The Specialist’s Paradox: Generalist AI May Better Organize Medical Knowledge
Abstract
1. Introduction
2. Materials and Methods
2.1. Data and Specialties
2.2. Embedding Models
2.3. Embedding and Clustering Procedure
2.4. Evaluation Metrics
2.5. Qualitative Analysis
2.6. Robustness-of-Clustering Analysis
3. Results
3.1. Clustering Performance Overview
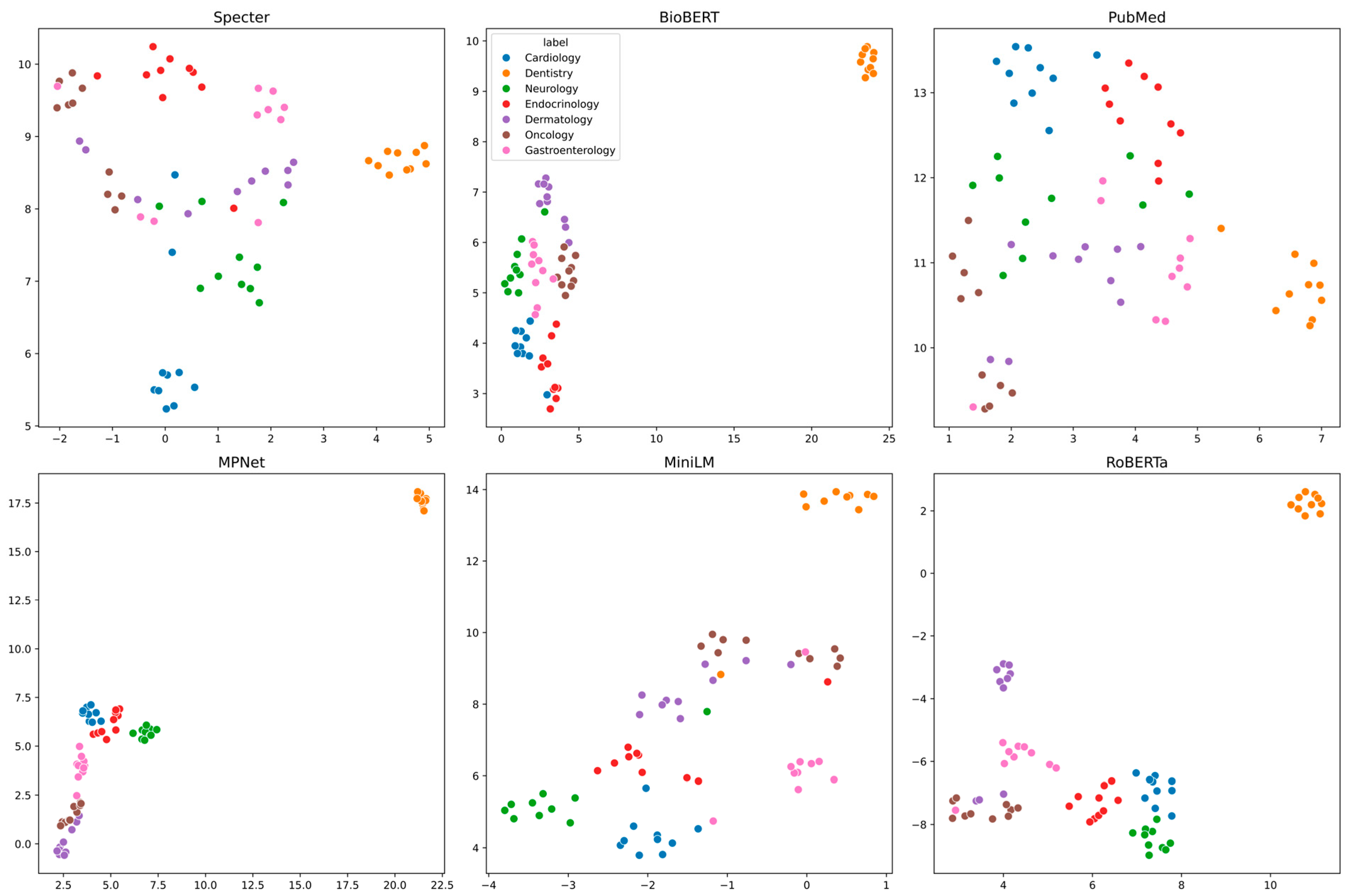
3.2. Visualization of Embedding Spaces
3.3. Quantitative Clustering Performance
3.4. Architectural Capacity and Semantic Coherence
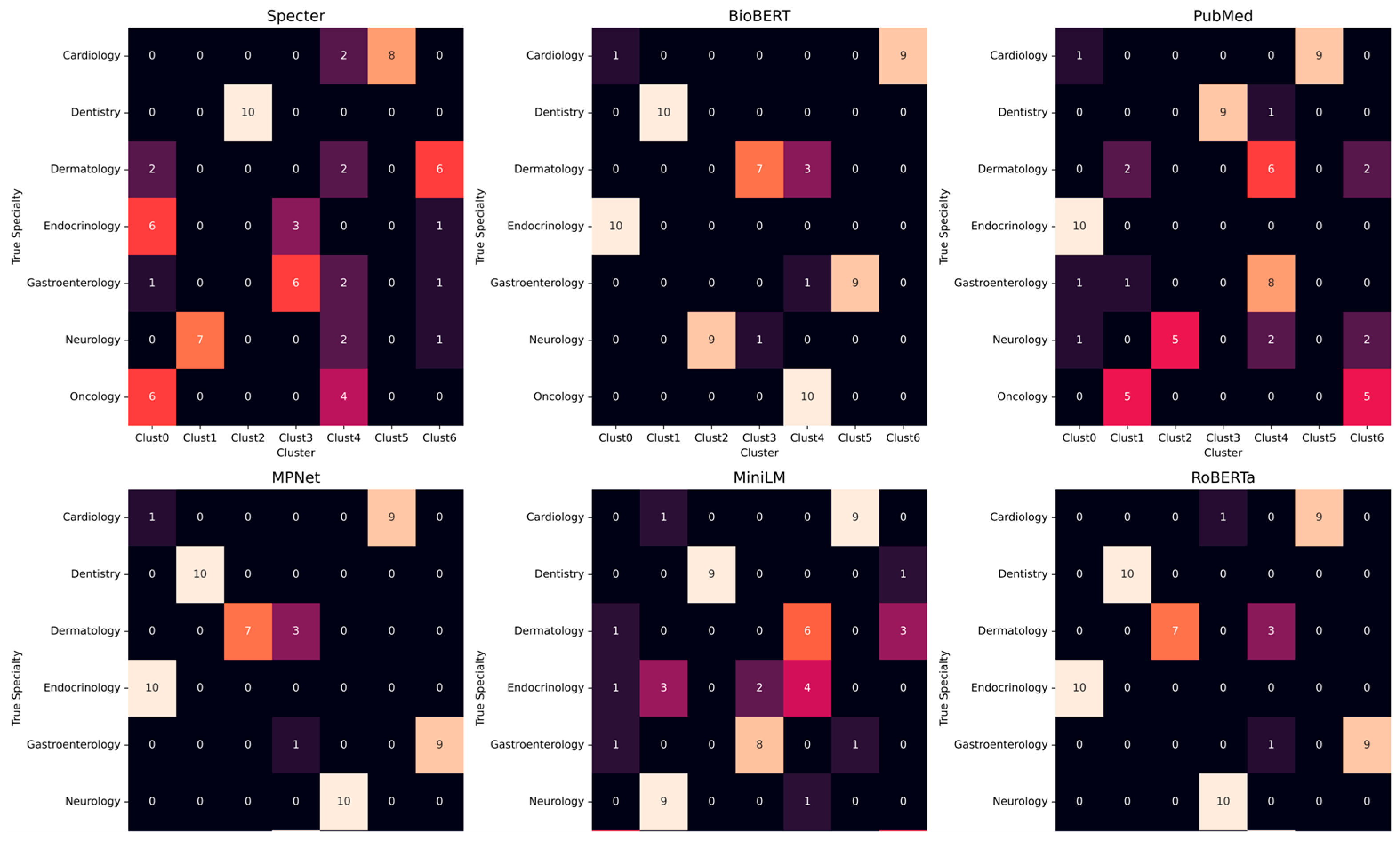
3.5. Cluster Composition and Misclassifications
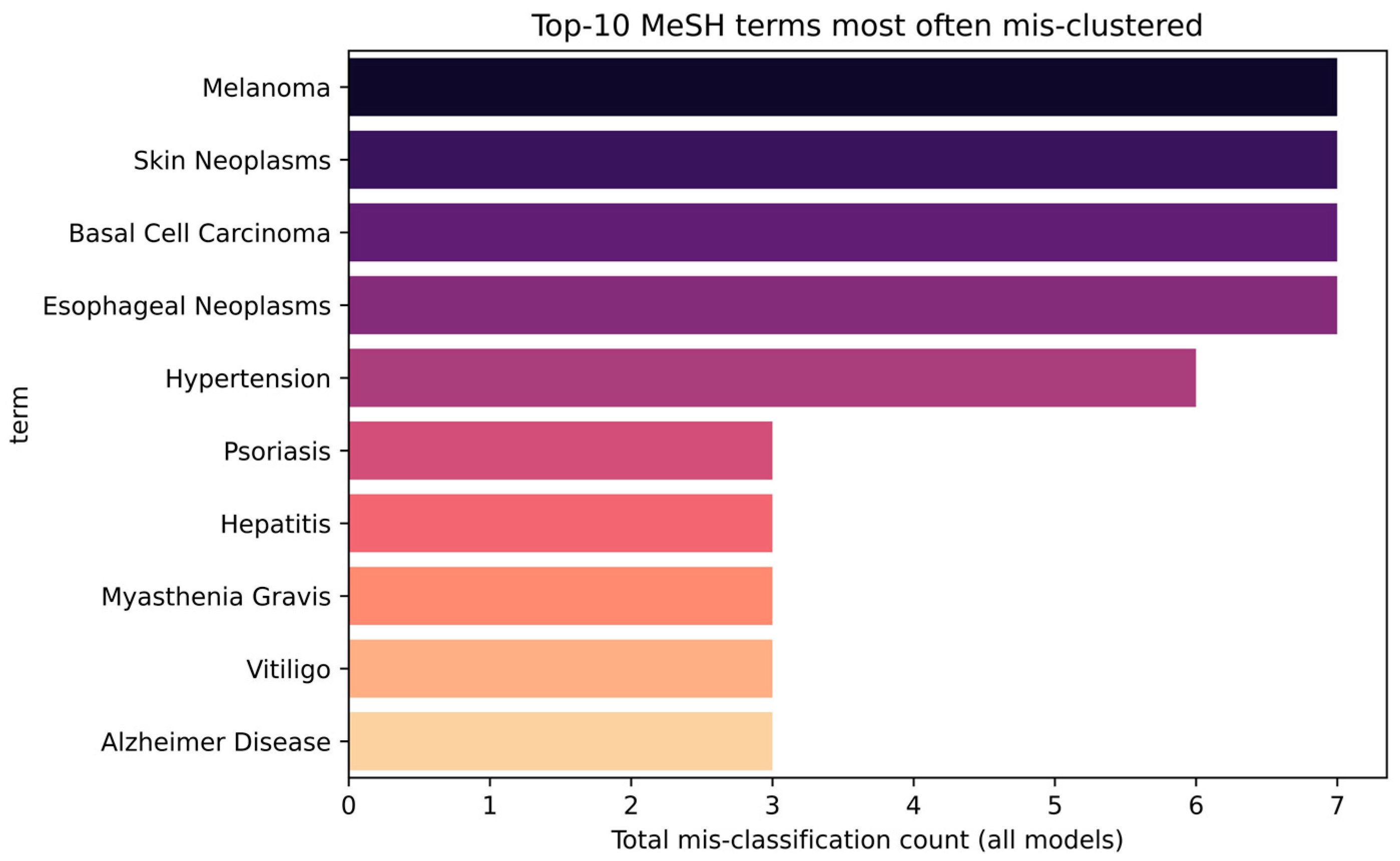
3.6. Analysis of Systematically Misclassified Terms
3.7. Model-Specific Error Fingerprints
3.8. Robustness of Clustering
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Term | True Specialty | Assigned Specialty | Model |
|---|---|---|---|
| Hypertension | Cardiology | Endocrinology | BioBERT |
| Basal Cell Carcinoma | Dermatology | Oncology | BioBERT |
| Melanoma | Dermatology | Oncology | BioBERT |
| Skin Neoplasms | Dermatology | Oncology | BioBERT |
| Esophageal Neoplasms | Gastroenterology | Oncology | BioBERT |
| Alzheimer Disease | Neurology | Dermatology | BioBERT |
| Hypertension | Cardiology | Endocrinology | PubMed |
| Periodontal Diseases | Dentistry | Gastroenterology | PubMed |
| Acne Vulgaris | Dermatology | Gastroenterology | PubMed |
| Basal Cell Carcinoma | Dermatology | Oncology | PubMed |
| Contact Dermatitis | Dermatology | Gastroenterology | PubMed |
| Dermatitis, Atopic | Dermatology | Gastroenterology | PubMed |
| Melanoma | Dermatology | Oncology | PubMed |
| Psoriasis | Dermatology | Gastroenterology | PubMed |
| Rosacea | Dermatology | Gastroenterology | PubMed |
| Skin Neoplasms | Dermatology | Oncology | PubMed |
| Urticaria | Dermatology | Gastroenterology | PubMed |
| Vitiligo | Dermatology | Oncology | PubMed |
| Esophageal Neoplasms | Gastroenterology | Oncology | PubMed |
| Hepatitis | Gastroenterology | Endocrinology | PubMed |
| Alzheimer Disease | Neurology | Endocrinology | PubMed |
| Amyotrophic Lateral Sclerosis | Neurology | Oncology | PubMed |
| Multiple Sclerosis | Neurology | Oncology | PubMed |
| Parkinson Disease | Neurology | Gastroenterology | PubMed |
| Peripheral Neuropathies | Neurology | Gastroenterology | PubMed |
| Hypertension | Cardiology | Endocrinology | MPNet |
| Basal Cell Carcinoma | Dermatology | Oncology | MPNet |
| Melanoma | Dermatology | Oncology | MPNet |
| Skin Neoplasms | Dermatology | Oncology | MPNet |
| Esophageal Neoplasms | Gastroenterology | Oncology | MPNet |
| Hypertension | Cardiology | Neurology | MiniLM |
| Malocclusion | Dentistry | Oncology | MiniLM |
| Basal Cell Carcinoma | Dermatology | Oncology | MiniLM |
| Melanoma | Dermatology | Oncology | MiniLM |
| Skin Neoplasms | Dermatology | Oncology | MiniLM |
| Vitiligo | Dermatology | Oncology | MiniLM |
| Adrenal Insufficiency | Endocrinology | Neurology | MiniLM |
| Cushing Syndrome | Endocrinology | Neurology | MiniLM |
| Diabetes Mellitus, Type 1 | Endocrinology | Gastroenterology | MiniLM |
| Diabetes Mellitus, Type 2 | Endocrinology | Gastroenterology | MiniLM |
| Hyperparathyroidism | Endocrinology | Dermatology | MiniLM |
| Hyperthyroidism | Endocrinology | Dermatology | MiniLM |
| Hypothyroidism | Endocrinology | Dermatology | MiniLM |
| Metabolic Syndrome | Endocrinology | Neurology | MiniLM |
| Polycystic Ovary Syndrome | Endocrinology | Oncology | MiniLM |
| Thyroid Diseases | Endocrinology | Dermatology | MiniLM |
| Esophageal Neoplasms | Gastroenterology | Oncology | MiniLM |
| Hepatitis | Gastroenterology | Cardiology | MiniLM |
| Myasthenia Gravis | Neurology | Dermatology | MiniLM |
| Atherosclerosis | Cardiology | Neurology | RoBERTa |
| Melanoma | Dermatology | Oncology | RoBERTa |
| Basal Cell Carcinoma | Dermatology | Oncology | RoBERTa |
| Skin Neoplasms | Dermatology | Oncology | RoBERTa |
| Esophageal Neoplasms | Gastroenterology | Oncology | RoBERTa |
| Angina Pectoris | Cardiology | Endocrinology | Bioclin |
| Heart Failure | Cardiology | Endocrinology | Bioclin |
| Hypertension | Cardiology | Endocrinology | Bioclin |
| Pericarditis | Cardiology | Dentistry | Bioclin |
| Malocclusion | Dentistry | Endocrinology | Bioclin |
| Acne Vulgaris | Dermatology | Endocrinology | Bioclin |
| Basal Cell Carcinoma | Dermatology | Oncology | Bioclin |
| Contact Dermatitis | Dermatology | Dentistry | Bioclin |
| Dermatitis, Atopic | Dermatology | Dentistry | Bioclin |
| Melanoma | Dermatology | Oncology | Bioclin |
| Psoriasis | Dermatology | Gastroenterology | Bioclin |
| Rosacea | Dermatology | Dentistry | Bioclin |
| Skin Neoplasms | Dermatology | Oncology | Bioclin |
| Urticaria | Dermatology | Dentistry | Bioclin |
| Vitiligo | Dermatology | Endocrinology | Bioclin |
| Esophageal Neoplasms | Gastroenterology | Oncology | Bioclin |
| Peptic Ulcer | Gastroenterology | Endocrinology | Bioclin |
| Alzheimer Disease | Neurology | Endocrinology | Bioclin |
| Amyotrophic Lateral Sclerosis | Neurology | Gastroenterology | Bioclin |
| Epilepsy | Neurology | Oncology | Bioclin |
| Migraine Disorders | Neurology | Endocrinology | Bioclin |
| Multiple Sclerosis | Neurology | Gastroenterology | Bioclin |
| Myasthenia Gravis | Neurology | Endocrinology | Bioclin |
| Parkinson Disease | Neurology | Gastroenterology | Bioclin |
| Peripheral Neuropathies | Neurology | Endocrinology | Bioclin |
| Stroke | Neurology | Dentistry | Bioclin |
| Traumatic Brain Injuries | Neurology | Dentistry | Bioclin |
Appendix B
| Model | Algorithm | Silhouette | DBI | ARI | NMI |
|---|---|---|---|---|---|
| BioBERT | Agglo-Average | 0.688 | 1.128 | 0.334 | 0.631 |
| BioBERT | Agglo-Ward | 0.394 | 0.457 | 0.75 | 0.842 |
| BioBERT | HDBSCAN | −0.280 | 0.135 | 0.094 | 0.348 |
| BioBERT | K-Means | 0.413 | 0.448 | 0.749 | 0.833 |
| Bioclin | Agglo-Average | 0.706 | 1.66 | 0.196 | 0.424 |
| Bioclin | Agglo-Ward | 0.208 | 0.727 | 0.272 | 0.464 |
| Bioclin | HDBSCAN | 0.276 | 0.611 | 0.098 | 0.326 |
| Bioclin | K-Means | 0.092 | 0.709 | 0.329 | 0.526 |
| MPNet | Agglo-Average | 0.852 | 0.916 | 0.52 | 0.738 |
| MPNet | Agglo-Ward | 0.583 | 0.467 | 0.78 | 0.877 |
| MPNet | HDBSCAN | 0.463 | 0.433 | 0.726 | 0.868 |
| MPNet | K-Means | 0.482 | 0.46 | 0.835 | 0.902 |
| MiniLM | Agglo-Average | 0.740 | 1.007 | 0.489 | 0.682 |
| MiniLM | Agglo-Ward | 0.607 | 0.439 | 0.687 | 0.799 |
| MiniLM | HDBSCAN | 0.938 | 0.248 | 0.074 | 0.29 |
| MiniLM | K-Means | 0.622 | 0.466 | 0.714 | 0.808 |
| PubMed | Agglo-Average | 0.707 | 3.357 | 0.24 | 0.477 |
| PubMed | Agglo-Ward | −0.011 | 0.701 | 0.513 | 0.683 |
| PubMed | HDBSCAN | 0.024 | 0.527 | 0.074 | 0.29 |
| PubMed | K-Means | −0.054 | 0.631 | 0.557 | 0.699 |
| RoBERTa | Agglo-Average | 0.772 | 1.184 | 0.429 | 0.664 |
| RoBERTa | Agglo-Ward | 0.269 | 0.413 | 0.748 | 0.851 |
| RoBERTa | HDBSCAN | 0.656 | 0.359 | 0.343 | 0.659 |
| RoBERTa | K-Means | 0.269 | 0.413 | 0.748 | 0.851 |
| Specter | Agglo-Average | 0.830 | 1.191 | 0.38 | 0.576 |
| Specter | Agglo-Ward | 0.354 | 0.535 | 0.495 | 0.669 |
| Specter | HDBSCAN | 0.044 | 0.609 | 0.173 | 0.502 |
| Specter | K-Means | 0.357 | 0.547 | 0.52 | 0.674 |
References
- Landhuis, E. Scientific Literature: Information Overload. Nature 2016, 535, 457–458. [Google Scholar] [CrossRef] [PubMed]
- Gurevitch, J.; Koricheva, J.; Nakagawa, S.; Stewart, G. Meta-Analysis and the Science of Research Synthesis. Nature 2018, 555, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Garritty, C.; Stevens, A.; Hamel, C.; Golfam, M.; Hutton, B.; Wolfe, D. Knowledge Synthesis in Evidence-Based Medicine. Semin. Nucl. Med. 2019, 49, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Sivarajkumar, S.; Mohammad, H.A.; Oniani, D.; Roberts, K.; Hersh, W.; Liu, H.; He, D.; Visweswaran, S.; Wang, Y. Clinical Information Retrieval: A Literature Review. J. Healthc. Inform. Res. 2024, 8, 313–352. [Google Scholar] [CrossRef] [PubMed]
- Chigbu, U.E.; Atiku, S.O.; Du Plessis, C.C. The Science of Literature Reviews: Searching, Identifying, Selecting, and Synthesising. Publications 2023, 11, 2. [Google Scholar] [CrossRef]
- Tamine, L.; Goeuriot, L. Semantic Information Retrieval on Medical Texts. ACM Comput. Surv. 2022, 54, 1–38. [Google Scholar] [CrossRef]
- Yun-tao, Z.; Ling, G.; Yong-cheng, W. An Improved TF-IDF Approach for Text Classification. J. Zhejiang Univ.-Sci. A 2005, 6, 49–55. [Google Scholar] [CrossRef]
- Jurafsky, D.; Martin, J.H. Vector Semantics and Embeddings. In Speech and Language Processing; Prentice Hall: Hoboken, NJ, USA, 2019; pp. 1–31. [Google Scholar]
- Haider, M.M.; Hossin, M.A.; Mahi, H.R.; Arif, H. Automatic Text Summarization Using Gensim Word2vec and K-Means Clustering Algorithm. In Proceedings of the 2020 IEEE Region 10 Symposium (TENSYMP), Dhaka, Bangladesh, 5–7 June 2020; pp. 283–286. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. GloVe: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Noh, J.; Kavuluru, R. Improved Biomedical Word Embeddings in the Transformer Era. J. Biomed. Inform. 2021, 120, 103867. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M. Transformers: State-of-the-Art Natural Language Processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics, Minneapolis, MN, USA, 2–7 June 2019; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019; pp. 4171–4186. [Google Scholar]
- Liu, Q.; Kusner, M.J.; Blunsom, P. A Survey on Contextual Embeddings. arXiv 2020, arXiv:2003.07278. [Google Scholar] [CrossRef]
- Ajallouda, L.; Najmani, K.; Zellou, A.; Benlahmar, E.H. Doc2Vec, SBERT, InferSent, and USE Which Embedding Technique for Noun Phrases? In Proceedings of the 2022 2nd International Conference on Innovative Research in Applied Science, Engineering and Technology (IRASET), Meknes, Morocco, 3–4 March 2022; pp. 1–5. [Google Scholar]
- Reimers, N.; Gurevych, I. Sentence-Bert: Sentence Embeddings Using Siamese Bert-Networks. arXiv 2019, arXiv:1908.10084. [Google Scholar] [CrossRef]
- Gu, X.; Liu, L.; Yu, H.; Li, J.; Chen, C.; Han, J. On the Transformer Growth for Progressive Bert Training. arXiv 2020, arXiv:2010.12562. [Google Scholar]
- Galli, C.; Cusano, C.; Guizzardi, S.; Donos, N.; Calciolari, E. Embeddings for Efficient Literature Screening: A Primer for Life Science Investigators. Metrics 2025, 1, 1. [Google Scholar] [CrossRef]
- Džuganová, B. Medical Language—A Unique Linguistic Phenomenon. JAHR 2019, 10, 129–145. [Google Scholar] [CrossRef]
- Hunter, L.; Cohen, K.B. Biomedical Language Processing: What’s Beyond PubMed? Mol. Cell 2006, 21, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Ngiam, J.; Peng, D.; Vasudevan, V.; Kornblith, S.; Le, Q.V.; Pang, R. Domain Adaptive Transfer Learning with Specialist Models. arXiv 2018, arXiv:1811.07056. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A Pre-Trained Biomedical Language Representation Model for Biomedical Text Mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Sabha, S.U.; Assad, A.; Din, N.M.U.; Bhat, M.R. From Scratch or Pretrained? An in-Depth Analysis of Deep Learning Approaches with Limited Data. Int. J. Syst. Assur. Eng. Manag. 2024, 1–10. [Google Scholar] [CrossRef]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans. Comput. Healthc. 2022, 3, 1–23. [Google Scholar] [CrossRef]
- Xia, Y.; Kim, J.; Chen, Y.; Ye, H.; Kundu, S.; Hao, C.C.; Talati, N. Understanding the Performance and Estimating the Cost of LLM Fine-Tuning. In Proceedings of the 2024 IEEE International Symposium on Workload Characterization (IISWC), Vancouver, BC, Canada, 15–17 September 2024; pp. 210–223. [Google Scholar]
- Kumar, K.; Ashraf, T.; Thawakar, O.; Anwer, R.M.; Cholakkal, H.; Shah, M.; Yang, M.-H.; Torr, P.H.S.; Khan, F.S.; Khan, S. Llm Post-Training: A Deep Dive into Reasoning Large Language Models. arXiv 2025, arXiv:2502.21321. [Google Scholar] [CrossRef]
- Sanchez Carmona, V.; Jiang, S.; Dong, B. A Multilevel Analysis of PubMed-Only BERT-Based Biomedical Models. In Proceedings of the 6th Clinical Natural Language Processing Workshop, Mexico City, Mexico, 20–21 June 2024; Naumann, T., Ben Abacha, A., Bethard, S., Roberts, K., Bitterman, D., Eds.; Association for Computational Linguistics: Mexico City, Mexico, 2024; pp. 105–110. [Google Scholar]
- Yang, D.; Zhang, Z.; Zhao, H. Learning Better Masking for Better Language Model Pre-Training. arXiv 2022, arXiv:2208.10806. [Google Scholar]
- Colangelo, M.T.; Meleti, M.; Guizzardi, S.; Calciolari, E.; Galli, C. A Comparative Analysis of Sentence Transformer Models for Automated Journal Recommendation Using PubMed Metadata. Big Data Cogn. Comput. 2025, 9, 67. [Google Scholar] [CrossRef]
- Lu, Z.; Kim, W.; Wilbur, W.J. Evaluation of Query Expansion Using MeSH in PubMed. Inf. Retr. Boston 2009, 12, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Siino, M. All-Mpnet at Semeval-2024 Task 1: Application of Mpnet for Evaluating Semantic Textual Relatedness. In Proceedings of the 18th International Workshop on Semantic Evaluation (SemEval-2024), Mexico City, Mexico, 20–21 June 2024; pp. 379–384. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. Roberta: A Robustly Optimized Bert Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Galli, C.; Donos, N.; Calciolari, E. Performance of 4 Pre-Trained Sentence Transformer Models in the Semantic Query of a Systematic Review Dataset on Peri-Implantitis. Information 2024, 15, 68. [Google Scholar] [CrossRef]
- Nuthakki, S.; Neela, S.; Gichoya, J.W.; Purkayastha, S. Natural Language Processing of MIMIC-III Clinical Notes for Identifying Diagnosis and Procedures with Neural Networks. arXiv 2019, arXiv:1912.12397. [Google Scholar] [CrossRef]
- Ling, Y. Bio+ Clinical BERT, BERT Base, and CNN Performance Comparison for Predicting Drug-Review Satisfaction. arXiv 2023, arXiv:2308.03782. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- McInnes, L.; Healy, J.; Astels, S. Hdbscan: Hierarchical Density Based Clustering. J. Open Source Softw. 2017, 2, 205. [Google Scholar] [CrossRef]
- Belyadi, H.; Haghighat, A. Unsupervised Machine Learning: Clustering Algorithms. In Machine Learning Guide for Oil and Gas Using Python; Elsevier: Amsterdam, The Netherlands, 2021; pp. 125–168. [Google Scholar]
- Singh, A.K.; Mittal, S.; Malhotra, P.; Srivastava, Y. V Clustering Evaluation by Davies-Bouldin Index(DBI) in Cereal Data Using K-Means. In Proceedings of the 2020 Fourth International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 11–13 March 2020; pp. 306–310. [Google Scholar]
- Warrens, M.J.; van der Hoef, H. Understanding the Adjusted Rand Index and Other Partition Comparison Indices Based on Counting Object Pairs. J. Classif. 2022, 39, 487–509. [Google Scholar] [CrossRef]
- McDaid, A.F.; Greene, D.; Hurley, N. Normalized Mutual Information to Evaluate Overlapping Community Finding Algorithms. arXiv 2011, arXiv:1110.2515. [Google Scholar]
- Ling, C.; Zhao, X.; Lu, J.; Deng, C.; Zheng, C.; Wang, J.; Chowdhury, T.; Li, Y.; Cui, H.; Zhang, X. Domain Specialization as the Key to Make Large Language Models Disruptive: A Comprehensive Survey. arXiv 2023, arXiv:2305.18703. [Google Scholar]
- Zhao, X.; Lu, J.; Deng, C.; Zheng, C.; Wang, J.; Chowdhury, T.; Yun, L.; Cui, H.; Xuchao, Z.; Zhao, T. Beyond One-Model-Fits-All: A Survey of Domain Specialization for Large Language Models. arXiv 2023, arXiv:2305.18703. [Google Scholar]
- Sinha, K.; Jia, R.; Hupkes, D.; Pineau, J.; Williams, A.; Kiela, D. Masked Language Modeling and the Distributional Hypothesis: Order Word Matters Pre-Training for Little. arXiv 2021, arXiv:2104.06644. [Google Scholar] [CrossRef]
- Soglia, S.; Pérez-Anker, J.; Lobos Guede, N.; Giavedoni, P.; Puig, S.; Malvehy, J. Diagnostics Using Non-Invasive Technologies in Dermatological Oncology. Cancers 2022, 14, 5886. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Reddy, S.S.; Banerjee, T. Various Dimension Reduction Techniques for High Dimensional Data Analysis: A Review. Artif. Intell. Rev. 2021, 54, 3473–3515. [Google Scholar] [CrossRef]
- Piperno, R.; Bacco, L.; Dell’Orletta, F.; Merone, M.; Pecchia, L. Cross-Lingual Distillation for Domain Knowledge Transfer with Sentence Transformers. Knowl. Based Syst. 2025, 311, 113079. [Google Scholar] [CrossRef]
- Kanakarajan, K.R.; Kundumani, B.; Abraham, A.; Sankarasubbu, M. BioSimCSE: BioMedical Sentence Embeddings Using Contrastive Learning. In Proceedings of the 13th International Workshop on Health Text Mining and Information Analysis (LOUHI), Abu Dhabi, United Arab Emirates, 7 December 2022; Lavelli, A., Holderness, E., Jimeno Yepes, A., Minard, A.-L., Pustejovsky, J., Rinaldi, F., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2022; pp. 81–86. [Google Scholar]
- Lim, S.; Kim, J. SAPBERT: Speaker-Aware Pretrained BERT for Emotion Recognition in Conversation. Algorithms 2022, 16, 8. [Google Scholar] [CrossRef]
- Harrison, C.J.; Sidey-Gibbons, C.J. Machine Learning in Medicine: A Practical Introduction to Natural Language Processing. BMC Med. Res. Methodol. 2021, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Tayefi, M.; Ngo, P.; Chomutare, T.; Dalianis, H.; Salvi, E.; Budrionis, A.; Godtliebsen, F. Challenges and Opportunities beyond Structured Data in Analysis of Electronic Health Records. WIREs Comput. Stat. 2021, 13, e1549. [Google Scholar] [CrossRef]
- Gao, T.; Yao, X.; Chen, D. Simcse: Simple Contrastive Learning of Sentence Embeddings. arXiv 2021, arXiv:2104.08821. [Google Scholar]
- Wang, K.; Reimers, N.; Gurevych, I. Tsdae: Using Transformer-Based Sequential Denoising Auto-Encoder for Unsupervised Sentence Embedding Learning. arXiv 2021, arXiv:2104.06979. [Google Scholar]
- O’Malley, K.J.; Cook, K.F.; Price, M.D.; Wildes, K.R.; Hurdle, J.F.; Ashton, C.M. Measuring Diagnoses: ICD Code Accuracy. Health Serv. Res. 2005, 40, 1620–1639. [Google Scholar] [CrossRef] [PubMed]
| Cardiology | Dentistry | Neurology | Endocrinology | Dermatology | Oncology | Gastroenterology |
|---|---|---|---|---|---|---|
| Myocardial Infarction | Dental Caries | Epilepsy | Diabetes Mellitus, Type 1 | Psoriasis | Breast Neoplasms | Gastrointestinal Diseases |
| Heart Failure | Periodontal Diseases | Parkinson Disease | Diabetes Mellitus, Type 2 | Dermatitis, Atopic | Lung Neoplasms | Inflammatory Bowel Diseases |
| Arrhythmias, Cardiac | Tooth Extraction | Alzheimer Disease | Thyroid Diseases | Acne Vulgaris | Leukemia | Crohn Disease |
| Coronary Artery Disease | Orthodontics | Multiple Sclerosis | Hyperthyroidism | Melanoma | Lymphoma | Ulcerative Colitis |
| Hypertension | Dental Implants | Migraine Disorders | Hypothyroidism | Basal Cell Carcinoma | Colorectal Neoplasms | Peptic Ulcer |
| Cardiomyopathies | Endodontics | Stroke | Adrenal Insufficiency | Contact Dermatitis | Prostatic Neoplasms | Hepatitis |
| Atherosclerosis | Malocclusion | Peripheral Neuropathies | Cushing Syndrome | Vitiligo | Sarcoma | Pancreatitis |
| Angina Pectoris | Prosthodontics | Amyotrophic Lateral Sclerosis | Hyperparathyroidism | Rosacea | Glioblastoma | Irritable Bowel Syndrome |
| Atrial Fibrillation | Oral Hygiene | Myasthenia Gravis | Polycystic Ovary Syndrome | Skin Neoplasms | Ovarian Neoplasms | Gastroesophageal Reflux |
| Model Name (in Paper) | Hugging Face ID | Base Architecture | Primary Pre-Training Corpus | Key Fine-Tuning Objective | Embedding Dim. |
|---|---|---|---|---|---|
| MPNet | all-mpnet-base-v2 | MPNet | General Web Text (>1 B sentence-pairs) | Contrastive Sentence-Pairs | 768 |
| RoBERTa | roberta-large (in S-T framework) | RoBERTa | General Web Text (BookCorpus, etc.) | MLM (base), then Contrastive | 768 |
| MiniLM | all-MiniLM-L6-v2 | Distilled BERT | General Web Text (>1 B sentence-pairs) | Contrastive Sentence-Pairs | 384 |
| BioBERT | pritamdeka/BioBERT-mnli-snli… | BERT | PubMed/PMC | MLM (base), then Contrastive on NLI/STS tasks | 768 |
| PubMedBERT | pritamdeka/S-PubMedBert-MS-MARCO | BERT | PubMed/PMC (from scratch) | MLM (base), then Passage Retrieval (MS-MARCO) | 768 |
| Model | Silhouette | DBI | ARI | NMI |
|---|---|---|---|---|
| BioBERT | 0.293 | 0.489 | 0.806 | 0.879 |
| PubMed | 0.104 | 0.777 | 0.506 | 0.674 |
| MPNet | 0.351 | 0.495 | 0.835 | 0.902 |
| MiniLM | 0.071 | 0.561 | 0.512 | 0.667 |
| RoBERTa | 0.396 | 0.451 | 0.835 | 0.902 |
| Bioclin | 0.152 | 0.743 | 0.350 | 0.534 |
| Model | Misclassified | Error Rate | Purity |
|---|---|---|---|
| MPNet | 5 | 7.14% | 92.86% |
| RoBERTa | 5 | 7.14% | 92.86% |
| BioBERT | 6 | 8.57% | 91.43% |
| PubMed | 19 | 27.14% | 72.86% |
| MiniLM | 19 | 27.14% | 72.86% |
| Bioclin | 27 | 38.57% | 61.43% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galli, C.; Colangelo, M.T.; Meleti, M.; Calciolari, E. The Specialist’s Paradox: Generalist AI May Better Organize Medical Knowledge. Algorithms 2025, 18, 451. https://doi.org/10.3390/a18070451
Galli C, Colangelo MT, Meleti M, Calciolari E. The Specialist’s Paradox: Generalist AI May Better Organize Medical Knowledge. Algorithms. 2025; 18(7):451. https://doi.org/10.3390/a18070451
Chicago/Turabian StyleGalli, Carlo, Maria Teresa Colangelo, Marco Meleti, and Elena Calciolari. 2025. "The Specialist’s Paradox: Generalist AI May Better Organize Medical Knowledge" Algorithms 18, no. 7: 451. https://doi.org/10.3390/a18070451
APA StyleGalli, C., Colangelo, M. T., Meleti, M., & Calciolari, E. (2025). The Specialist’s Paradox: Generalist AI May Better Organize Medical Knowledge. Algorithms, 18(7), 451. https://doi.org/10.3390/a18070451










