A Data-Driven Intelligent Methodology for Developing Explainable Diagnostic Model for Febrile Diseases
Abstract
:1. Introduction
- The integration of ML models (Random Forest, XGBoost, MLP) with LIME and ChatGPT for explainability in diagnosing six febrile diseases;
- A comparative evaluation of ML models, demonstrating their effectiveness in multi-label classification for febrile diseases;
- The validation of model outputs with explainability techniques to ensure clinicians can understand and trust AI-driven diagnoses.
2. Methodology
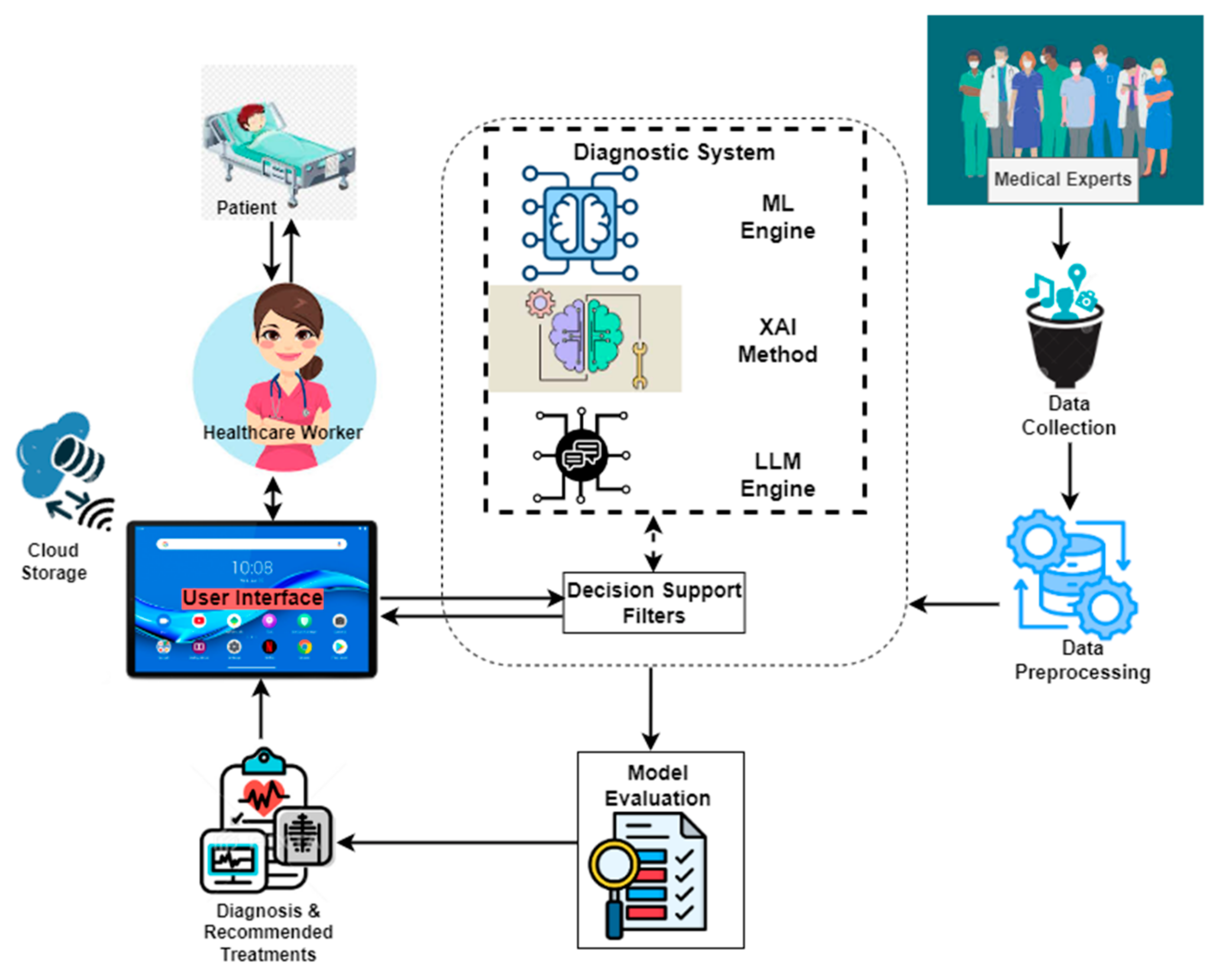
2.1. Enhanced Diagnostic Framework
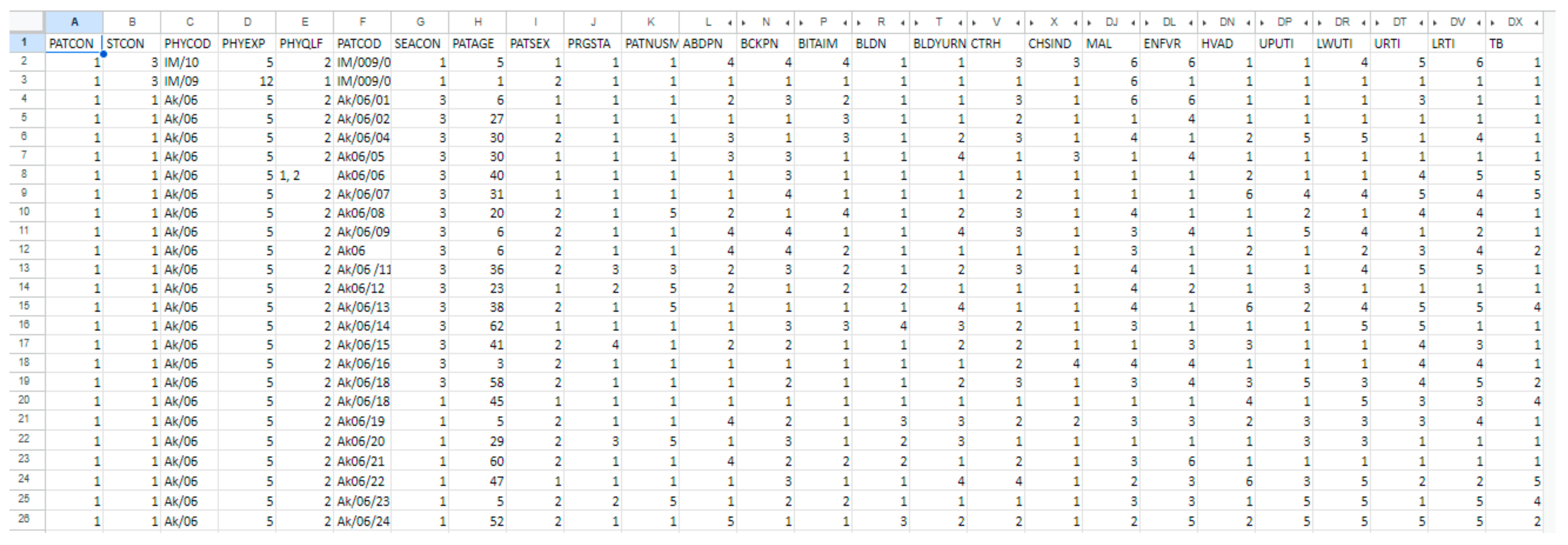
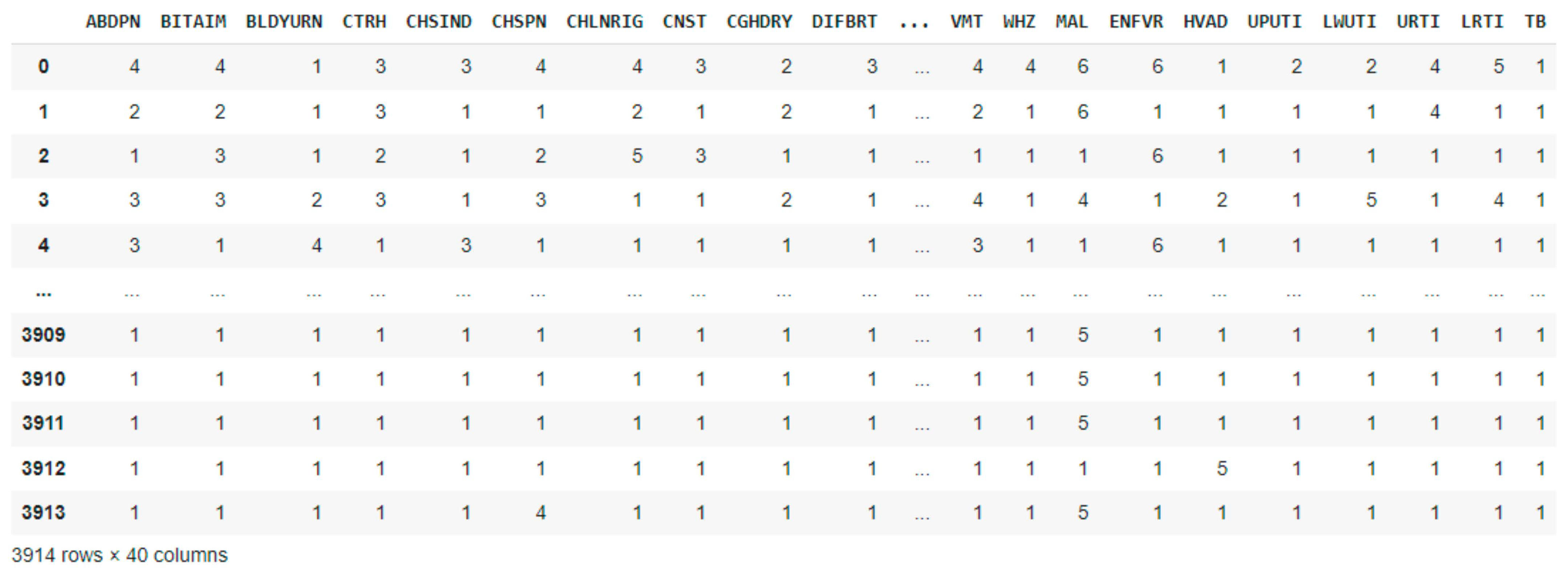
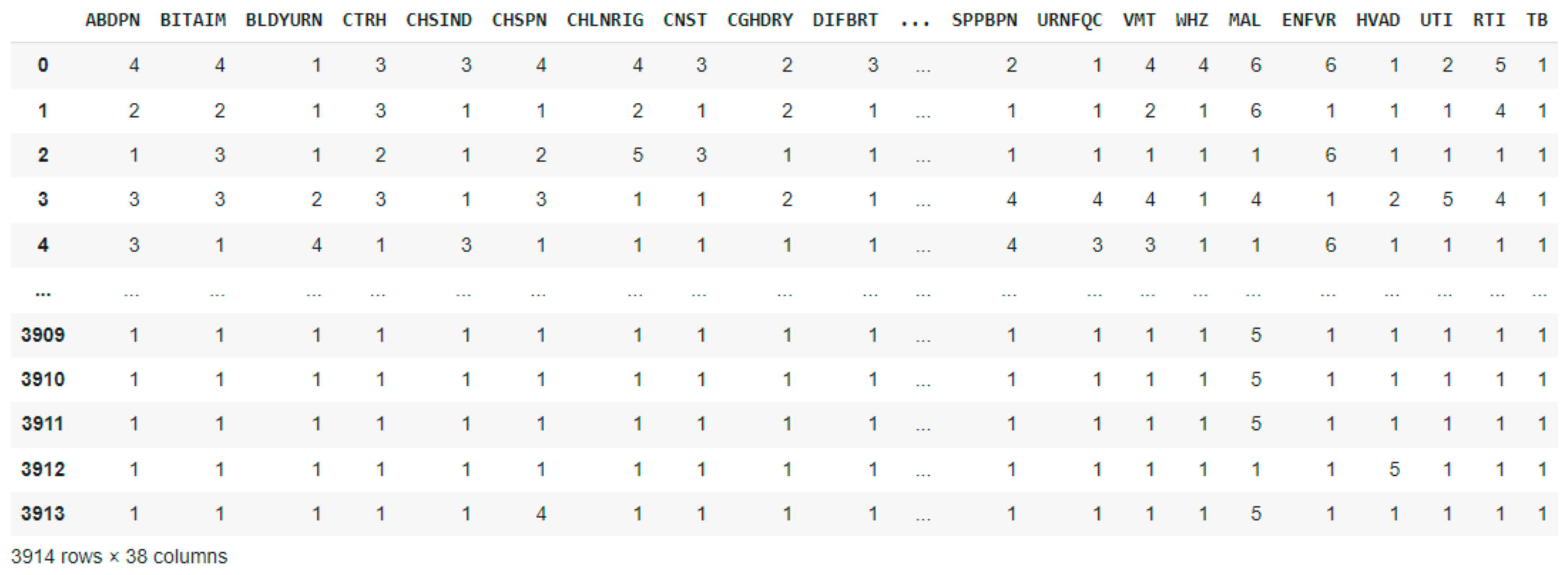
2.2. Dataset Description and Preprocessing
| Algorithm 1. Explainable AI-based febrile disease diagnosis with hyperparameter tuning pseudocode |
| BEGIN # Step 1: Dataset Preparation Load dataset D = {X, Y} Preprocess dataset Split dataset into training and testing: D_train, D_test ← split(D, 0.8) # Step 2: Severity Combination Using Max Operation FOR each patient record DO M(U, L) ← max(U, L) # Combine severity scores # Step 3: Hyperparameter Tuning and Model Training # Random Forest (RF) RF_params = {max_depth: [None, 10, 20], n_estimators: [100, 200, 300]} best_RF ← GridSearchCV(RandomForest, RF_params, D_train) # Multi-layer Perceptron (MLP) MLP_params = {activation: [“relu”, “tanh”, “logistic”], alpha: [0.0001, 0.001, 0.01], hidden_layer_sizes: [(100,), (50, 50), (50, 25, 10)]} best_MLP ← GridSearchCV(MLP, MLP_params, D_train) # XGBoost XGB_params = {n_estimators: [100, 200, 300], max_depth: [3, 5, 7]} best_XGB ← GridSearchCV(XGBoost, XGB_params, D_train) # Step 4: Model Selection Evaluate best_RF, best_MLP, best_XGB on D_test Select best_model ← model with highest F1 score # Step 5: Interpretability with LIME FOR each prediction in D_test DO explanation ← LIME(best_model, X_test) # Step 6: ChatGPT Integration for Enhanced Explanation FOR each patient DO P ← {best_model_prediction, explanation} response ← ChatGPT(P) # Step 7: Deployment FOR each patient DO Display {best_model_prediction, M(U, L), explanation, response} END |
2.3. Integration of Explainable AI
2.4. System Implementation
- User Interaction: Healthcare professionals and patients access the system through a mobile interface, designed using Figma and developed with Flet-0.24.0 for an intuitive user experience;
- Data Input and Processing: Users provide symptom details, which are processed by the backend, built using PythonAnywhere for online hosting.
- Machine Learning Diagnosis: The input symptoms are fed into a trained ML model (Random Forest), and this model generates disease predictions based on the given symptoms;
- Explainability Layer: To enhance interpretability, LIME-0.2.0.1 is applied to highlight the most important symptoms influencing the diagnosis. Additionally, ChatGPT processes the model’s outputs to generate human-readable explanations;
- Database Management: Patient records and diagnostic results are securely stored and retrieved using MySQL-9.0, which ensures compliance with medical data privacy regulations;
- Result Presentation: The final diagnosis, along with explanations from LIME and ChatGPT, is presented to the user in a clear and understandable format, aiding clinical decision-making.
2.5. Model Performance Metrics
3. Results and Discussion
4. Conclusions
- Pilot testing in clinical environments—A real-world evaluation is necessary to assess the model’s usability, effectiveness, and integration within existing healthcare workflows;
- Dataset expansion—Including more diverse populations, particularly pediatric patients under five years old, and incorporating additional febrile diseases such as hemorrhagic fevers will enhance generalizability;
- Handling imbalanced data—Diseases like tuberculosis and HIV/AIDS had fewer records in the dataset, affecting model performance. Future iterations should explore data augmentation or ensemble techniques to address this limitation;
- Continuous model updates—The system should be retrained periodically with new patient data to reflect emerging disease trends and improve diagnostic accuracy over time;
- Mobile and cloud deployment—Deploying the model via mobile health (mHealth) apps will facilitate accessibility, particularly in low-resource regions where medical infrastructure is limited.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Premaratna, R. Dealing with acute febrile illness in the resource-poor tropics. Trop. Med. Surg. 2013, 1, 101. [Google Scholar] [CrossRef]
- Butcher, L. Prognosis? Misdiagnosis! The High Price of Getting It Wrong. Manag. Care 2019, 28, 32–36. [Google Scholar] [PubMed]
- Attai, K.; Amannejad, Y.; Vahdat Pour, M.; Obot, O.; Uzoka, F.M. A systematic review of applications of machine learning and other soft computing techniques for the diagnosis of tropical diseases. Trop. Med. Infect. Dis. 2022, 7, 398. [Google Scholar] [CrossRef] [PubMed]
- Bagam, N. Applications of Machine Learning in Healthcare Data Analysis. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2020, 6, 373–386. [Google Scholar] [CrossRef]
- Naveed, M.A. Transforming healthcare through artificial intelligence and machine learning. Pak. J. Health Sci. 2023, 4, 1. [Google Scholar] [CrossRef]
- Kupusinac, A.; Doroslovački, R. An Overview of the Algorithmic Diagnostics Methodology: A Big Data Approach. In Proceedings of the 2018 Zooming Innovation in Consumer Technologies Conference (ZINC), Novi Sad, Serbia, 30–31 May 2018; pp. 104–105. [Google Scholar] [CrossRef]
- Wu, W.; Zhou, H. Data-driven diagnosis of cervical cancer with support vector machine-based approaches. IEEE Access 2017, 5, 25189–25195. [Google Scholar] [CrossRef]
- Gupta, D.; Kose, U.; Le Nguyen, B.; Bhattacharyya, S. Artificial Intelligence for Data-Driven Medical Diagnosis; De Gruyter: Berlin, Germany, 2021. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, T.; Zhang, K.H. Data-driven decision-making for precision diagnosis of digestive diseases. Biomed. Eng. Online 2023, 22, 87. [Google Scholar]
- Arif, M.S.; Mukheimer, A.; Asif, D. Enhancing the Early Detection of Chronic Kidney Disease: A Robust Machine Learning Model. Big Data Cogn. Comput. 2023, 7, 144. [Google Scholar] [CrossRef]
- Asif, D.; Bibi, M.; Arif, M.S.; Mukheimer, A. Enhancing Heart Disease Prediction through Ensemble Learning Techniques with Hyperparameter Optimization. Algorithms 2023, 16, 308. [Google Scholar] [CrossRef]
- Hu, J.; Perer, A.; Wang, F. Data-Driven Analytics for Personalized Healthcare. In Healthcare Information Management Systems: Cases, Strategies, and Solutions; Springer: Berlin, Germany, 2016; pp. 529–554. [Google Scholar] [CrossRef]
- Melnykova, N.; Shakhovska, N.; Gregus, M.; Melnykov, V.; Zakharchuk, M.; Vovk, O. Data-driven analytics for personalized medical decision-making. Mathematics 2020, 8, 1211. [Google Scholar] [CrossRef]
- Mendhe, D.; Dogra, A.; Nair, D.S.; Punitha, S.; Preetha, D.S.; Babu, G.T. AI-Enabled Data-Driven Approaches for Personalized Medicine and Healthcare Analytics. In Proceedings of the 2024 Ninth International Conference on Science Technology Engineering and Mathematics (ICONSTEM), Chennai, India, 4–5 April 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Ivanović, M.; Autexier, S.; Kokkonidis, M. AI Approaches in Processing and Using Data in Personalized Medicine. In Proceedings of the Symposium on Advances in Databases and Information Systems, Turin, Italy, 5–8 September 2022. [Google Scholar] [CrossRef]
- Ekanayake, I.U.; Meddage, D.P.; Rathnayake, U.S. A novel approach to explain the black-box nature of machine learning in compressive strength predictions of concrete using Shapley additive explanations (SHAP). Case Stud. Constr. Mater. 2022, 16, e01059. [Google Scholar] [CrossRef]
- Kulaklıoğlu, D. Explainable AI: Enhancing Interpretability of Machine Learning Models. Hum.-Comput. Interact. 2024, 8, 91. [Google Scholar] [CrossRef]
- Alblooshi, M.; Alhajeri, H.; Almatrooshi, M.; Alaraj, M. Unlocking Transparency in Credit Scoring: Leveraging XGBoost with XAI for Informed Business Decision-Making. In Proceedings of the 2024 International Conference on Artificial Intelligence, Computer, Data Sciences and Applications (ACDSA), Mahe, Seychelles, 1–2 February 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Quinn, T.P.; Jacobs, S.; Senadeera, M.; Le, V.; Coghlan, S. The Three Ghosts of Medical AI: Can the Black Box Present Deliver? Artif. Intell. Med. 2020, 124, 102158. [Google Scholar] [CrossRef] [PubMed]
- Inukonda, J.; Rajasekhara Reddy Tetala, V.; Hallur, J. Explainable Artificial Intelligence (XAI) in Healthcare: Enhancing Transparency and Trust. Int. J. Multidiscip. Res. 2024, 6, 30010. [Google Scholar] [CrossRef]
- Huang, S.; Mamidanna, S.; Jangam, S.; Zhou, Y.; Gilpin, L. Can Large Language Models Explain Themselves? A Study of LLM-Generated Self-Explanations. arXiv 2023, arXiv:2310.11207. [Google Scholar] [CrossRef]
- Hsu, C.; Wu, I.; Liu, S. Decoding AI Complexity: SHAP Textual Explanations via LLM for Improved Model Transparency. In Proceedings of the 2024 International Conference on Consumer Electronics—Taiwan (ICCE-Taiwan), Taching, China, 9–11 July 2024; pp. 197–198. [Google Scholar] [CrossRef]
- University of Uyo Teaching Hospital; Mount Royal University. NFRF Project Patient Dataset with Febrile Diseases [Data Set]; Zenodo: Meyrin, Switzerland, 2024. [Google Scholar] [CrossRef]
- Bellman, R.E.; Zadeh, L.A. Decision-making in a fuzzy environment. Manag. Sci. 1970, 17, B-141. [Google Scholar] [CrossRef]
- Murphy, A.; Moore, C. Random Forest (Machine Learning). 2019. Available online: https://radiopaedia.org/articles/67772 (accessed on 23 March 2025). [CrossRef]
- Yadav, D.C.; Pal, S. Analysis of Heart Disease Using Parallel and Sequential Ensemble Methods with Feature Selection Techniques. Int. J. Big Data Anal. Healthc. 2021, 6, 40–56. [Google Scholar] [CrossRef]
- Yu, H.; Samuels, D.C.; Zhao, Y.; Guo, Y. Architectures and Accuracy of Artificial Neural Network for Disease Classification from Omics Data. BMC Genom. 2019, 20, 167. [Google Scholar] [CrossRef]
- Ribeiro, M.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- Thakkar, P. Drug Classification Using Black-Box Models and Interpretability. Int. J. Res. Appl. Sci. Eng. Technol. 2021, 9, 1518–1529. [Google Scholar] [CrossRef]
- Sirriani, J.; Sezgin, E.; Claman, D.M.; Linwood, S. Medical Text Prediction and Suggestion Using Generative Pretrained Transformer Models with Dental Medical Notes. Methods Inf. Med. 2022, 61, 195–200. [Google Scholar] [CrossRef]
- Kumar, T.; Kait, R.; Ankita; Rani, S. Possibilities and Pitfalls of Generative Pre-Trained Transformers in Healthcare. In Proceedings of the 2023 International Conference on Advanced Computing & Communication Technologies (ICACCTech), Banur, India, 23–24 December 2023; pp. 37–44. [Google Scholar] [CrossRef]
- Erickson, B.J.; Kitamura, F. Magician’s corner: 9. Performance metrics for machine learning models. Radiol. Artif. Intell. 2021, 3, e200126. [Google Scholar] [CrossRef] [PubMed]
- Barracloug, P.A.; Were, C.M.; Mwangakala, H.; Fehringer, G.; Ohanya, D.O.; Agola, H.; Nandi, P. Artificial Intelligence System for Malaria Diagnosis. Int. J. Adv. Comput. Sci. Appl. 2024, 15, 100806. [Google Scholar] [CrossRef]
- La-Ariandi, H.; Setyanto, A.; Sudarmawan, S. Classification of Malaria Types Using Naïve Bayes Classification. J. Indones. Sos. Teknol. 2024, 5, 2311–2327. [Google Scholar] [CrossRef]
- Bhuiyan, M.A.; Rad, S.S.; Johora, F.T.; Islam, A.; Hossain, M.I.; Khan, A.A. Prediction of Typhoid Using Machine Learning and ANN Prior to Clinical Test. In Proceedings of the 2023 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 23–25 January 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–7. [Google Scholar]
- Awotunde, J.B.; Imoize, A.L.; Salako, D.P.; Farhaoui, Y. An Enhanced Medical Diagnosis System for Malaria and Typhoid Fever Using Genetic Neuro-Fuzzy System. In Proceedings of the International Conference on Artificial Intelligence and Smart Environment, Errachidia, Morocco, 24–26 November 2022; Springer International Publishing: Cham, Switzerland, 2022; pp. 173–183. [Google Scholar]
- Odion, P.O.; Ogbonnia, E.O. Web-Based Diagnosis of Typhoid and Malaria Using Machine Learning. Niger. Def. Acad. J. Mil. Sci. Interdiscip. Stud. 2022, 1, 89–103. [Google Scholar]
- Apanisile, T.; Ayeni, J.A. Development of an Extended Medical Diagnostic System for Typhoid and Malaria Fever. Artif. Intell. Adv. 2023, 5, 28–40. [Google Scholar] [CrossRef]
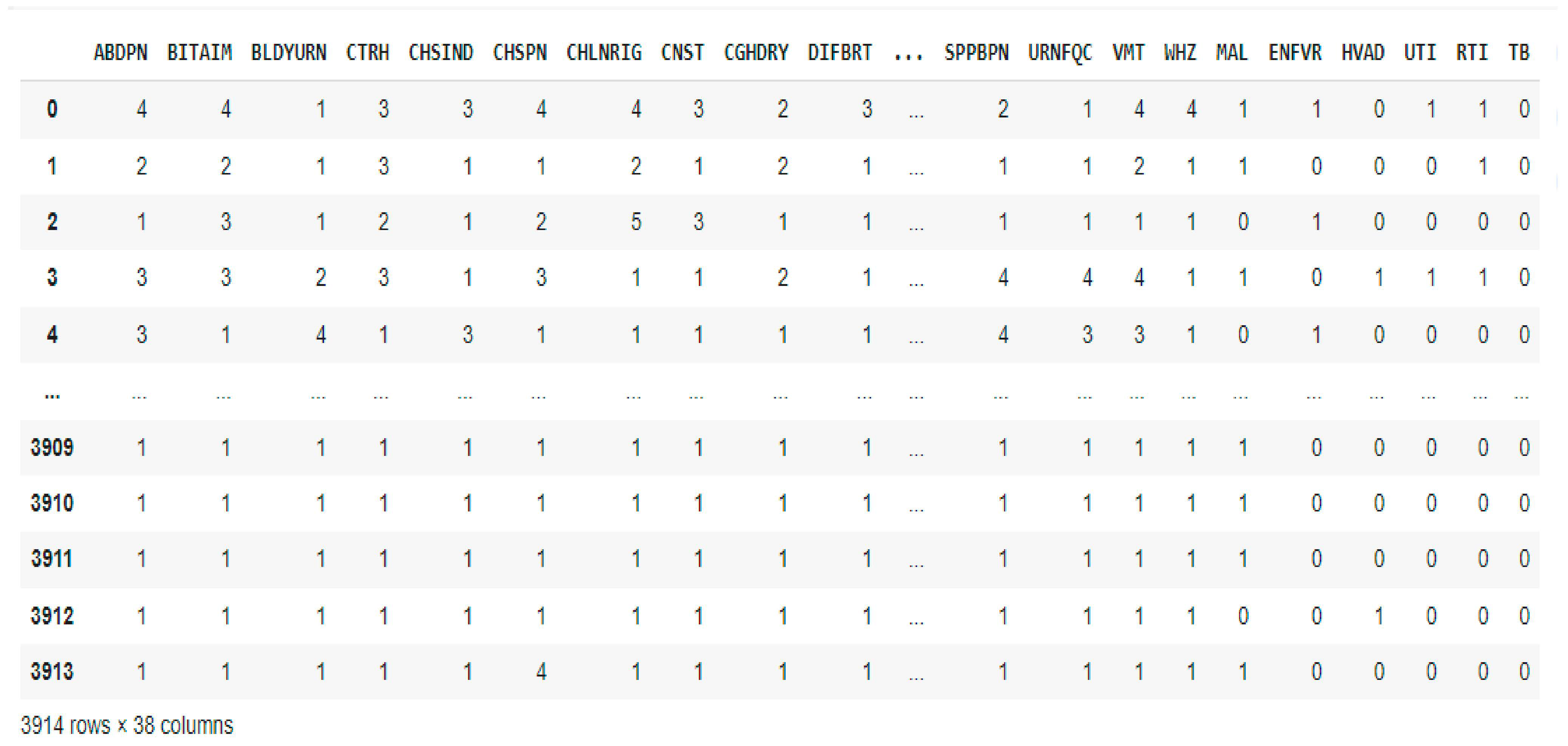

| Age Range | Male | Female | Pregnant Women | No | Nursing Mothers | No |
|---|---|---|---|---|---|---|
| <5 years | 534 | 419 | 1st trimester | 139 | 0–3 months | 27 |
| 5 years to 12 years | 346 | 323 | 2nd trimester | 184 | 4–6 months | 35 |
| 13 years to 19 years | 150 | 213 | 3rd trimester | 86 | 7–9 months | 28 |
| 20 years to 64 years | 1012 | 1605 | Over 9 months | 63 | ||
| 65 years and above | 133 | 135 | ||||
| Total | 2175 | 2695 | Total | 409 | Total | 153 |
| SN | Symptom/Disease | Abbreviation | SN | Symptom/Disease | Abbreviation |
|---|---|---|---|---|---|
| 1 | Abdominal pains | ABDPN | 33 | Muscle and body pain | MSCBDYPN |
| 2 | Back pain | BCKPN | 34 | Mouth ulcer | MUTUCR |
| 3 | Bitter taste in mouth | BITAIM | 35 | Nausea | NUS |
| 4 | Bleeding | BLDN | 36 | Night sweats | NGTSWT |
| 5 | Bloody urine | BLDYURN | 37 | Pain behind the eyes | PNBHEYE |
| 6 | Catarrh | CTRH | 38 | Upper back pain (loin) | UPBCKPN |
| 7 | Chest indraw | CHSIND | 39 | Painful urination | PNFLURNTN |
| 8 | Chest pain | CHSPN | 40 | Peritonitis | PERTN |
| 9 | Chills and rigors | CHLNRIG | 41 | Red eyes | REDEYE |
| 10 | Cloudy urine | CLDYURN | 42 | Red eyes, face, tongue | REDEYEFCTNG |
| 11 | Constipation | CNST | 43 | Sensitivity to light | SENLHT |
| 12 | Cough (initial dry) | CGHDRY | 44 | Shock | SHK |
| 13 | Diarrhea | DRH | 45 | Skin rash | SKNRSH |
| 14 | Difficulty breathing | DIFBRT | 46 | Sore throat | SRTRT |
| 15 | Dizziness | DIZ | 47 | Suprapubic pains | SPPBPN |
| 16 | Dry cough | DRYCGH | 48 | Urinary frequency | URNFQC |
| 17 | Fatigue | FTG | 49 | Vomiting | VMT |
| 18 | Fever | FVR | 50 | Wheezing | WHZ |
| 19 | High persistent fever | HGPSFVR | 51 | Malaria | MAL |
| 20 | High-grade fever | HGGDFVR | 52 | Typhoid fever | ENFVR |
| 21 | Stepwise rise fever | SWRFVR | 53 | HIV and AIDS | HVAD |
| 22 | Sudden onset fever | SUDONFVR | 54 | Upper urinary tract infection | UPUTI |
| 23 | Low-grade fever | LWGDFVR | 55 | Lower urinary tract infection | LWUTI |
| 24 | Foul breath | FOLBRT | 56 | Upper respiratory tract infection | URTI |
| 25 | Body itching | BDYICH | 57 | Lower respiratory tract infection | LRTI |
| 26 | Generalized body pain | GENBDYPN | 58 | Tuberculosis | TB |
| 27 | Generalized rashes | GENRSH | 59 | Lassa fever | LASFVR |
| 28 | Headaches | HDACH | 60 | Yellow fever | YELFVR |
| 29 | Intestinal bleeding and perforation | INTBLEPRF | 61 | Dengue fever | DENFVR |
| 30 | Joint swelling | JNTSWL | |||
| 31 | Lethargy | LTG | |||
| 32 | Lymph node swelling | LMPNDSWL |
| Symptom/Disease | Abbreviation | Symptom/Disease | Abbreviation | ||
|---|---|---|---|---|---|
| 1 | Abdominal pains | ABDPN | 20 | Headaches | HDACH |
| 2 | Bitter taste in mouth | BITAIM | 21 | Lethargy | LTG |
| 3 | Bloody urine | BLDYURN | 22 | Lymph node swelling | LMPNDSWL |
| 4 | Catarrh | CTRH | 23 | Muscle and body pain | MSCBDYPN |
| 5 | Chest indraw | CHSIND | 24 | Mouth ulcer | MUTUCR |
| 6 | Chest pain | CHSPN | 25 | Nausea | NUS |
| 7 | Chills and rigors | CHLNRIG | 26 | Night sweats | NGTSWT |
| 8 | Constipation | CNST | 27 | Painful urination | PNFLURNTN |
| 9 | Cough (initial dry) | CGHDRY | 28 | Sore throat | SRTRT |
| 10 | Difficulty breathing | DIFBRT | 29 | Suprapubic pains | SPPBPN |
| 11 | Dry cough | DRYCGH | 30 | Urinary frequency | URNFQC |
| 12 | Fatigue | FTG | 31 | Vomiting | VMT |
| 13 | Fever | FVR | 32 | Wheezing | WHZ |
| 14 | High-grade fever | HGGDFVR | 33 | Malaria | MAL |
| 15 | Stepwise rise fever | SWRFVR | 34 | Typhoid fever | ENFVR |
| 16 | Low-grade fever | LWGDFVR | 35 | HIV and AIDS | HVAD |
| 17 | Foul breath | FOLBRT | 36 | Urinary tract infection | UTI |
| 18 | Generalized body pain | GENBDYPN | 37 | Respiratory tract infection | RTI |
| 19 | Generalized rashes | GENRSH | 38 | Tuberculosis | TB |
| Disease | Total Cases | Training Set (80%) | Test Set (20%) |
|---|---|---|---|
| Malaria | 2719 | 2175 | 544 |
| Typhoid fever | 1157 | 926 | 231 |
| HIV/AIDS | 424 | 339 | 85 |
| Urinary tract infection | 907 | 726 | 181 |
| Respiratory tract infection | 1094 | 875 | 219 |
| Tuberculosis | 381 | 305 | 76 |
| MAL | ENFVR | HVAD | UTI | RTI | TB | ||
|---|---|---|---|---|---|---|---|
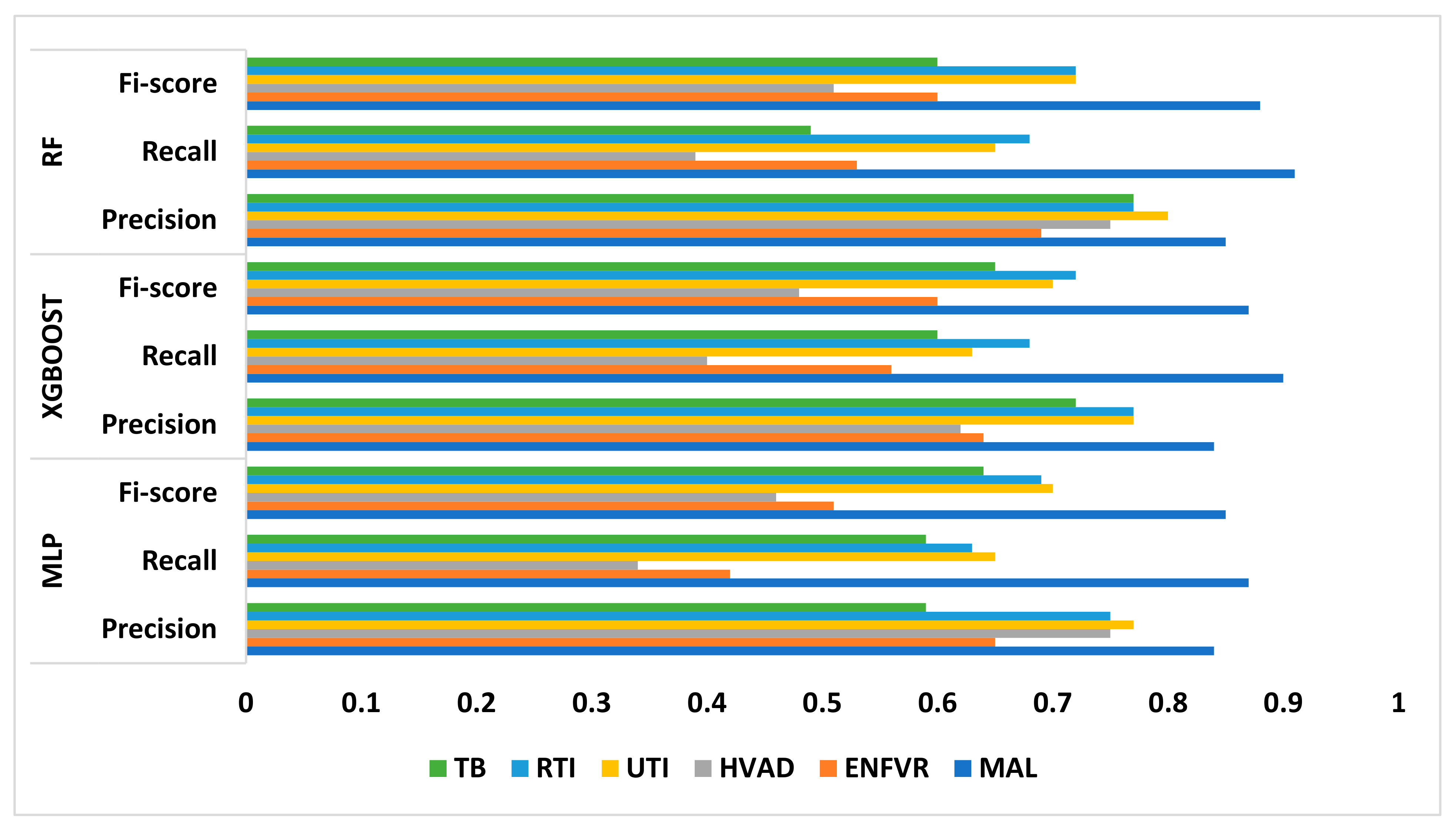
| MLP | Precision | 0.84 | 0.65 | 0.75 | 0.77 | 0.75 | 0.59 |
| Recall | 0.87 | 0.42 | 0.34 | 0.65 | 0.63 | 0.59 | |
| F1 score | 0.85 | 0.51 | 0.46 | 0.70 | 0.69 | 0.64 | |
| XGBoost | Precision | 0.84 | 0.64 | 0.62 | 0.77 | 0.77 | 0.72 |
| Recall | 0.90 | 0.56 | 0.40 | 0.63 | 0.68 | 0.60 | |
| F1 score | 0.87 | 0.60 | 0.48 | 0.70 | 0.72 | 0.65 | |
| RF | Precision | 0.85 | 0.69 | 0.75 | 0.80 | 0.77 | 0.77 |
| Recall | 0.91 | 0.53 | 0.39 | 0.65 | 0.68 | 0.49 | |
| F1 score | 0.88 | 0.60 | 0.51 | 0.72 | 0.72 | 0.60 |
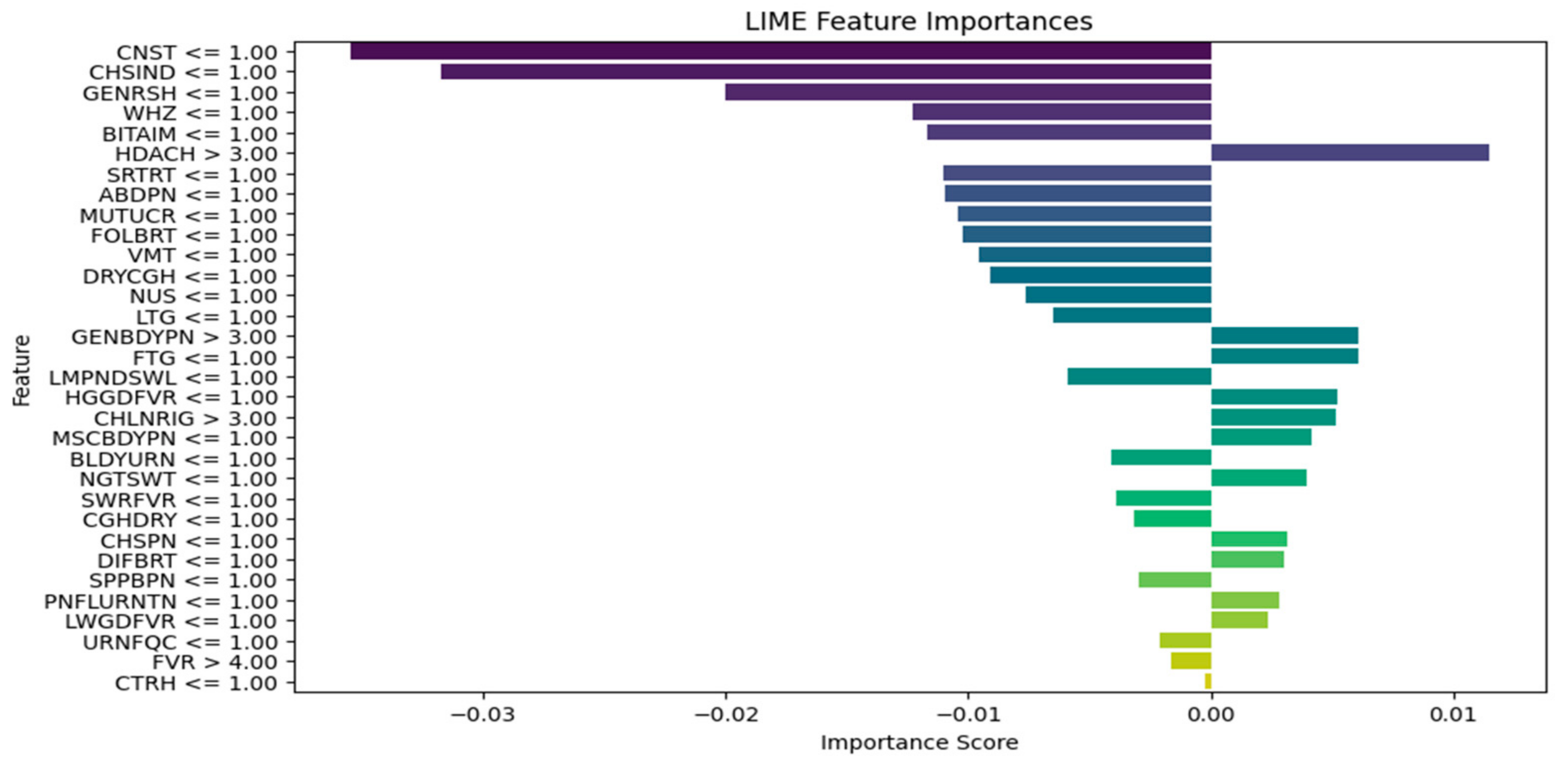
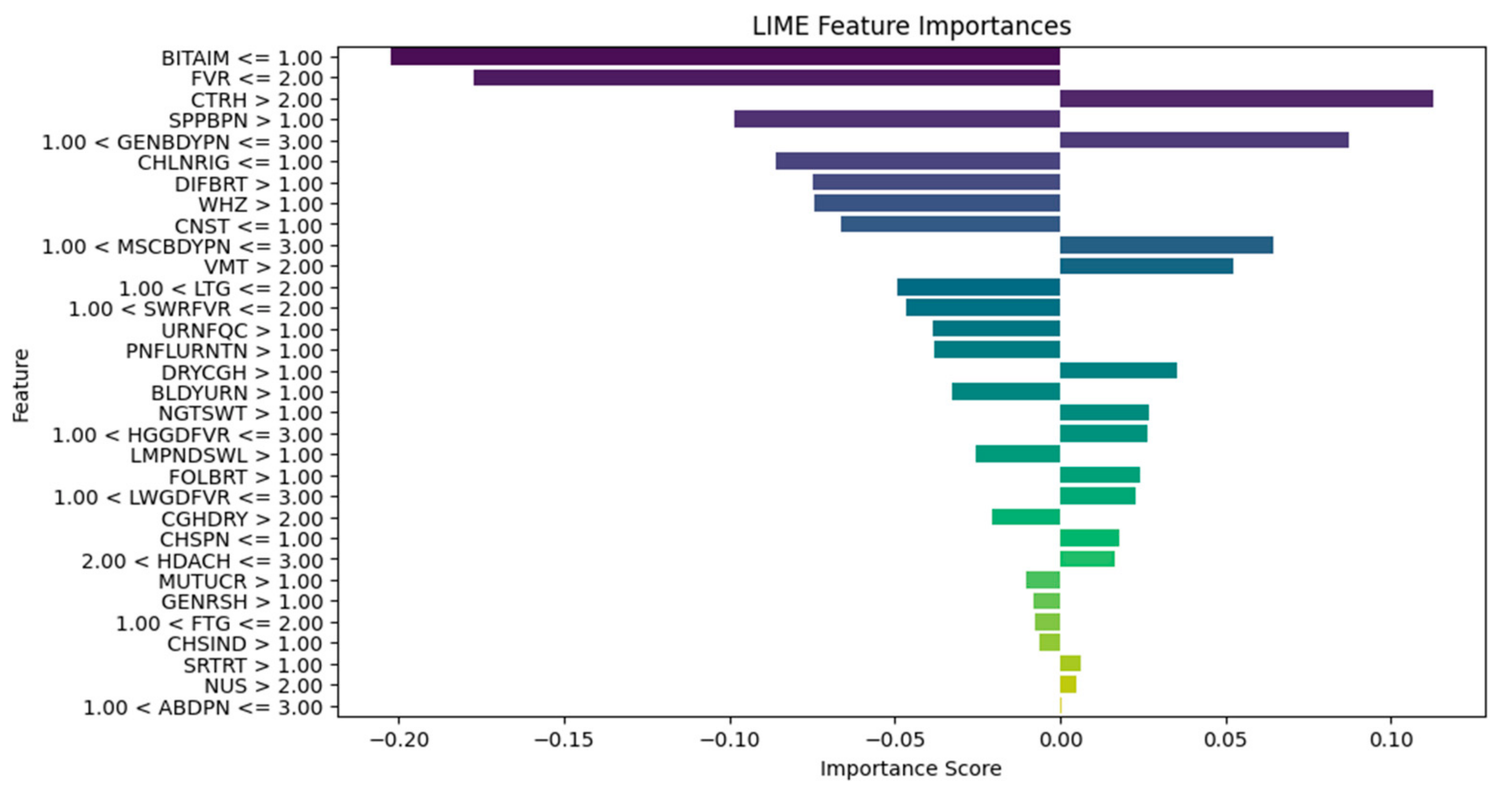
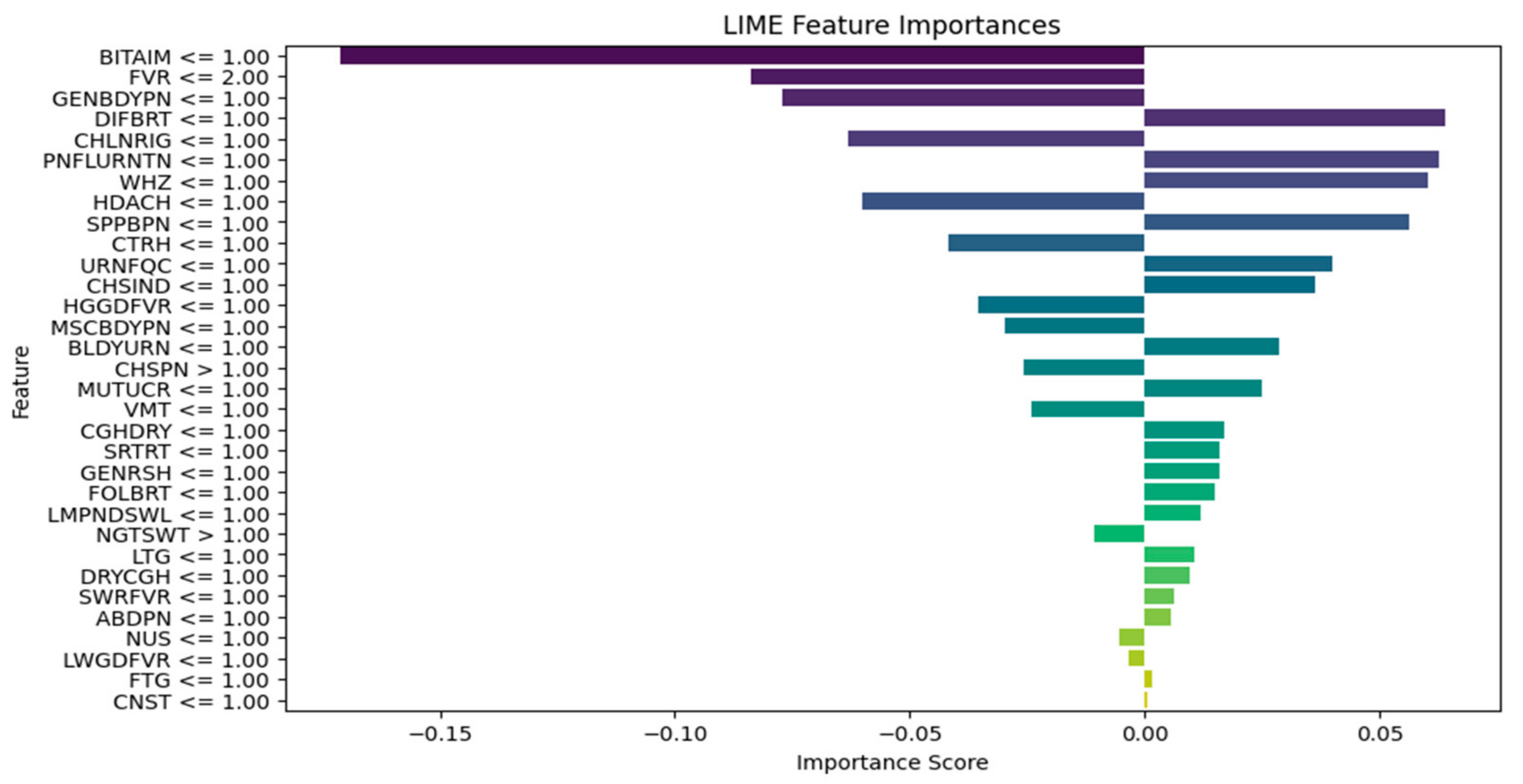
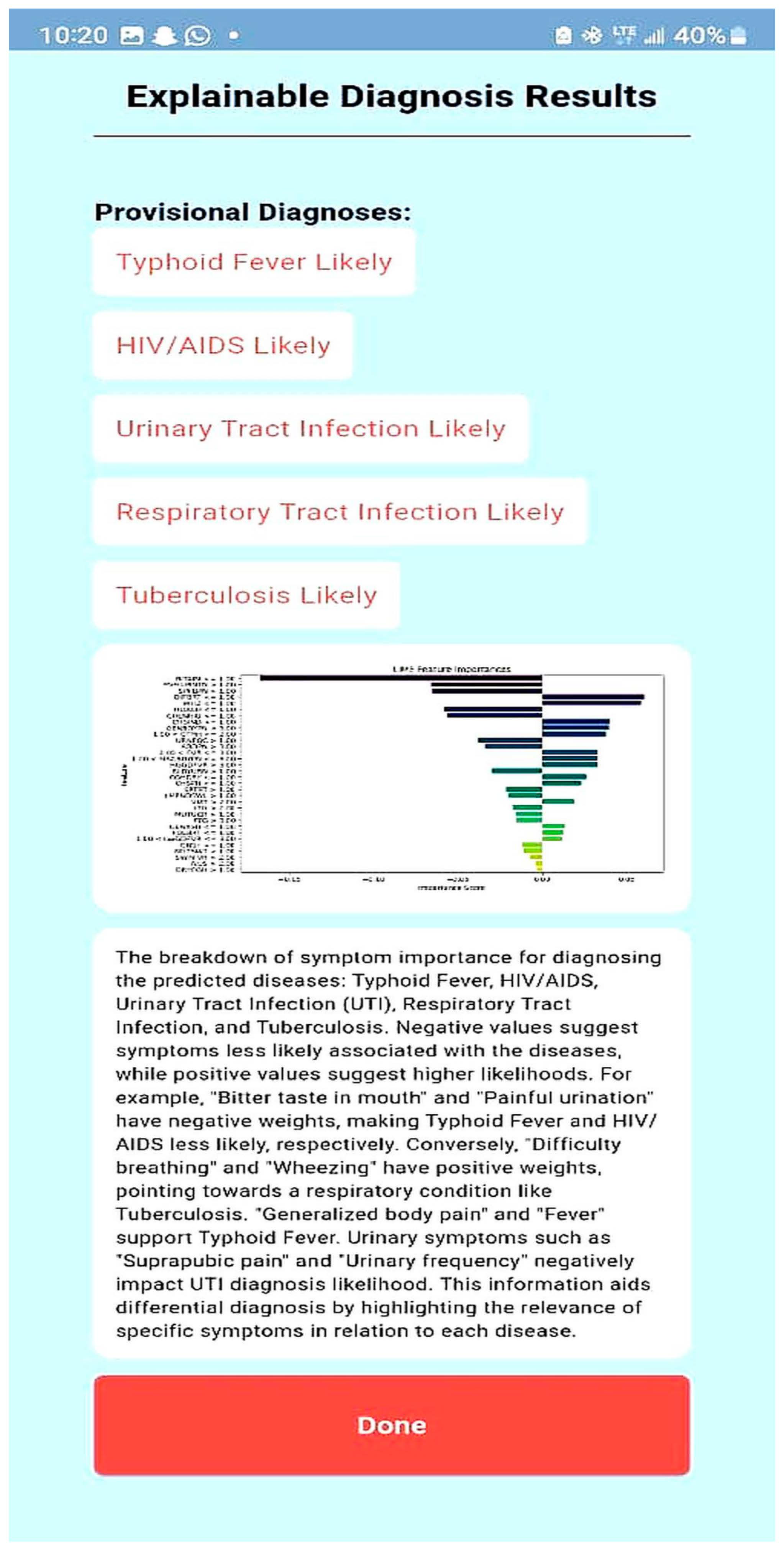
| Explain the LIME results below (disease or diseases and significant symptoms) to a physician with not more than 300 words: Predictions [“Typhoid Fever Likely”, “HIV/AIDS Likely”, “Urinary Tract Infection Likely”, “Respiratory Tract Infection Likely”, “Tuberculosis Likely”] LIME Explanation; BITTER TASTE IN MOUTH <= 1.00; −0.16758919765052108 Painful Urination > 1.00; −0.06605868111597245 Suprapubic_Pain > 1.00; −0.06550467141471979 Difficulty Breathing <= 1.00; 0.06071678705925383 Wheezing <= 1.00; 0.0590462921972035 Headache <= 1.00; −0.05826547312462451 CHILLS AND RIGORS <= 1.00; −0.05630271637802242 CHEST INDRAW <= 1.00; 0.04070095788472542 Generalized Body Pain > 3.00; 0.03974979336103006 1.00 < CATARRH <= 2.00; 0.03816070620416529 Urinary_Frequency > 1.00; −0.03770207672685268 ABDOMINAL PAIN > 3.00; −0.033902377938511155 2.00 < Fever <= 3.00; 0.032999053123995245 1.00 < Muscle and Body Pain <= 3.00; 0.03295097711760973 HGGDFever > 3.00; 0.032768000129691194 BLOODY URINE > 1.00; −0.029955841393233477 Cough (Initial Dry) <= 1.00; 0.026364166193042264 CHEST PAIN <= 1.00; 0.02354889167461192 Sore_Throat > 1.00; −0.021402483028506374 Lymph Node Swelling > 1.00; −0.0202552321969518 Vomiting > 2.00; 0.019057721531348118 Lethargy > 2.00; −0.017371438160508137 Mouth Ulcer > 1.00; −0.015856144250446513 Fatigue > 3.00; −0.015122895567807396 Generalized Rash <= 1.00; 0.014081682731125898 Foul Breath <= 1.00; 0.01260963643914623 1.00 < LWGDFever <= 3.00; 0.012187835432654306 CONSTIPATION <= 1.00; −0.012145436356090885 Night Sweat > 1.00; −0.010556292961671602 SWRFever > 2.00; −0.007451681601225083 Nausea > 2.00; −0.00406587833157149 Dry Cough > 1.00; −0.003064108573649665 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amannah, C.; Attai, K.F.; Uzoka, F.-M. A Data-Driven Intelligent Methodology for Developing Explainable Diagnostic Model for Febrile Diseases. Algorithms 2025, 18, 190. https://doi.org/10.3390/a18040190
Amannah C, Attai KF, Uzoka F-M. A Data-Driven Intelligent Methodology for Developing Explainable Diagnostic Model for Febrile Diseases. Algorithms. 2025; 18(4):190. https://doi.org/10.3390/a18040190
Chicago/Turabian StyleAmannah, Constance, Kingsley Friday Attai, and Faith-Michael Uzoka. 2025. "A Data-Driven Intelligent Methodology for Developing Explainable Diagnostic Model for Febrile Diseases" Algorithms 18, no. 4: 190. https://doi.org/10.3390/a18040190
APA StyleAmannah, C., Attai, K. F., & Uzoka, F.-M. (2025). A Data-Driven Intelligent Methodology for Developing Explainable Diagnostic Model for Febrile Diseases. Algorithms, 18(4), 190. https://doi.org/10.3390/a18040190









