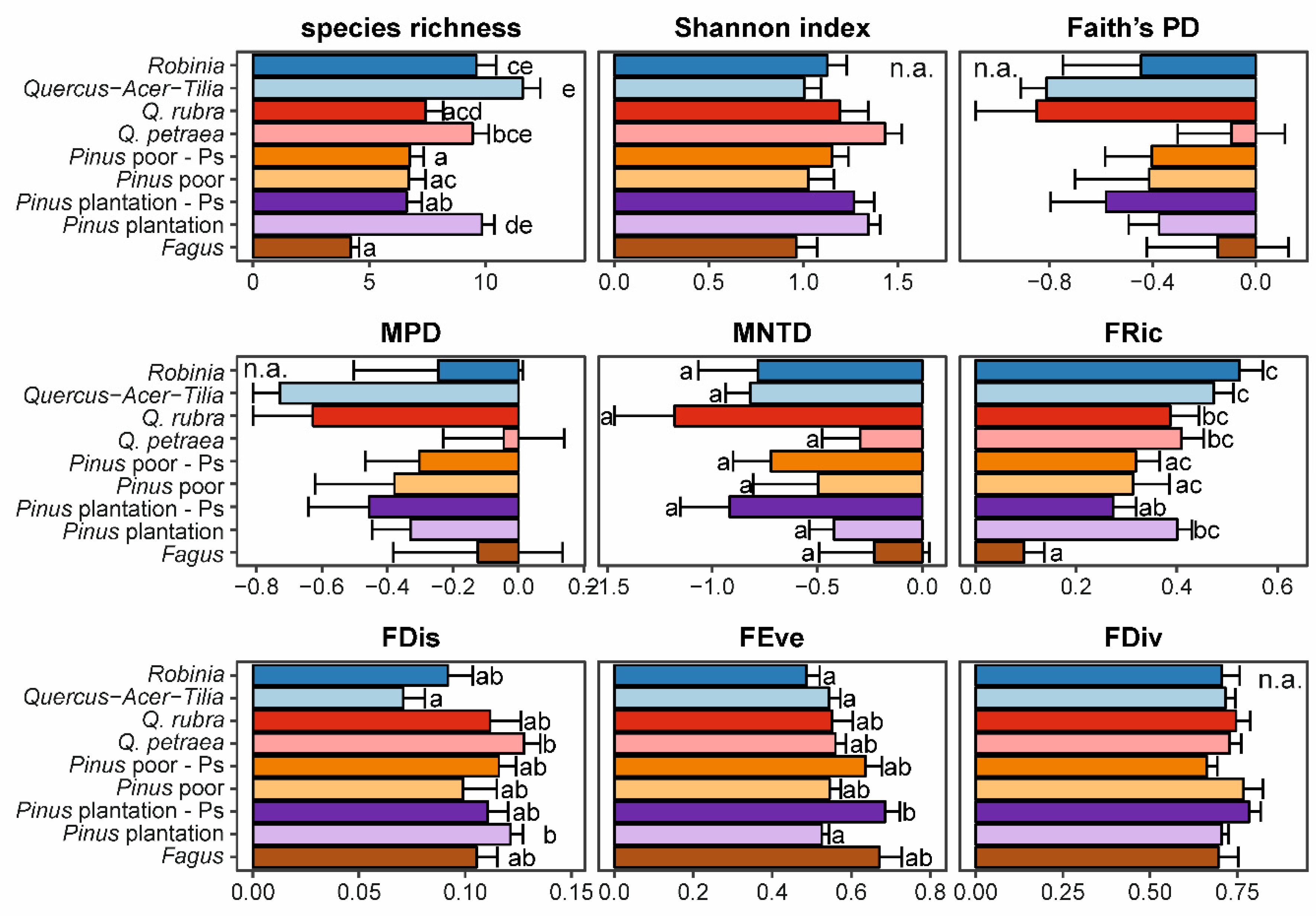
3.2. Impact of Alien Tree Species on Natural Regeneration Diversity
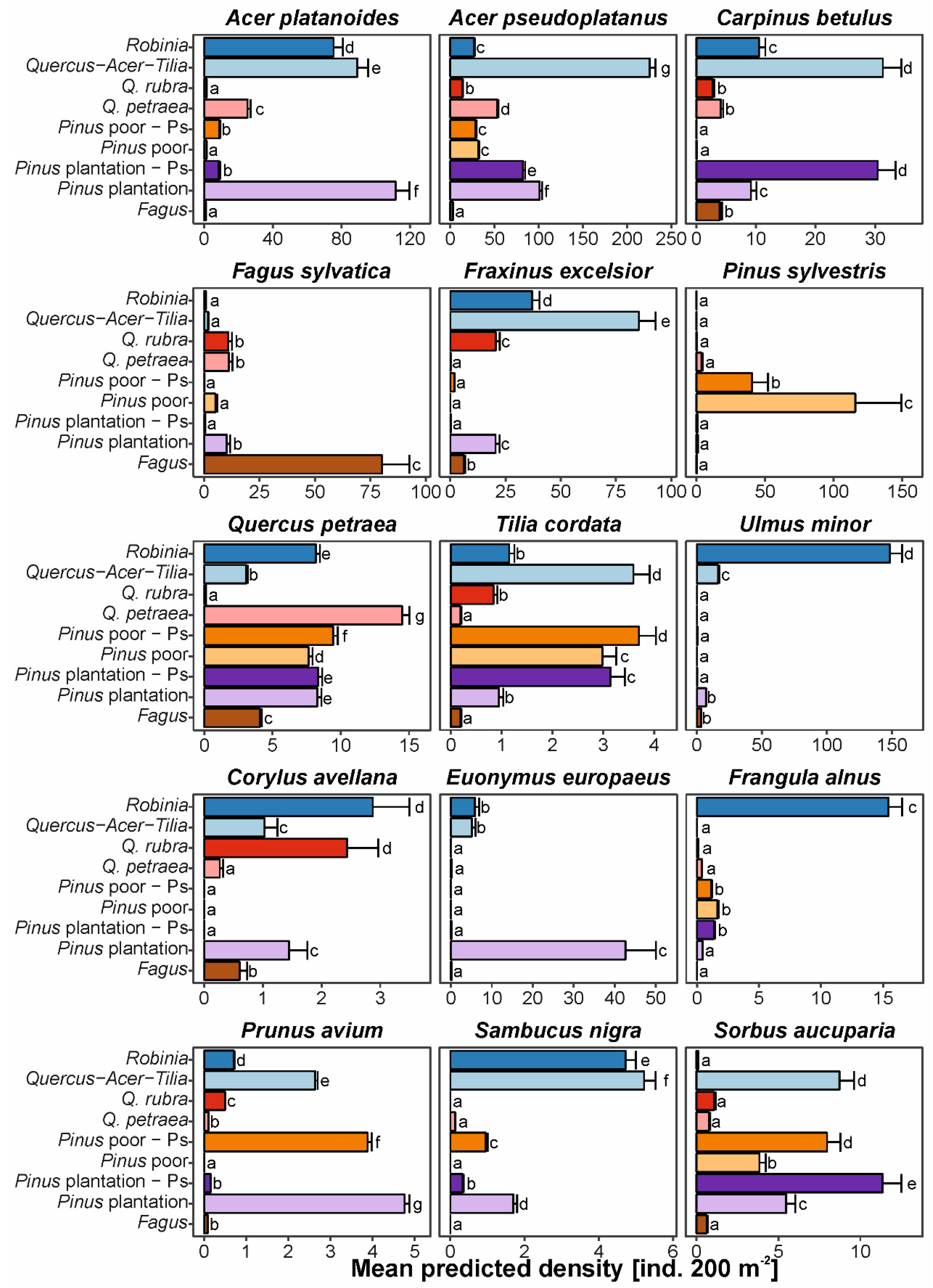
3.3. Impacts on Natural Regeneration Densities of the Most Frequent Native Tree Species
4. Discussion
4.1. Continuity in Species Composition of Natural Regeneration
4.2. Low and Context-Dependent Impacts of Particular Invasive Species on Natural Regeneration Diversities
4.3. Decreasing Ability of Native Trees to Regenerate
4.4. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Spatial Distribution of Forest Types within Blocks and Species Frequency within Studied Forest Types
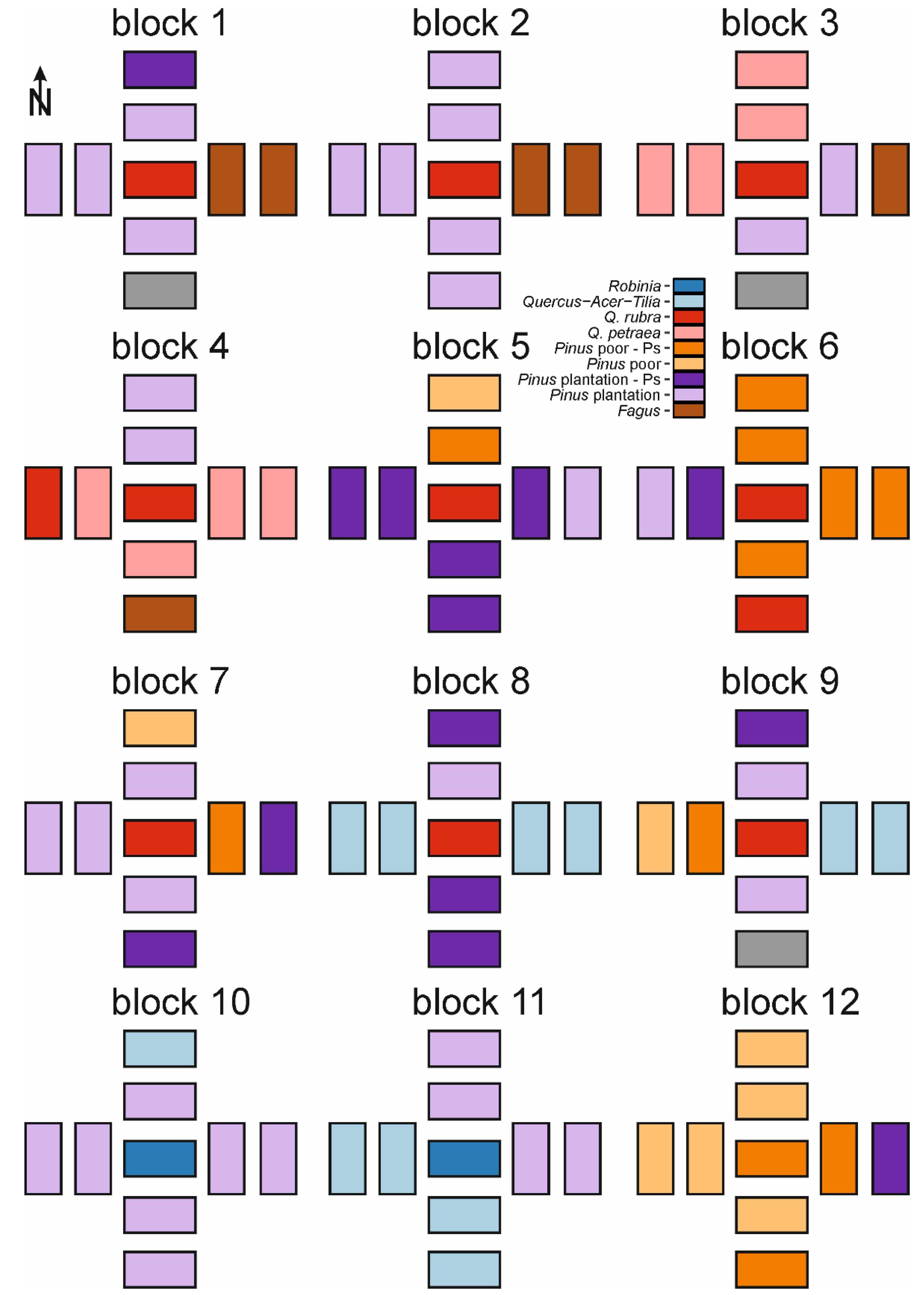
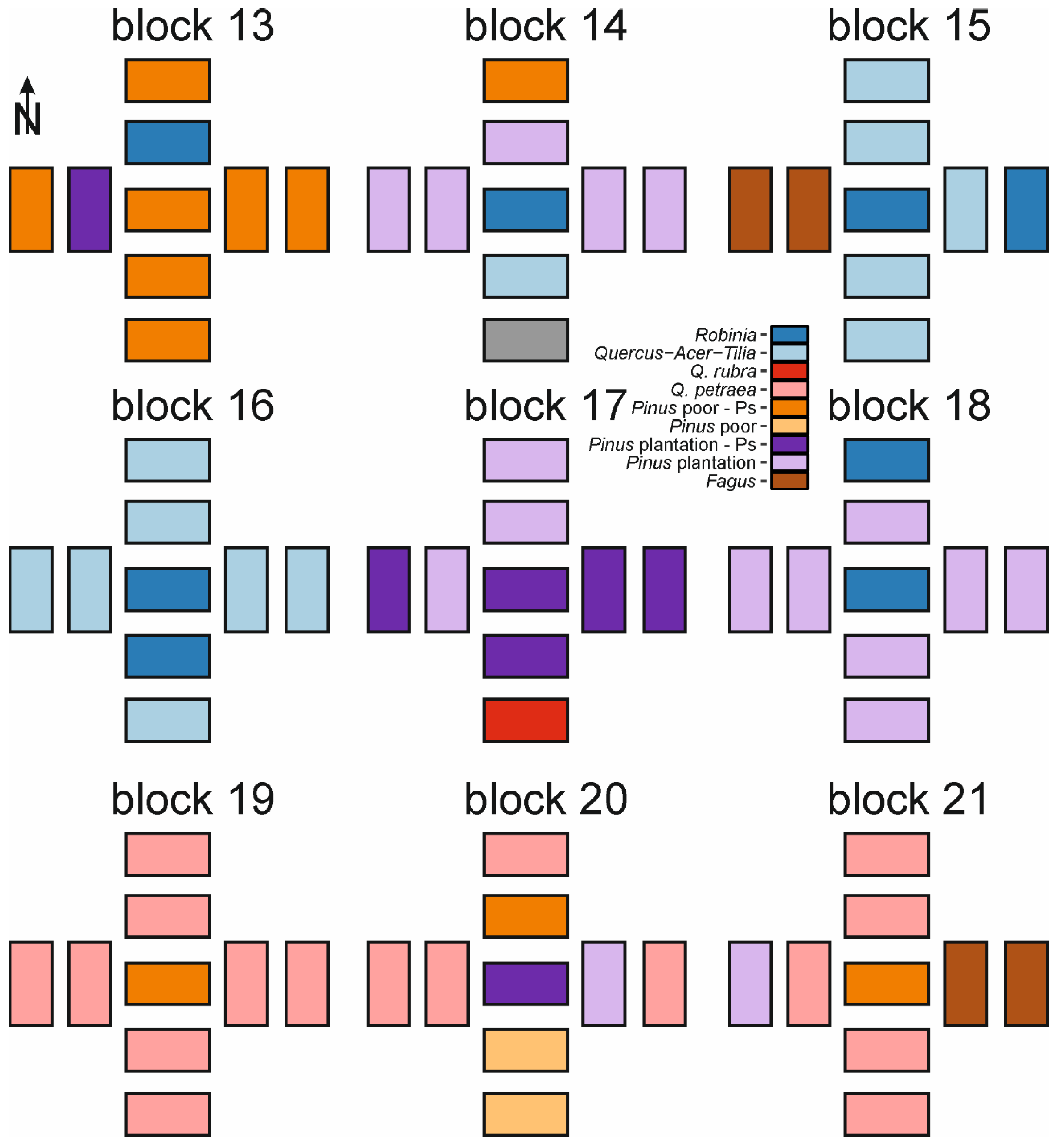
| Species | Fagus (n = 10) | Pinus plantation (n = 52) | Pinus plantation-Ps (n = 20) | Pinus poor (n = 10) | Pinus poor-Ps (n = 23) | Q. petraea (n = 25) | Q. rubra (n = 12) | Quercus-Acer-Tilia (n = 24) | Robinia (n = 10) |
|---|---|---|---|---|---|---|---|---|---|
| Acer campestre L. | 0.0 | 3.8 | 15.0 | 10.0 | 21.7 | 0.0 | 0.0 | 25.0 | 10.0 |
| Acer negundo L. | 0.0 | 1.9 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 0.0 |
| Acer platanoides L. | 20.0 | 69.2 | 40.0 | 40.0 | 26.1 | 28.0 | 25.0 | 100.0 | 100.0 |
| Acer pseudoplatanus L. | 60.0 | 90.4 | 100.0 | 100.0 | 87.0 | 72.0 | 83.3 | 83.3 | 100.0 |
| Aesculus hippocastanum L. | 0.0 | 1.9 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 |
| Amelanchier spicata K.Koch | 0.0 | 7.7 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 |
| Berberis aquifolium Pursh | 0.0 | 7.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 10.0 |
| Betula pendula Roth | 0.0 | 0.0 | 15.0 | 10.0 | 39.1 | 12.0 | 0.0 | 4.2 | 0.0 |
| Betula pubescens Ehrh. | 0.0 | 0.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Carpinus betulus L. | 20.0 | 61.5 | 30.0 | 30.0 | 8.7 | 28.0 | 25.0 | 33.3 | 60.0 |
| Cornus sanguinea L. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 29.2 | 0.0 |
| Corylus avellana L. | 20.0 | 23.1 | 0.0 | 0.0 | 0.0 | 12.0 | 8.3 | 8.3 | 20.0 |
| Cotoneaster lucidus Schltdl. | 0.0 | 9.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 20.0 |
| Crataegus laevigata DC. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 |
| Crataegus monogyna Jacq. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 |
| Crataegus rhipidophylla Gand. | 0.0 | 7.7 | 10.0 | 0.0 | 4.3 | 4.0 | 8.3 | 12.5 | 20.0 |
| Euonymus europaeus L. | 10.0 | 17.3 | 5.0 | 0.0 | 0.0 | 4.0 | 0.0 | 50.0 | 90.0 |
| Fagus sylvatica L. | 100.0 | 38.5 | 15.0 | 40.0 | 4.3 | 68.0 | 33.3 | 45.8 | 70.0 |
| Frangula alnus Mill. | 0.0 | 15.4 | 40.0 | 50.0 | 43.5 | 16.0 | 8.3 | 0.0 | 10.0 |
| Fraxinus excelsior L. | 40.0 | 48.1 | 15.0 | 0.0 | 8.7 | 4.0 | 8.3 | 58.3 | 70.0 |
| Juniperus communis L. | 0.0 | 3.8 | 10.0 | 10.0 | 0.0 | 0.0 | 8.3 | 0.0 | 0.0 |
| Ligustrum vulgare L. | 0.0 | 7.7 | 0.0 | 0.0 | 4.3 | 0.0 | 0.0 | 8.3 | 10.0 |
| Lonicera xylosteum L. | 10.0 | 19.2 | 15.0 | 0.0 | 0.0 | 4.0 | 0.0 | 20.8 | 20.0 |
| Malus sylvestris Mill. | 0.0 | 1.9 | 25.0 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 |
| Pinus sylvestris L. | 10.0 | 5.8 | 25.0 | 60.0 | 47.8 | 36.0 | 16.7 | 0.0 | 0.0 |
| Populus tremula L. | 0.0 | 1.9 | 0.0 | 20.0 | 8.7 | 4.0 | 0.0 | 0.0 | 10.0 |
| Prunus avium (L.) L. | 10.0 | 26.9 | 10.0 | 0.0 | 8.7 | 12.0 | 8.3 | 29.2 | 20.0 |
| Prunus domestica L. | 20.0 | 42.3 | 50.0 | 10.0 | 26.1 | 16.0 | 8.3 | 37.5 | 60.0 |
| Prunus mahaleb L. | 0.0 | 5.8 | 5.0 | 0.0 | 4.3 | 8.0 | 0.0 | 0.0 | 0.0 |
| Prunus padus L. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 |
| Prunus serotina Ehrh. | 40.0 | 48.1 | 100.0 | 80.0 | 100.0 | 76.0 | 58.3 | 37.5 | 30.0 |
| Prunus spinosa L. | 0.0 | 7.7 | 0.0 | 10.0 | 0.0 | 0.0 | 8.3 | 4.2 | 0.0 |
| Pyrus communis L. | 0.0 | 11.5 | 25.0 | 10.0 | 21.7 | 8.0 | 0.0 | 12.5 | 10.0 |
| Quercus petraea (Matt.) Liebl. | 80.0 | 73.1 | 100.0 | 100.0 | 95.7 | 96.0 | 16.7 | 54.2 | 70.0 |
| Quercus robur L. | 0.0 | 5.8 | 15.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 10.0 |
| Quercus rubra L. | 50.0 | 61.5 | 90.0 | 50.0 | 60.9 | 28.0 | 83.3 | 25.0 | 10.0 |
| Rhamnus cathartica L. | 0.0 | 0.0 | 5.0 | 0.0 | 4.3 | 0.0 | 0.0 | 0.0 | 20.0 |
| Ribes alpinum L. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 |
| Ribes rubrum L. | 0.0 | 7.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 40.0 |
| Ribes uva-crispa L. | 0.0 | 13.5 | 5.0 | 10.0 | 0.0 | 4.0 | 0.0 | 4.2 | 0.0 |
| Robinia pseudoacacia L. | 30.0 | 48.1 | 35.0 | 0.0 | 8.7 | 4.0 | 8.3 | 37.5 | 60.0 |
| Rosa canina L. | 0.0 | 5.8 | 15.0 | 10.0 | 17.4 | 8.0 | 0.0 | 4.2 | 20.0 |
| Sambucus nigra L. | 0.0 | 32.7 | 25.0 | 0.0 | 17.4 | 8.0 | 0.0 | 62.5 | 90.0 |
| Sorbus aucuparia L. | 30.0 | 40.4 | 90.0 | 40.0 | 78.3 | 48.0 | 33.3 | 12.5 | 10.0 |
| Sorbus intermedia (Ehrh.) Pers. | 0.0 | 3.8 | 15.0 | 10.0 | 8.7 | 8.0 | 0.0 | 0.0 | 10.0 |
| Symphoricarpos albus (L.) C.Koch | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 |
| Syringa vulgaris L. | 0.0 | 1.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.2 | 10.0 |
| Taxus baccata L. | 0.0 | 5.8 | 20.0 | 0.0 | 4.3 | 0.0 | 0.0 | 4.2 | 10.0 |
| Tilia cordata Mill. | 10.0 | 34.6 | 55.0 | 10.0 | 39.1 | 8.0 | 16.7 | 37.5 | 20.0 |
| Tilia platyphyllos Scop. | 0.0 | 9.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.3 | 0.0 |
| Ulmus glabra Huds. | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 33.3 | 20.0 |
| Ulmus minor Mill. | 10.0 | 34.6 | 10.0 | 0.0 | 8.7 | 4.0 | 0.0 | 54.2 | 90.0 |
| Viburnum opulus L. | 0.0 | 1.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
References
- Baraloto, C.; Goldberg, D.E.; Bonal, D. Performance trade-offs among tropical tree seedlings in contrasting microhabitats. Ecology 2005, 86, 2461–2472. [Google Scholar] [CrossRef]
- Gazda, A.; Kościelniak, P.; Hardy, M.; Muter, E.; Kędra, K.; Bodziarczyk, J.; Frączek, M.; Chwistek, K.; Różański, W.; Szwagrzyk, J. Upward expansion of distribution ranges of tree species: Contrasting results from two national parks in Western Carpathians. Sci. Total Environ. 2019, 653, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Niinemets, Ü.; Valladares, F. Tolerance to shade, drought, and waterlogging of temperate Northern Hemisphere trees and shrubs. Ecol. Monogr. 2006, 76, 521–547. [Google Scholar] [CrossRef]
- Legras, E.C.; Vander Wall, S.B.; Board, D.I. The role of germination microsite in the establishment of sugar pine and Jeffrey pine seedlings. For. Ecol. Manag. 2010, 260, 806–813. [Google Scholar] [CrossRef]
- Canham, C.D.; Murphy, L. The demography of tree species response to climate: Seedling recruitment and survival. Ecosphere 2016, 7, e01424. [Google Scholar] [CrossRef]
- Lorimer, C.G.; Chapman, J.W.; Lambert, W.D. Tall Understorey Vegetation as a Factor in the Poor Development of Oak Seedlings Beneath Mature Stands. J. Ecol. 1994, 82, 227–237. [Google Scholar] [CrossRef]
- Dech, J.P.; Robinson, L.M.; Nosko, P. Understorey plant community characteristics and natural hardwood regeneration under three partial harvest treatments applied in a northern red oak (Quercus rubra L.) stand in the Great Lakes-St. Lawrence forest region of Canada. For. Ecol. Manag. 2008, 256, 760–773. [Google Scholar] [CrossRef]
- Packer, A.; Clay, K. Soil pathogens and Prunus serotina seedling and sapling growth near conspecific trees. Ecology 2003, 84, 108–119. [Google Scholar] [CrossRef]
- Bakys, R.; Vasaitis, R.; Barklund, P.; Ihrmark, K.; Stenlid, J. Investigations concerning the role of Chalara fraxinea in declining Fraxinus excelsior. Plant Pathol. 2009, 58, 284–292. [Google Scholar] [CrossRef]
- Kain, M.; Battaglia, L.; Royo, A.; Carson, W.P. Over-browsing in Pennsylvania creates a depauperate forest dominated by an understory tree: Results from a 60-year-old deer exclosure. J. Torrey Biol. Soc. 2011, 138, 322–326. [Google Scholar] [CrossRef]
- Iszkuło, G.; Nowak-Dyjeta, K.; Sekiewicz, M. Influence of initial light intensity and deer browsing on Taxus baccata saplings: A six years field study. Dendrobiology 2014, 71, 93–99. [Google Scholar] [CrossRef]
- Orwig, D.A.; Cogbill, C.V.; Foster, D.R.; O’Keefe, J.F. Variations in Old-Growth Structure and Definitions: Forest Dynamics on Wachusett Mountain, Massachusetts. Ecol. Appl. 2001, 11, 437–452. [Google Scholar] [CrossRef]
- Prach, K.; Pyšek, P.; Bastl, M. Spontaneous vegetation succession in human-disturbed habitats: A pattern across seres. Appl. Veg. Sci. 2001, 4, 83–88. [Google Scholar] [CrossRef]
- Kowarik, I.; Hiller, A.; Planchuelo, G.; Seitz, B.; von der Lippe, M.; Buchholz, S. Emerging Urban Forests: Opportunities for Promoting the Wild Side of the Urban Green Infrastructure. Sustainability 2019, 11, 6318. [Google Scholar] [CrossRef]
- Parker, G.R.; Leopold, D.J.; Eichenberger, J.K. Tree dynamics in an old-growth, deciduous forest. For. Ecol. Manag. 1985, 11, 31–57. [Google Scholar] [CrossRef]
- Orczewska, A.; Czortek, P.; Jaroszewicz, B. The impact of salvage logging on herb layer species composition and plant community recovery in Białowieża Forest. Biodivers. Conserv. 2019, 28, 3407–3428. [Google Scholar] [CrossRef]
- Jaroszewicz, B.; Cholewińska, O.; Gutowski, J.M.; Samojlik, T.; Zimny, M.; Latałowa, M. Białowieża Forest—A Relic of the High Naturalness of European Forests. Forests 2019, 10, 849. [Google Scholar] [CrossRef]
- Clarke, P.J.; Keith, D.A.; Vincent, B.E.; Letten, A.D. Post-grazing and post-fire vegetation dynamics: Long-term changes in mountain bogs reveal community resilience. J. Veg. Sci. 2015, 26, 278–290. [Google Scholar] [CrossRef]
- Pröll, G.; Darabant, A.; Gratzer, G.; Katzensteiner, K. Unfavourable microsites, competing vegetation and browsing restrict post-disturbance tree regeneration on extreme sites in the Northern Calcareous Alps. Eur. J. For. Res. 2015, 134, 293–308. [Google Scholar] [CrossRef]
- Czortek, P.; Ratyńska, H.; Dyderski, M.K.; Jagodziński, A.M.; Orczewska, A.; Jaroszewicz, B. Cessation of livestock grazing and windthrow drive a shift in plant species composition in the Western Tatra Mts. Tuexenia 2018, 37, 177–196. [Google Scholar]
- Holmgren, M.; Lin, C.-Y.; Murillo, J.E.; Nieuwenhuis, A.; Penninkhof, J.; Sanders, N.; Bart, T.; Veen, H.; Vasander, H.; Vollebregt, M.E.; et al. Positive shrub–tree interactions facilitate woody encroachment in boreal peatlands. J. Ecol. 2015, 103, 58–66. [Google Scholar] [CrossRef]
- Jagodziński, A.M.; Horodecki, P.; Rawlik, K.; Dyderski, M.K. Do understorey or overstorey traits drive tree encroachment on a drained raised bog? Plant Biol. 2017, 19, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Brundu, G.; Richardson, D.M. Planted forests and invasive alien trees in Europe: A Code for managing existing and future plantings to mitigate the risk of negative impacts from invasions. NeoBiota 2016, 30, 5–47. [Google Scholar] [CrossRef]
- Hejda, M.; Hanzelka, J.; Kadlec, T.; Štrobl, M.; Pyšek, P.; Reif, J. Impacts of an invasive tree across trophic levels: Species richness, community composition and resident species’ traits. Divers. Distrib. 2017, 23, 997–1007. [Google Scholar] [CrossRef]
- Niinemets, Ü. A review of light interception in plant stands from leaf to canopy in different plant functional types and in species with varying shade tolerance. Ecol. Res. 2010, 25, 693–714. [Google Scholar] [CrossRef]
- Dyderski, M.K.; Jagodziński, A.M. Similar Impacts of Alien and Native Tree Species on Understory Light Availability in a Temperate Forest. Forests 2019, 10, 951. [Google Scholar] [CrossRef]
- Castro-Díez, P.; Godoy, O.; Alonso, A.; Gallardo, A.; Saldaña, A. What explains variation in the impacts of exotic plant invasions on the nitrogen cycle? A meta-analysis. Ecol. Lett. 2014, 17, 1–12. [Google Scholar] [CrossRef]
- Mueller, K.E.; Hobbie, S.E.; Chorover, J.; Reich, P.B.; Eisenhauer, N.; Castellano, M.J.; Chadwick, O.A.; Dobies, T.; Hale, C.M.; Jagodziński, A.M.; et al. Effects of litter traits, soil biota, and soil chemistry on soil carbon stocks at a common garden with 14 tree species. Biogeochemistry 2015, 123, 313–327. [Google Scholar] [CrossRef]
- Castro-Díez, P.; Vaz, A.S.; Silva, J.S.; Van Loo, M.; Alonso, Á.; Aponte, C.; Bayón, Á.; Bellingham, P.J.; Chiuffo, M.C.; DiManno, N.; et al. Global effects of non-native tree species on multiple ecosystem services. Biol. Rev. 2019, 94, 1477–1501. [Google Scholar] [CrossRef]
- Gentili, R.; Ferrè, C.; Cardarelli, E.; Montagnani, C.; Bogliani, G.; Citterio, S.; Comolli, R. Comparing Negative Impacts of Prunus serotina, Quercus rubra and Robinia pseudoacacia on Native Forest Ecosystems. Forests 2019, 10, 842. [Google Scholar] [CrossRef]
- Stanek, M.; Stefanowicz, A.M. Invasive Quercus rubra negatively affected soil microbial communities relative to native Quercus robur in a semi-natural forest. Sci. Total Environ. 2019, 696, 133977. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.E.; Eisenhauer, N.; Reich, P.B.; Hobbie, S.E.; Chadwick, O.A.; Chorover, J.; Dobies, T.; Hale, C.M.; Jagodziński, A.M.; Kałucka, I.; et al. Light, earthworms, and soil resources as predictors of diversity of 10 soil invertebrate groups across monocultures of 14 tree species. Soil Biol. Biochem. 2016, 92, 184–198. [Google Scholar] [CrossRef]
- Kohyt, J.; Skubała, P. Oribatid mite (Acari: Oribatida) communities reveal the negative impact of the red oak (Quercus rubra L.) on soil fauna in Polish commercial forests. Pedobiologia 2019, 79, 150594. [Google Scholar] [CrossRef]
- Dickie, I.A.; Bolstridge, N.; Cooper, J.A.; Peltzer, D.A. Co-invasion by Pinus and its mycorrhizal fungi. New Phytol. 2010, 187, 475–484. [Google Scholar] [CrossRef]
- Kałucka, I.L.; Jagodziński, A.M. Successional traits of ectomycorrhizal fungi in forest reclamation after surface mining and agricultural disturbances: A review. Dendrobiology 2016, 76, 91–104. [Google Scholar] [CrossRef]
- Woziwoda, B.; Staniaszek-Kik, M.; Stefańska-Krzaczek, E. Assemblages of native bryophytes in secondary forests with introduced Quercus rubra. Nord. J. Bot. 2017, 35, 111–120. [Google Scholar] [CrossRef]
- Jagodziński, A.M.; Wierzcholska, S.; Dyderski, M.K.; Horodecki, P.; Rusińska, A.; Gdula, A.K.; Kasprowicz, M. Tree species effects on bryophyte guilds on a reclaimed post-mining site. Ecol. Eng. 2018, 110, 117–127. [Google Scholar] [CrossRef]
- Taylor, K.T.; Maxwell, B.D.; Pauchard, A.; Nuñez, M.A.; Rew, L.J. Native versus non-native invasions: Similarities and differences in the biodiversity impacts of Pinus contorta in introduced and native ranges. Divers. Distrib. 2016, 22, 578–588. [Google Scholar] [CrossRef]
- Woziwoda, B.; Dyderski, M.K.; Jagodziński, A.M. Effects of land use change and Quercus rubra introduction on Vaccinium myrtillus performance in Pinus sylvestris forests. For. Ecol. Manag. 2019, 440, 1–11. [Google Scholar] [CrossRef]
- Šibíková, M.; Jarolímek, I.; Hegedüšová, K.; Májeková, J.; Mikulová, K.; Slabejová, D.; Škodová, I.; Zaliberová, M.; Medvecká, J. Effect of planting alien Robinia pseudoacacia trees on homogenization of Central European forest vegetation. Sci. Total Environ. 2019, 687, 1164–1175. [Google Scholar] [CrossRef]
- González-Muñoz, N.; Castro-Díez, P.; Godoy, O. Lack of superiority of invasive over co-occurring native riparian tree seedling species. Biol. Invasions 2014, 16, 269–281. [Google Scholar] [CrossRef]
- Kawaletz, H.; Mölder, I.; Annighöfer, P.; Terwei, A.; Zerbe, S.; Ammer, C. Back to the roots: How do seedlings of native tree species react to the competition by exotic species? Ann. For. Sci. 2014, 71, 337–347. [Google Scholar] [CrossRef]
- Lockwood, J.L.; Cassey, P.; Blackburn, T. The role of propagule pressure in explaining species invasions. Trends Ecol. Evol. 2005, 20, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Jagodziński, A.M.; Dyderski, M.K.; Horodecki, P.; Rawlik, K. Limited dispersal prevents Quercus rubra invasion in a 14-species common garden experiment. Divers. Distrib. 2018, 24, 403–414. [Google Scholar] [CrossRef]
- Terwei, A.; Zerbe, S.; Zeileis, A.; Annighöfer, P.; Kawaletz, H.; Mölder, I.; Ammer, C. Which are the factors controlling tree seedling establishment in North Italian floodplain forests invaded by non-native tree species? For. Ecol. Manag. 2013, 304, 192–203. [Google Scholar] [CrossRef]
- Terwei, A.; Zerbe, S.; Mölder, I.; Annighöfer, P.; Kawaletz, H.; Ammer, C. Response of floodplain understorey species to environmental gradients and tree invasion: A functional trait perspective. Biol. Invasions 2016, 18, 2951–2973. [Google Scholar] [CrossRef]
- Woziwoda, B.; Dyderski, M.K.; Kobus, S.; Parzych, A.; Jagodziński, A.M. Natural regeneration and recruitment of native Quercus robur and introduced Q. rubra in European oak-pine mixed forests. For. Ecol. Manag. 2019, 449, 117473. [Google Scholar] [CrossRef]
- Lichstein, J.W.; Grau, H.R.; Aragón, R.; Ejrnæs, R. Recruitment limitation in secondary forests dominated by an exotic tree. J. Veg. Sci. 2004, 15, 721–728. [Google Scholar] [CrossRef]
- Peña, E.; Hidalgo, M.; Langdon, B.; Pauchard, A. Patterns of spread of Pinus contorta Dougl. ex Loud. invasion in a Natural Reserve in southern South America. For. Ecol. Manag. 2008, 256, 1049–1054. [Google Scholar] [CrossRef]
- Fuentes-Ramírez, A.; Pauchard, A.; Cavieres, L.A.; García, R.A. Survival and growth of Acacia dealbata vs. native trees across an invasion front in south-central Chile. For. Ecol. Manag. 2011, 261, 1003–1009. [Google Scholar]
- Gorchov, D.L.; Trisel, D.E. Competitive effects of the invasive shrub, Lonicera maackii (Rupr.) Herder (Caprifoliaceae), on the growth and survival of native tree seedlings. Plant Ecol. 2003, 166, 13–24. [Google Scholar] [CrossRef]
- Wagner, V.; Chytrý, M.; Jiménez-Alfaro, B.; Pergl, J.; Hennekens, S.; Biurrun, I.; Knollová, I.; Berg, C.; Vassilev, K.; Rodwell, J.S.; et al. Alien plant invasions in European woodlands. Divers. Distrib. 2017, 23, 969–981. [Google Scholar] [CrossRef]
- Brus, R.; Pötzelsberger, E.; Lapin, K.; Brundu, G.; Orazio, C.; Straigyte, L.; Hasenauer, H. Extent, distribution and origin of non-native forest tree species in Europe. Scand. J. For. Res. 2019, 34, 533–544. [Google Scholar] [CrossRef]
- Starfinger, U.; Kowarik, I.; Rode, M.; Schepker, H. From desirable ornamental plant to pest to accepted addition to the flora?—The perception of an alien tree species through the centuries. Biol. Invasions 2003, 5, 323–335. [Google Scholar] [CrossRef]
- Cierjacks, A.; Kowarik, I.; Joshi, J.; Hempel, S.; Ristow, M.; von der Lippe, M.; Weber, E. Biological Flora of the British Isles: Robinia pseudoacacia. J. Ecol. 2013, 101, 1623–1640. [Google Scholar] [CrossRef]
- Woziwoda, B.; Potocki, M.; Sagan, J.; Zasada, M.; Tomusiak, R.; Wilczynski, S. Commercial forestry as a vector of alien tree species-the case of Quercus rubra L. introduction in Poland. Balt. For. 2014, 20, 131–141. [Google Scholar]
- Jagodziński, A.M.; Dyderski, M.K.; Rawlik, M.; Banaszczak, P. Plantation of coniferous trees modifies risk and size of Padus serotina (Ehrh.) Borkh. invasion—Evidence from a Rogów Arboretum case study. For. Ecol. Manag. 2015, 357, 84–94. [Google Scholar]
- Jagodziński, A.M.; Dyderski, M.K.; Horodecki, P.; Knight, K.S.; Rawlik, K.; Szmyt, J. Light and propagule pressure affect invasion intensity of Prunus serotina in a 14-tree species forest common garden experiment. NeoBiota 2019, 46, 1–21. [Google Scholar] [CrossRef]
- Horodecki, P.; Jagodziński, A.M. Tree species effects on litter decomposition in pure stands on afforested post-mining sites. For. Ecol. Manag. 2017, 406, 1–11. [Google Scholar] [CrossRef]
- Bieberich, J.; Lauerer, M.; Aas, G. Acorns of introduced Quercus rubra are neglected by European Jay but spread by mice. Ann. For. Res. 2016, 59, 249–258. [Google Scholar] [CrossRef]
- Myczko, Ł.; Dylewski, Ł.; Zduniak, P.; Sparks, T.H.; Tryjanowski, P. Predation and dispersal of acorns by European Jay (Garrulus glandarius) differs between a native (Pedunculate Oak Quercus robur) and an introduced oak species (Northern Red Oak Quercus rubra) in Europe. For. Ecol. Manag. 2014, 331, 35–39. [Google Scholar] [CrossRef]
- Rice, S.K.; Westerman, B.; Federici, R. Impacts of the exotic, nitrogen-fixing black locust (Robinia pseudoacacia) on nitrogen-cycling in a pine–oak ecosystem. Plant Ecol. 2004, 174, 97–107. [Google Scholar] [CrossRef]
- Vítková, M.; Müllerová, J.; Sádlo, J.; Pergl, J.; Pyšek, P. Black locust (Robinia pseudoacacia) beloved and despised: A story of an invasive tree in Central Europe. For. Ecol. Manag. 2017, 384, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Dyderski, M.K.; Jagodziński, A.M. Drivers of invasive tree and shrub natural regeneration in temperate forests. Biol. Invasions 2018, 20, 2363–2379. [Google Scholar] [CrossRef]
- Woziwoda, B.; Kopeć, D.; Witkowski, J. The negative impact of intentionally introduced Quercus rubra L. on a forest community. Acta Soc. Bot. Pol. 2014, 83, 39–49. [Google Scholar] [CrossRef]
- Purcel, A. Obce gatunki drzew i krzewów w Wielkopolskim Parku Narodowym—ich występowanie i rola w biocenozach Parku. Morena 2009, 14, 35–191. [Google Scholar]
- Nowak, G.; Kara, M.; Bernat, Z.; Cykowiak, Z. Wybrane zagadnienia z planu ochrony ekosystemów leśnych Wielkopolskiego Parku Narodowego. Morena 2000, 7, 85–129. [Google Scholar]
- Dyderski, M.K.; Jagodziński, A.M. Seedling survival of Prunus serotina Ehrh., Quercus rubra L. and Robinia pseudoacacia L. in temperate forests of Western Poland. For. Ecol. Manag. 2019, 450, 117498. [Google Scholar] [CrossRef]
- Reich, P.B.; Oleksyn, J.; Modrzynski, J.; Mrozinski, P.; Hobbie, S.E.; Eissenstat, D.M.; Chorover, J.; Chadwick, O.A.; Hale, C.M.; Tjoelker, M.G. Linking litter calcium, earthworms and soil properties: A common garden test with 14 tree species. Ecol. Lett. 2005, 8, 811–818. [Google Scholar] [CrossRef]
- Bank Danych o Lasach. Available online: http://www.bdl.lasy.gov.pl/ (accessed on 31 January 2017).
- GBIF. Global Biodiversity Information Facility. Available online: http://www.gbif.org/ (accessed on 16 April 2020).
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- Hill, M.O.; Gauch, H.G. Detrended correspondence analysis: An improved ordination technique. Vegetatio 1980, 42, 47–58. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Michin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Henry, M.; Stevens, H.; et al. “Vegan” 2.3.3.—Community Ecology Package. Available online: https://cran.r-project.org/package=vegan (accessed on 16 April 2020).
- Jin, Y.; Qian, H.V. PhyloMaker: An R package that can generate very large phylogenies for vascular plants. Ecography 2019, 42, 1353–1359. [Google Scholar] [CrossRef]
- Tsirogiannis, C.; Sandel, B. PhyloMeasures: A package for computing phylogenetic biodiversity measures and their statistical moments. Ecography 2016, 39, 709–714. [Google Scholar] [CrossRef]
- Kleyer, M.; Bekker, R.M.; Knevel, I.C.; Bakker, J.P.; Thompson, K.; Sonnenschein, M.; Poschlod, P.; Van Groenendael, J.M.; Klimeš, L.; Klimešová, J.; et al. The LEDA Traitbase: A database of life-history traits of the Northwest European flora. J. Ecol. 2008, 96, 1266–1274. [Google Scholar] [CrossRef]
- Klotz, S.; Kühn, I.; Durka, W. BIOLFLOR—Eine Datenbank zu biologisch-ökologischen Merkmalen der Gefäßpflanzen in Deutschland. Schriftenreihe für Vegetationskunde; Schriftenreihe für Vegetationskunde; Bundesamt für Naturschutz: Bonn, Germany, 2002. [Google Scholar]
- Enquist, B.J.; Condit, R.; Peet, R.K.; Schildhauer, M.; Thiers, B.M. Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. PeerJ Prepr. 2016, 4, e2615v2. [Google Scholar]
- Zanne, A.E.; Lopez-Gonzalez, G.; Coomes, D.A.; Ilic, J.; Jansen, S.; Lewis, S.L.; Miller, R.B.; Swenson, N.G.; Wiemann, M.C.; Chave, J. Data from: Towards a worldwide wood economics spectrum. 2019. Available online: https://datadryad.org/stash/dataset/doi:10.5061/dryad.234 (accessed on 16 April 2020). [CrossRef]
- Ellenberg, H.; Leuschner, C. Vegetation Mitteleuropas mit den Alpen in ökologischer, Dynamischer und Historischer Sicht; UTB: Stuttgart, Germany, 2010. [Google Scholar]
- Bretz, F.; Hothorn, T.; Westfall, P.H. Multiple Comparisons Using R; CRC Press: Boca Raton, FL, USA, 2011; ISBN 978-1-58488-574-0. [Google Scholar]
- Zeileis, A.; Kleiber, C.; Jackman, S. Regression Models for Count Data in R. J. Stat. Softw. 2008, 27, 1–21. [Google Scholar] [CrossRef]
- Sitko, A.; Biecek, P. The Merging Path Plot: Adaptive fusing of k-groups with likelihood-based model selection. arXiv 2017, arXiv:1709.04412. [Google Scholar]
- Biecek, P. DALEX: Explainers for Complex Predictive Models in R. J. Mach. Learn. Res. 2018, 19, 3245–3249. [Google Scholar]
- Lepš, J.; Hadincová, V. How reliable are our vegetation analyses? J. Veg. Sci. 1992, 3, 119–124. [Google Scholar] [CrossRef]
- Holeksa, J.; Woźniak, G. Biased vegetation patterns and detection of vegetation changes using phytosociological databases. A case study in the forests of the Babia Góra National Park (the West Carpathians, Poland). Phytocoenologia 2005, 35, 1–18. [Google Scholar] [CrossRef]
- Bigelow, S.W.; Canham, C.D. Community organization of tree species along soil gradients in a north-eastern USA forest. J. Ecol. 2002, 90, 188–200. [Google Scholar] [CrossRef]
- Barney, J.N.; Tekiela, D.R.; Barrios-Garcia, M.N.; Dimarco, R.D.; Hufbauer, R.A.; Leipzig-Scott, P.; Nuñez, M.A.; Pauchard, A.; Pyšek, P.; Vítková, M.; et al. Global Invader Impact Network (GIIN): Toward standardized evaluation of the ecological impacts of invasive plants. Ecol. Evol. 2015, 5, 2878–2889. [Google Scholar] [CrossRef] [PubMed]
- Dyderski, M.K.; Jagodziński, A.M. Low impact of disturbance on ecological success of invasive tree and shrub species in temperate forests. Plant Ecol. 2018, 219, 1369–1380. [Google Scholar] [CrossRef]
- Lososová, Z.; Chytrý, M.; Danihelka, J.; Tichý, L.; Ricotta, C. Biotic homogenization of urban floras by alien species: The role of species turnover and richness differences. J. Veg. Sci. 2016, 27, 452–459. [Google Scholar] [CrossRef]
- Olden, J.D.; Comte, L.; Giam, X. The Homogocene: A research prospectus for the study of biotic homogenisation. NeoBiota 2018, 37, 23–36. [Google Scholar] [CrossRef]
- Pairon, M.; Chabrerie, O.; Casado, C.M.; Jacquemart, A.-L. Sexual regeneration traits linked to black cherry (Prunus serotina Ehrh.) invasiveness. Acta Oecologica 2006, 30, 238–247. [Google Scholar] [CrossRef]
- Dylewski, Ł.; Kurek, P.; Wiatrowska, B.; Jerzak, L.; Tryjanowski, P. Man-made perching sites—Electricity pylons accelerate fleshy-fruited plants succession in farmlands. Flora 2017, 231, 51–56. [Google Scholar] [CrossRef]
- Wróbel, A.; Zwolak, R. Habitat-dependent seed dispersal of an introduced tree species by native rodents. For. Ecol. Manag. 2019, 433, 563–568. [Google Scholar] [CrossRef]
- González-Moreno, P.; Diez, J.M.; Ibáñez, I.; Font, X.; Vilà, M. Plant invasions are context-dependent: Multiscale effects of climate, human activity and habitat. Divers. Distrib. 2014, 20, 720–731. [Google Scholar] [CrossRef]
- Dyderski, M.K.; Jagodziński, A.M. Context-Dependence of Urban Forest Vegetation Invasion Level and Alien Species’ Ecological Success. Forests 2019, 10, 26. [Google Scholar] [CrossRef]
- Sapsford, S.J.; Brandt, A.J.; Davis, K.T.; Peralta, G.; Dickie, I.A.; Gibson, R.D.; Green, J.L.; Hulme, P.E.; Nuñez, M.A.; Orwin, K.H.; et al. Towards a framework for understanding the context-dependence of impacts of non-native tree species. Funct. Ecol. 2020. [Google Scholar] [CrossRef]
- Zerbe, S.; Wirth, P. Non-indigenous plant species and their ecological range in Central European pine (Pinus sylvestris L.) forests. Ann. For. Sci. 2006, 63, 189–203. [Google Scholar] [CrossRef]
- Felton, A.; Lindbladh, M.; Brunet, J.; Fritz, Ö. Replacing coniferous monocultures with mixed-species production stands: An assessment of the potential benefits for forest biodiversity in northern Europe. For. Ecol. Manag. 2010, 260, 939–947. [Google Scholar] [CrossRef]
- Knight, K.S.; Oleksyn, J.; Jagodziński, A.M.; Reich, P.B.; Kasprowicz, M. Overstorey tree species regulate colonization by native and exotic plants: A source of positive relationships between understorey diversity and invasibility. Divers. Distrib. 2008, 14, 666–675. [Google Scholar] [CrossRef]
- Jordano, P.; Schupp, E.W. Seed disperser effectiveness: The quantity component and patterns of seed rain for Prunus mahaleb. Ecol. Monogr. 2000, 70, 591–615. [Google Scholar] [CrossRef]
- Closset-Kopp, D.; Chabrerie, O.; Valentin, B.; Delachapelle, H.; Decocq, G. When Oskar meets Alice: Does a lack of trade-off in r/K-strategies make Prunus serotina a successful invader of European forests? For. Ecol. Manag. 2007, 247, 120–130. [Google Scholar] [CrossRef]
- Halarewicz, A.; Żołnierz, L. Changes in the understorey of mixed coniferous forest plant communities dominated by the American black cherry (Prunus serotina Ehrh.). For. Ecol. Manag. 2014, 313, 91–97. [Google Scholar] [CrossRef]
- Piwczyński, M.; Puchałka, R.; Ulrich, W. Influence of tree plantations on the phylogenetic structure of understorey plant communities. For. Ecol. Manag. 2016, 376, 231–237. [Google Scholar] [CrossRef]
- Stagne, E.E.; Shea, K.L. Effects of Deer Browsing, Fabric Mats, and Tree Shelters on Quercus rubra Seedlings. Restor. Ecol. 1998, 6, 29–34. [Google Scholar]
| Type | n | DIFN | Soil pH | Soil C:N | Dominant Tree Species |
|---|---|---|---|---|---|
| Fagus | 10 | 0.024 ± 0.003 (0.012–0.045) | 4.06 ± 0.10 (3.64–4.64) | 40.85 ± 2.94 (29.89–58.55) | Fagus sylvatica |
| Pinus plantation | 52 | 0.031 ± 0.005 (0.007–0.251) | 4.31 ± 0.10 (3.71–6.65) | 38.71 ± 0.91 (26.42–57.15) | Pinus sylvestris, Acer pseudoplatanus, Quercus petraea |
| Pinus plantation-Ps | 20 | 0.039 ± 0.006 (0.010–0.111) | 4.13 ± 0.06 (3.78–4.86) | 37.31 ± 1.04 (29.43–48.93) | P. sylvestris, A. pseudoplatanus, Q. petraea |
| Pinus poor | 10 | 0.102 ± 0.018 (0.024–0.168) | 3.93 ± 0.04 (3.76–4.16) | 40.93 ± 1.27 (34.75–47.57) | P. sylvestris |
| Pinus poor-Ps | 23 | 0.075 ± 0.009 (0.019–0.151) | 4.20 ± 0.07 (3.78–5.06) | 43.08 ± 1.25 (31.91–54.44) | P. sylvestris |
| Q. petraea | 25 | 0.057 ± 0.006 (0.017–0.120) | 3.91 ± 0.04 (3.56–4.32) | 42.77 ± 1.16 (30.26–51.32) | Q. petraea |
| Q. rubra | 12 | 0.039 ± 0.005 (0.013–0.078) | 4.21 ± 0.12 (3.67–5.16) | 48.52 ± 2.26 (32.75–61.92) | Q. rubra |
| Quercus-Acer-Tilia | 24 | 0.024 ± 0.001 (0.007–0.034) | 5.64 ± 0.20 (3.95–7.10) | 36.77 ± 1.13 (26.10–47.06) | Q. petraea, A. platanoides, A. pseuodoplatanus, Tilia cordata, Carpinus betulus |
| Robinia | 10 | 0.020 ± 0.002 (0.014–0.035) | 5.66 ± 0.36 (3.79–6.89) | 29.19 ± 1.24 (25.38–35.49) | Robinia pseudoacacia, Ulmus minor |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyderski, M.K.; Jagodziński, A.M. Impact of Invasive Tree Species on Natural Regeneration Species Composition, Diversity, and Density. Forests 2020, 11, 456. https://doi.org/10.3390/f11040456
Dyderski MK, Jagodziński AM. Impact of Invasive Tree Species on Natural Regeneration Species Composition, Diversity, and Density. Forests. 2020; 11(4):456. https://doi.org/10.3390/f11040456
Chicago/Turabian StyleDyderski, Marcin K., and Andrzej M. Jagodziński. 2020. "Impact of Invasive Tree Species on Natural Regeneration Species Composition, Diversity, and Density" Forests 11, no. 4: 456. https://doi.org/10.3390/f11040456
APA StyleDyderski, M. K., & Jagodziński, A. M. (2020). Impact of Invasive Tree Species on Natural Regeneration Species Composition, Diversity, and Density. Forests, 11(4), 456. https://doi.org/10.3390/f11040456





