Transcriptome Sequencing and Expression Analysis of Genes Related to Anthocyanin Biosynthesis in Leaves of Malus ‘Profusion’ Infected by Japanese Apple Rust
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total Anthocyanin Content
2.3. Anthocyanin Biosynthesis-Related Compounds Content
2.4. RNA Extraction, Library Construction, and De Novo Transcriptome Assembly
2.5. Gene Identification, Gene Annotation, and Analysis of DEGs
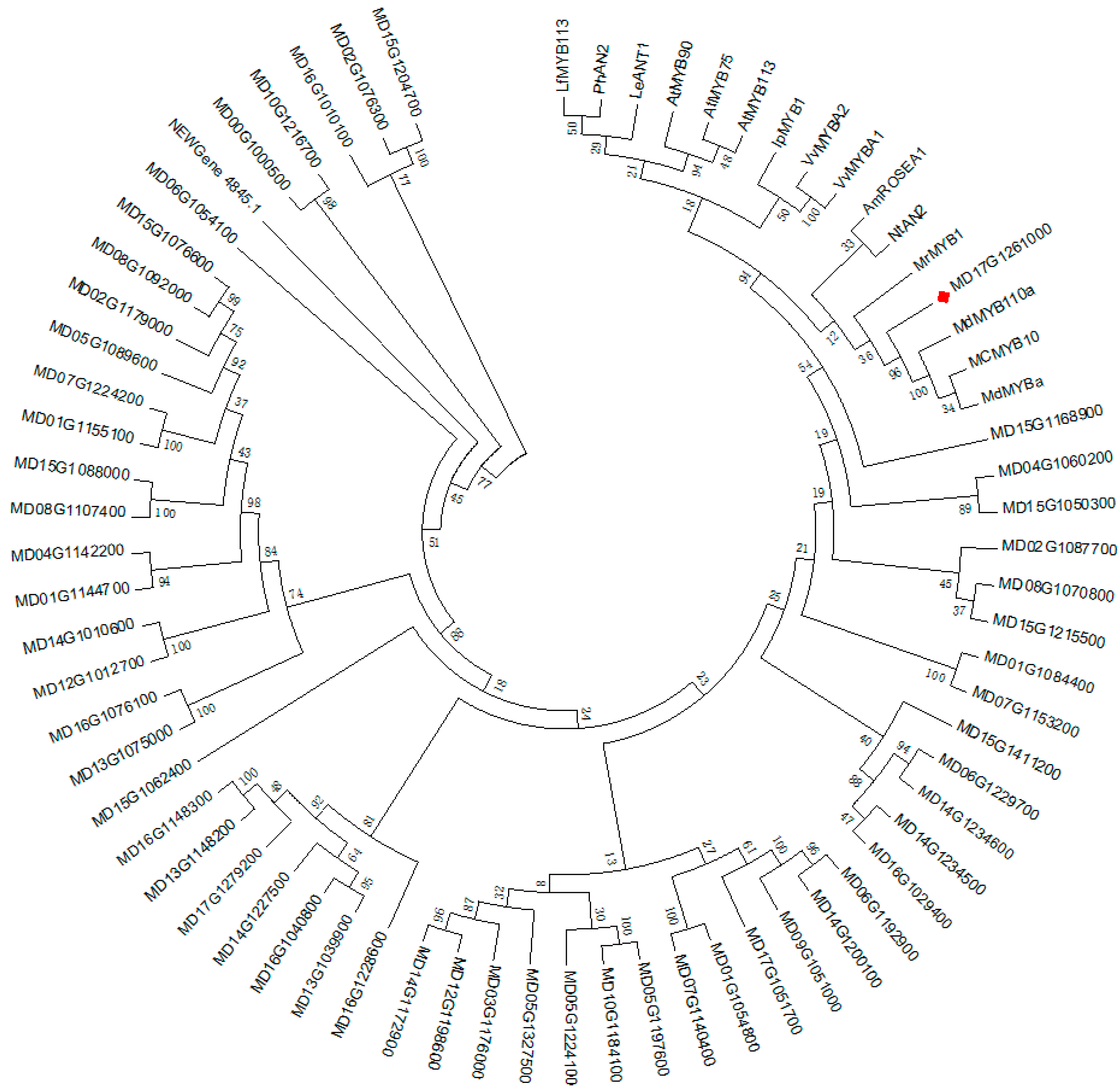
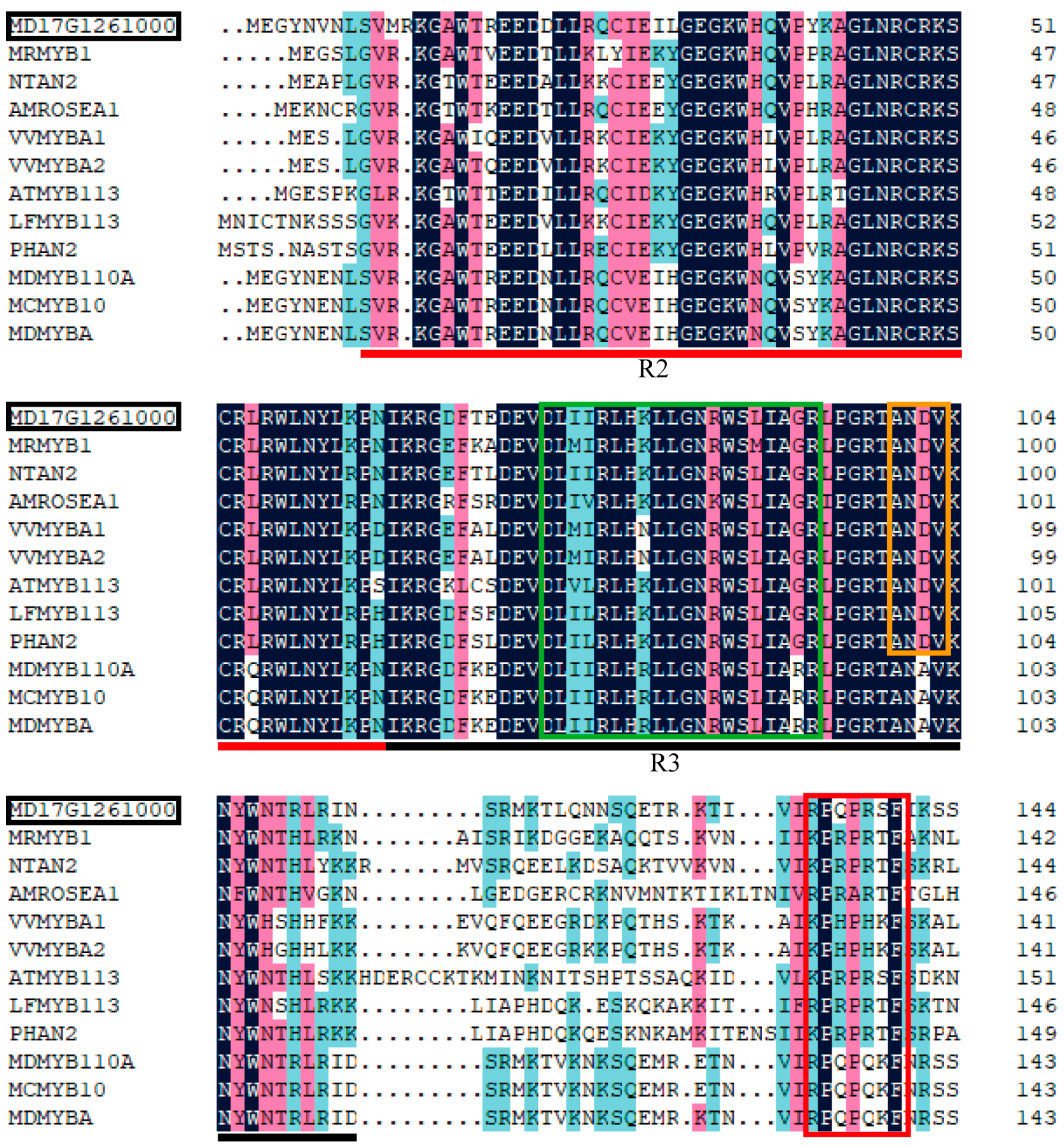
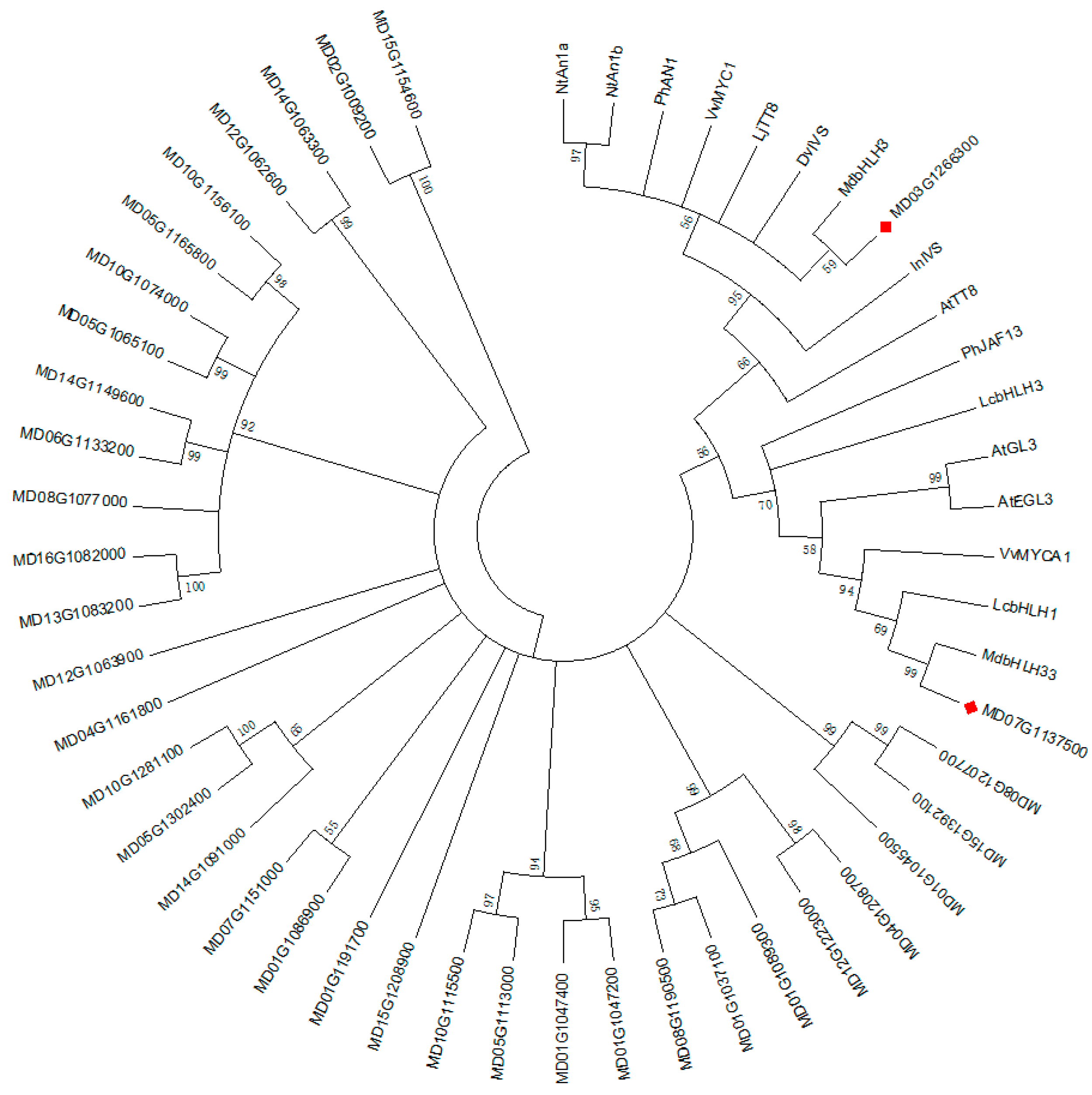
2.6. Phylogenetic Analysis and Sequence Alignment
2.7. Quantitative Real Time PCR (qRT-PCR) Analysis
2.8. Data Analysis
3. Results
3.1. Analysis of Total Anthocyanin Content and Composition
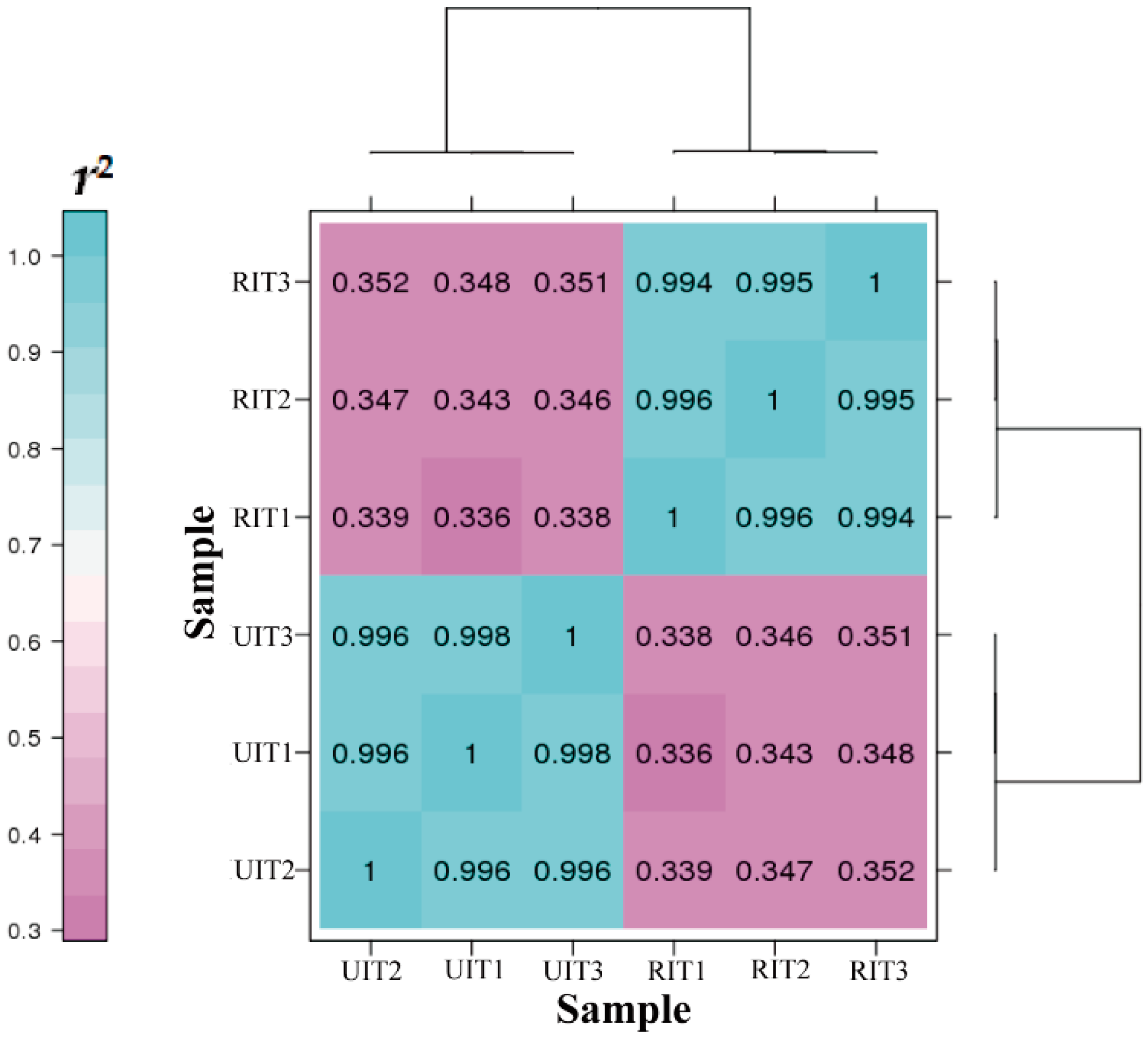
3.2. Library Construction and Transcriptome Sequencing
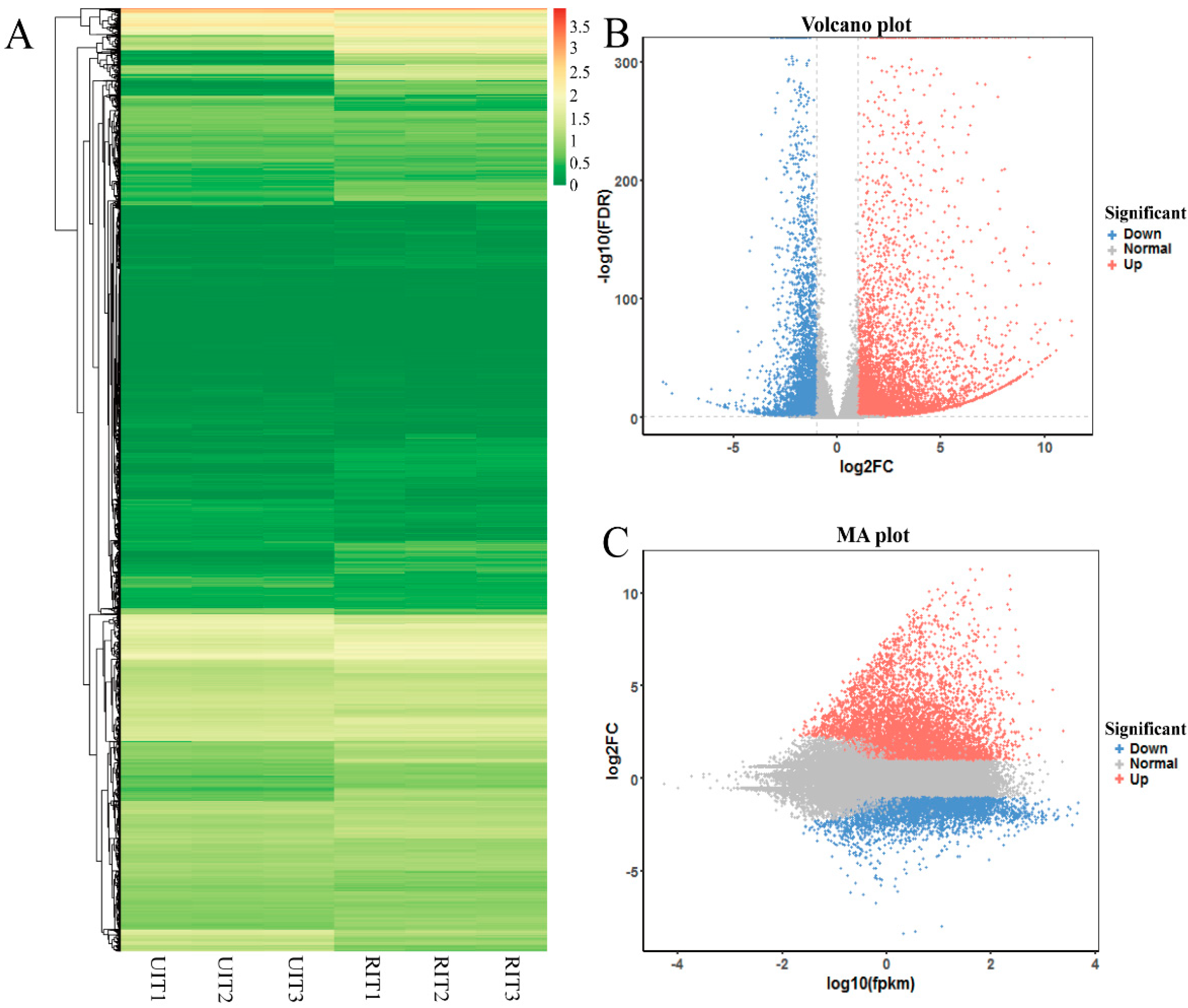
3.3. Annotation and Identification of DEGs
3.4. Analysis of DEGs
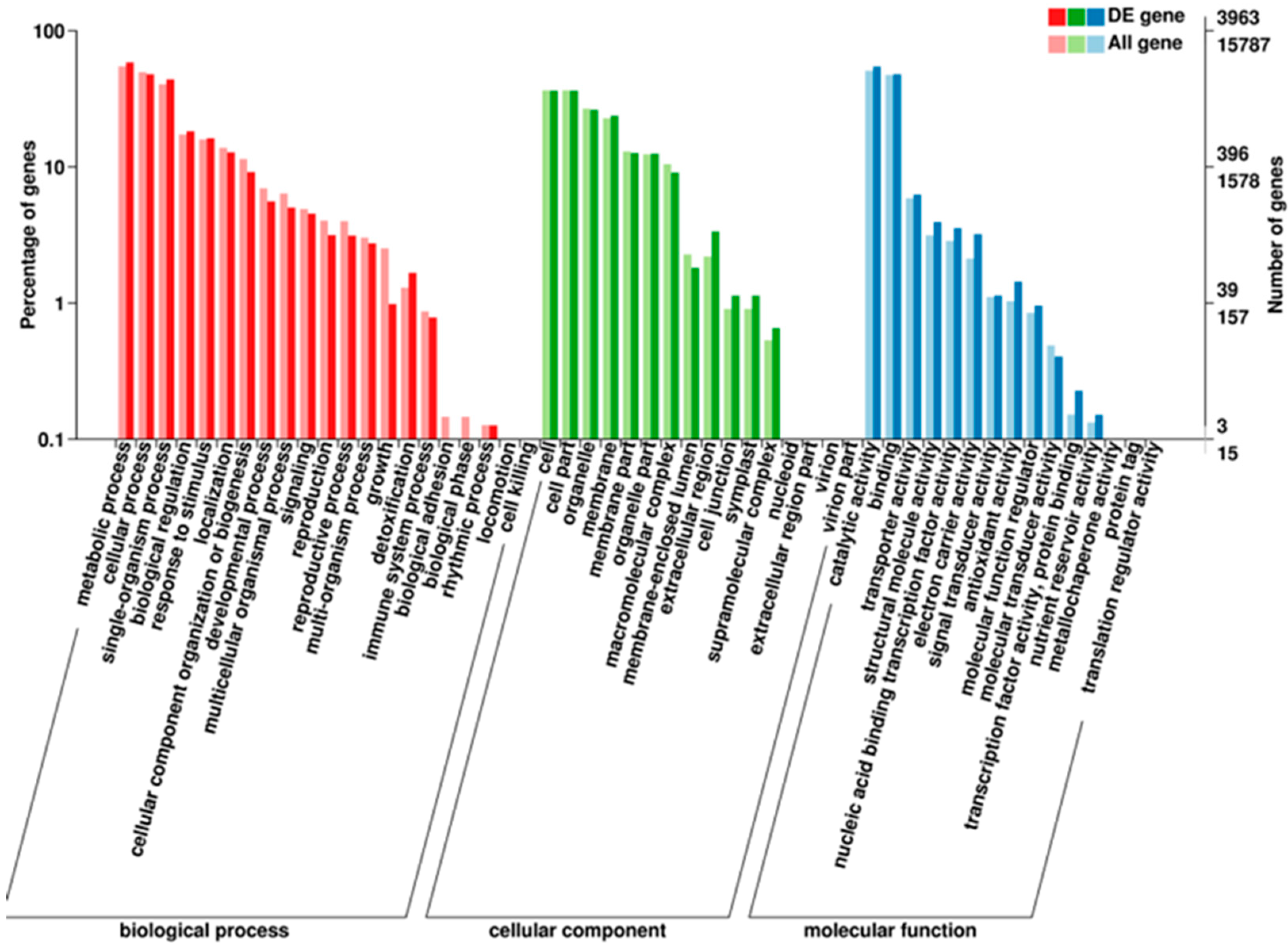
3.4.1. GO Enrichment Analysis of the DEGs
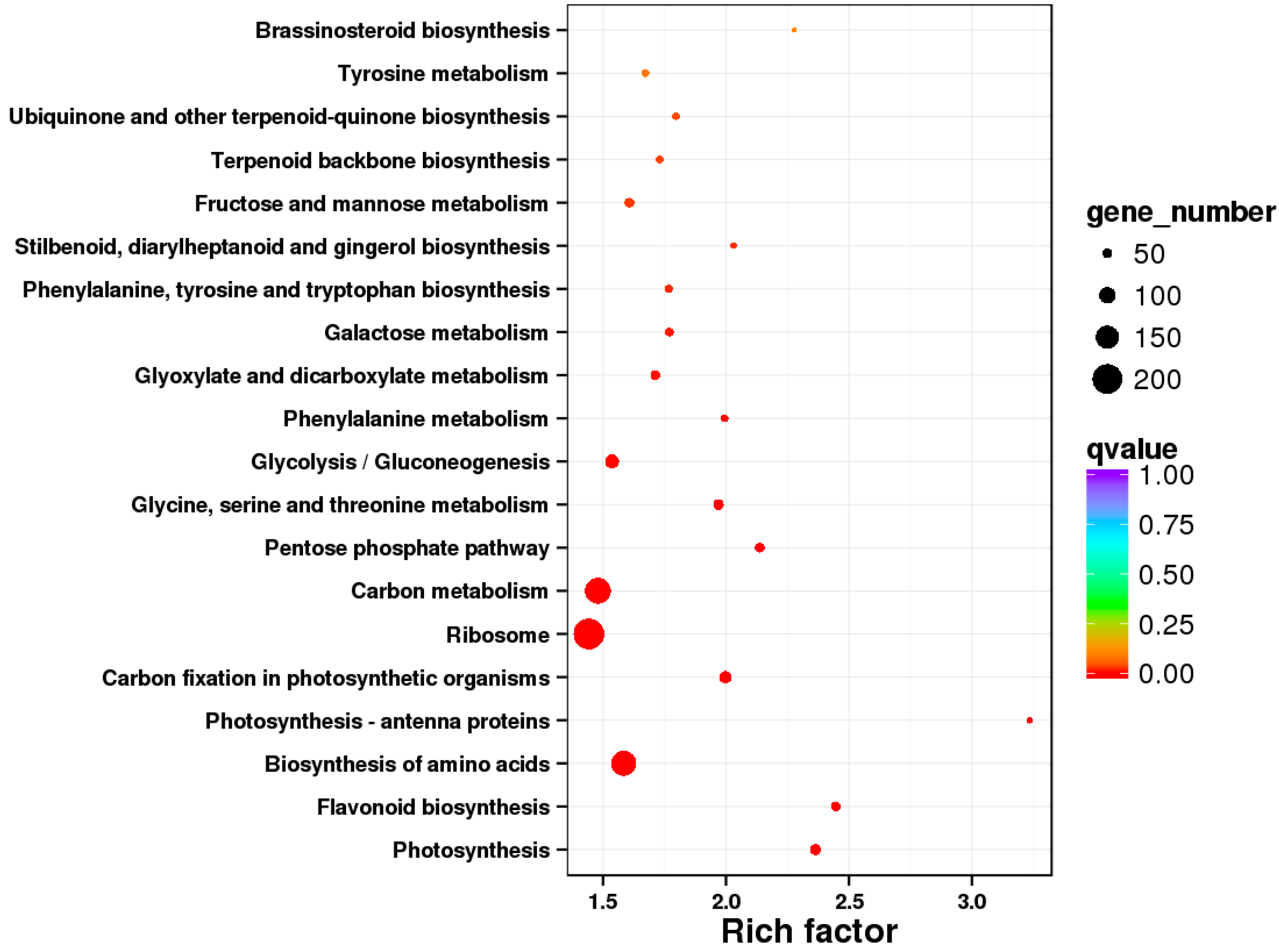
3.4.2. KEGG Enrichment Analysis of the DEGs
3.4.3. Analysis of Transcription Factors in Identified DEGs
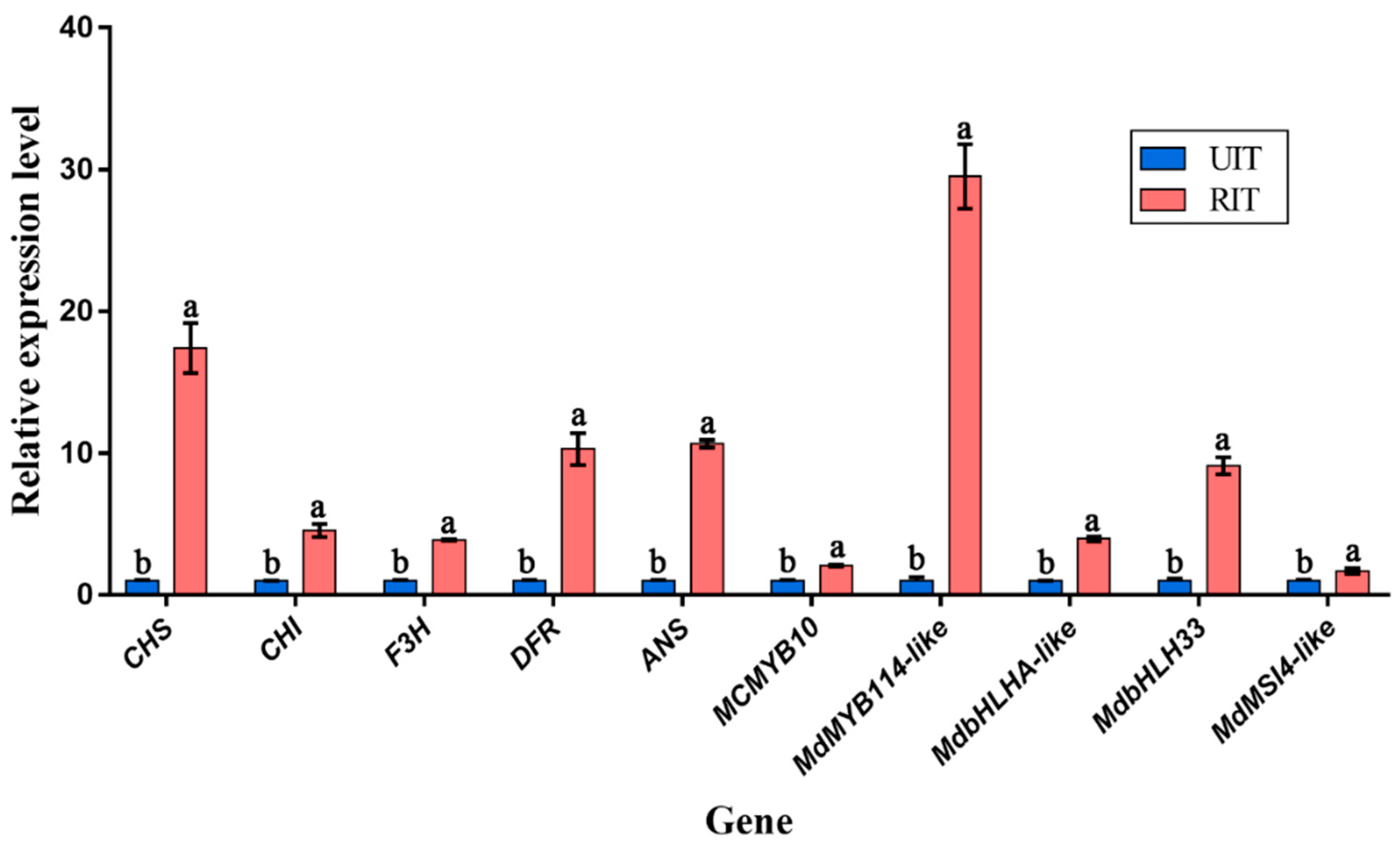
3.5. Quantitative Real-Time PCR (qRT-PCR)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef]
- Lee, D.; Ahn, S.; Cho, H.Y.; Yun, H.Y.; Park, J.H.; Lim, J.; Lee, J.; Kwon, S.W. Metabolic response induced by parasitic plant-fungus interactions hinder amino sugar and nucleotide sugar metabolism in the host. Sci. Rep. 2016, 6, 37434. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Cao, B.; Tian, C.; Liang, Y. Development and Characterization of Novel Genic-SSR Markers in Apple-Juniper Rust Pathogen Gymnosporangium yamadae (Pucciniales: Pucciniaceae) Using Next-Generation Sequencing. Int. J. Mol. Sci. 2018, 19, 1178. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, Q.; Bu, Y.; Luo, R.; Hao, S.; Zhang, J.; Tian, J.; Yao, Y. Flavonoid Accumulation Plays an Important Role in the Rust Resistance of Malus Plant Leaves. Front. Plant Sci. 2017, 8, 1286. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Duan, J.; Huo, D.; Shi, Q.; Niu, L.; Zhang, Y. Transcriptomic Analysis Reveals Transcription Factors Related to Leaf Anthocyanin Biosynthesis in Paeonia qiui. Molecules 2017, 22, 2186. [Google Scholar] [CrossRef]
- Yuan, Y.; Chiu, L.W.; Li, L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 2009, 230, 1141–1153. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; Van Houwelingen, A.M.; de Vos, R.C.; Jonker, H.H.; Xu, W.; Routaboul, J.-M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Song, T.; Li, K.; Wu, T.; Wang, Y.; Zhang, X.; Xu, X.; Yao, Y.; Han, Z. Identification of new regulators through transcriptome analysis that regulate anthocyanin biosynthesis in apple leaves at low temperatures. PLoS ONE 2019, 14, e0210672. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Laima, C.; Remigijus, D.; Pranas, V.K.; Antanas, S. Determination of the total phenolic and anthocyanin contents and antimicrobial activity of Viburnum opulus fruit juice. Plant Foods Hum. Nutr. 2012, 67, 256–261. [Google Scholar]
- Zhao, X.; Zhang, C.; Guigas, C.; Ma, Y.; Corrales, M.; Tauscher, B.; Hu, X. Composition, antimicrobial activity, and antiproliferative capacity of anthocyanin extracts of purple corn (Zea mays L.) from China. Eur. Food Res. Technol. 2009, 228, 759–765. [Google Scholar] [CrossRef]
- Szankowski, I.; Flachowsky, H.; Li, H.; Halbwirth, H.; Treutter, D.; Regos, I.; Hanke, M.V.; Stich, K.; Fischer, T.C. Shift in polyphenol profile and sublethal phenotype caused by silencing of anthocyanidin synthase in apple (Malus sp.). Planta 2009, 229, 681–692. [Google Scholar] [CrossRef]
- Flachowsky, H.; Szankowski, I.; Fischer, T.C.; Richter, K.; Peil, A.; Höfer, M.; Dörschel, C.; Schmoock, S.; Gau, A.E.; Halbwirth, H.; et al. Transgenic apple plants overexpressing the Lc gene of maize show an altered growth habit and increased resistance to apple scab and fire blight. Planta 2010, 231, 623–635. [Google Scholar] [CrossRef]
- Li, H.; Flachowsky, H.; Fischer, T.C.; Hanke, M.; Forkmann, G.; Treutter, D.; Schwab, W.; Hoffmann, T.; Szankowski, I. Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 2007, 226, 1243–1254. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef]
- Wei, H.; Chen, X.; Zong, X.; Shu, H.; Gao, D.; Liu, Q. Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in the Red and Yellow Fruits of Sweet Cherry (Prunus avium L.). PLoS ONE 2015, 10, e121164. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. Enrichment Analysis for Gene Ontology; R Package Version 2.8; Bioconductor: Buffalo, NY, USA, 2010. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liu, Y.; Kui, L.; Deng, C.; Warran, B.; Wang, L.; Yu, B.; Yang, H.; Wang, J.; Espley, R.V.; Zhang, J.; et al. Comparative Transcriptome Analysis of White and Purple Potato to Identify Genes Involved in Anthocyanin Biosynthesis. PLoS ONE 2015, 10, e0129148. [Google Scholar] [CrossRef]
- Rong, J.; Ji, T.; Song, T.; Jie, Z.; Yao, Y. The Malus crabapple transcription factor McMYB10 regulates anthocyanin biosynthesis during petal coloration. Sci. Hortic. 2014, 166, 42–49. [Google Scholar]
- Liu, X.F.; Yin, X.R.; Allan, A.C.; Kui, L.W.; Shi, Y.N.; Huang, Y.J.; Ferguson, I.B.; Xu, C.J.; Chen, K.S. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tissue Organ Cult. 2013, 115, 285–298. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Chen, X.; Tang, Y.; Bai, Y.; Chen, Y. Agrobacterium-mediated Transformation of Anthocyanin Regulatory Gene LC-NtAn2 into Tobacco. Acta Agric. Boreali Occident. Sin. 2013, 22, 50–55. [Google Scholar]
- Sun, S.; Gugger, P.F.; Wang, Q.; Chen, J. Identification of a R2R3-MYB gene regulating anthocyanin biosynthesis and relationships between its variation and flower color difference in lotus (Nelumbo Adans.). PeerJ 2016, 4, e2369. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Kui, L.W.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar]
- Peng, T.; Moriguchi, T. The molecular network regulating the coloration in apple. Sci. Hortic. 2013, 163, 1–9. [Google Scholar] [CrossRef]
- Bai, Y.; Pattanaik, S.; Patra, B.; Werkman, J.R.; Xie, C.H.; Yuan, L. Flavonoid-related basic helix-loop-helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 2011, 234, 363–375. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Tian, J.; Li, K.; Zhang, S.; Zhang, J.; Song, T.; Zhu, Y.; Yao, Y. The Structure and Methylation Level of the McMYB10 Promoter Determine the Leaf Color of Malus Crabapple. Hortscience 2017, 52, 520–526. [Google Scholar] [CrossRef]
- Kawamura, Y.; Takenaka, S.; Hase, S.; Kubota, M.; Ichinose, Y.; Takahashi, H. Enhanced defense responses in Arabidopsis induced by the cell wall protein fractions from Pythium oligandrum require SGT1, RAR1, NPR1 and JAR1. Plant Cell Physiol. 2009, 50, 924–934. [Google Scholar] [CrossRef]
- Vorwerk, S.; Somerville, S.; Somerville, C. The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 2004, 9, 203–209. [Google Scholar] [CrossRef]
- Dubovskiy, I.M.; Whitten, M.M.; Kryukov, V.Y.; Yaroslavtseva, O.N.; Grizanova, E.V.; Butt, T.M. More than a colour change: Insect melanism, disease resistance and fecundity. Proc. R. Soc. B Biol. Sci. 2013, 280. [Google Scholar] [CrossRef]
- Fogelman, E.; Tanami, S.; Ginzberg, I. Anthocyanin synthesis in native and wound periderms of potato. Physiol. Plant. 2015, 153, 616–626. [Google Scholar] [CrossRef]
- Kawahigashi, H.; Kasuga, S.; Sawada, Y.; Yonemaru, J.; Ando, T.; Matsumoto, T. The sorghum gene for leaf color changes upon wounding (P) encodes a flavanone 4-reductase in the 3-deoxyanthocyanidin biosynthesis pathway. G3 Genes Genomes Genet. 2016, 6, 1439–1447. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Schaefer, H.M.; Rentzsch, M.; Breuer, M. Anthocyanins reduce fungal growth in fruits. Nat. Prod. Commun. 2008, 3, 1267–1272. [Google Scholar] [CrossRef]
- Wen, H.; Kang, J.; Li, D.; Wen, W.; Yang, F.; Hu, H.; Liu, C. Antifungal activities of anthocyanins from purple sweet potato in the presence of food preservatives. Food Sci. Biotechnol. 2016, 25, 165–171. [Google Scholar] [CrossRef]
- Tellez, P.; Rojas, E.; Bael, S.V. Red Coloration in Young Tropical Leaves Associated with Reduced Fungal Pathogen Damage. Biotropica 2016, 48, 150–153. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Lan, L.X.; Sheng, Z.M.; Xiang, L. The research progress on plant anthocyanin synthetase ANS gene. Plant Physiol. J. 2016, 52, 817–827. [Google Scholar]
- Li, K.; Zhang, J.; Kang, Y.; Chen, M.; Song, T.; Geng, H.; Tian, J.; Yao, Y. McMYB10 Modulates the Expression of a Ubiquitin Ligase, McCOP1 During Leaf Coloration in Crabapple. Front. Plant Sci. 2018, 9, 704–718. [Google Scholar] [CrossRef]
- Wan, H.; Zhang, J.; Song, T.; Tian, J.; Yao, Y. Promotion of flavonoid biosynthesis in leaves and calli of ornamental crabapple (Malus sp.) by high carbon to nitrogen ratios. Front. Plant Sci. 2015, 6, 673. [Google Scholar] [CrossRef]


| Compound | ||
|---|---|---|
| Catechin | 72.63 1.00 b | 188.45 0.45 a |
| Chlorogenic acid | 513.59 12.56 b | 1947.46 142.48 a |
| Coumaric acid | 107.24 2.94 b | 171.26 4.82 a |
| Cyanidin-3-galactoside chloride | 44.71 ± 0.48 b | 706.59 ± 7.52 a |
| Dihydromyricetin | 281.39 7.62 b | 384.08 16.89 a |
| Dihydroquercetin | 1485.13 72.57 b | 3525.75 61.70 a |
| Epicatechin | 54.89 2.45 b | 268.37 0.41 a |
| Eriodictyol | 271.32 7.08 a | 278.33 8.13 a |
| Naringenin | 1222.09 31.85 b | 1915.27 100.36 a |
| Phloretin | 10,404.03 116.34 b | 28,553.27 576.97 a |
| Proanthocyanin B1 | 92.67 2.47 b | 123.94 ± 3.19 a |
| Proanthocyanin B2 | 4.56 0.15 b | 38.50 0.53 a |
| Sample Name | Clean Reads | Clean Bases | Q30 | GC Content |
|---|---|---|---|---|
| RIT1 | 28,534,074 | 8,533,976,408 | 94.10% | 50.04% |
| RIT2 | 22,858,851 | 6,835,393,636 | 93.71% | 50.01% |
| RIT3 | 27,789,895 | 8,305,725,370 | 93.40% | 49.84% |
| UIT1 | 32,429,472 | 9,701,511,274 | 93.58% | 46.84% |
| UIT2 | 27,360,624 | 8,189,892,014 | 93.46% | 47.18% |
| UIT3 | 25,312,509 | 7,573,777,970 | 93.97% | 46.79% |
| DEG Set | Total 1 | COG | GO | KEGG | KOG | NR | Pfam | Swiss-Prot |
|---|---|---|---|---|---|---|---|---|
| RIT vs. UIT | 9749 | 4242 | 3950 | 3609 | 4978 | 9628 | 8204 | 7818 |
| Genes 2 | ||||||||
| Annotated (%) | 100.0 | 43.5 | 40.5 | 37.0 | 51.1 | 98.8 | 84.2 | 80.2 |
| GO_ID | Term | Annotated | Significant 1 | D Value 2 |
|---|---|---|---|---|
| GO: 0015979 | photosynthesis | 242 | 152 | 3.60 × 10−20 |
| GO: 0055114 | oxidation-reduction process | 1726 | 603 | 6.90 × 10−13 |
| GO: 0009765 | photosynthesis, light harvesting | 35 | 26 | 7.50 × 10−11 |
| GO: 0006098 | pentose phosphate shunt | 103 | 52 | 2.60 × 10−10 |
| GO: 0010027 | thylakoid membrane organization | 97 | 51 | 6.40 × 10−10 |
| GO: 0019252 | starch biosynthetic process | 77 | 41 | 1.90 × 10−8 |
| GO: 0000023 | maltose metabolic process | 62 | 35 | 1.90 × 10−8 |
| GO: 0010207 | photosystem II assembly | 65 | 34 | 2.10 × 10−7 |
| GO: 0035304 | regulation of protein dephosphorylation | 35 | 23 | 7.00 × 10−7 |
| GeneID | log2 Ratio (RIT/UIT) | FDR | Annotation |
|---|---|---|---|
| MD13G1285100 | 3.10 | 0 | chalcone synthase [Malus domestica Borkh.] |
| MD04G1003300 | 4.16 | 0 | chalcone synthase [Sorbus aucuparia Linn.] |
| MD01G1118100 | 2.10 | 2.91 × 10−42 | chalcone isomerase [Pyrus pyrifolia (Burm. f.) Nakai.] |
| MD15G1353800 | 2.52 | 0 | flavanone 3-hydroxylase [Prunus avium Linn.] |
| MD15G1024100 | 3.00 | 0 | dihydroflavonol 4-reductase [Malus domestica Borkh.] |
| MD03G1001100 | 3.12 | 0 | leucoanthocyanidin dioxygenase [Malus domestica Borkh.] |
| MD06G1071600 | 2.69 | 0 | leucoanthocyanidin dioxygenase [Malus domestica Borkh.] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Wang, Y.; Meng, J.; Zhang, X.; Zhou, J.; Han, M.; Yang, C.; Gan, L.; Li, H. Transcriptome Sequencing and Expression Analysis of Genes Related to Anthocyanin Biosynthesis in Leaves of Malus ‘Profusion’ Infected by Japanese Apple Rust. Forests 2019, 10, 665. https://doi.org/10.3390/f10080665
Liu P, Wang Y, Meng J, Zhang X, Zhou J, Han M, Yang C, Gan L, Li H. Transcriptome Sequencing and Expression Analysis of Genes Related to Anthocyanin Biosynthesis in Leaves of Malus ‘Profusion’ Infected by Japanese Apple Rust. Forests. 2019; 10(8):665. https://doi.org/10.3390/f10080665
Chicago/Turabian StyleLiu, Pengyuan, Yilin Wang, Jiaxin Meng, Xian Zhang, Jing Zhou, Meiling Han, Chen Yang, Lingxin Gan, and Houhua Li. 2019. "Transcriptome Sequencing and Expression Analysis of Genes Related to Anthocyanin Biosynthesis in Leaves of Malus ‘Profusion’ Infected by Japanese Apple Rust" Forests 10, no. 8: 665. https://doi.org/10.3390/f10080665
APA StyleLiu, P., Wang, Y., Meng, J., Zhang, X., Zhou, J., Han, M., Yang, C., Gan, L., & Li, H. (2019). Transcriptome Sequencing and Expression Analysis of Genes Related to Anthocyanin Biosynthesis in Leaves of Malus ‘Profusion’ Infected by Japanese Apple Rust. Forests, 10(8), 665. https://doi.org/10.3390/f10080665




