GATransformer: A Graph Attention Network-Based Transformer Model to Generate Explainable Attentions for Brain Tumor Detection
Abstract
1. Introduction
2. Literature Review
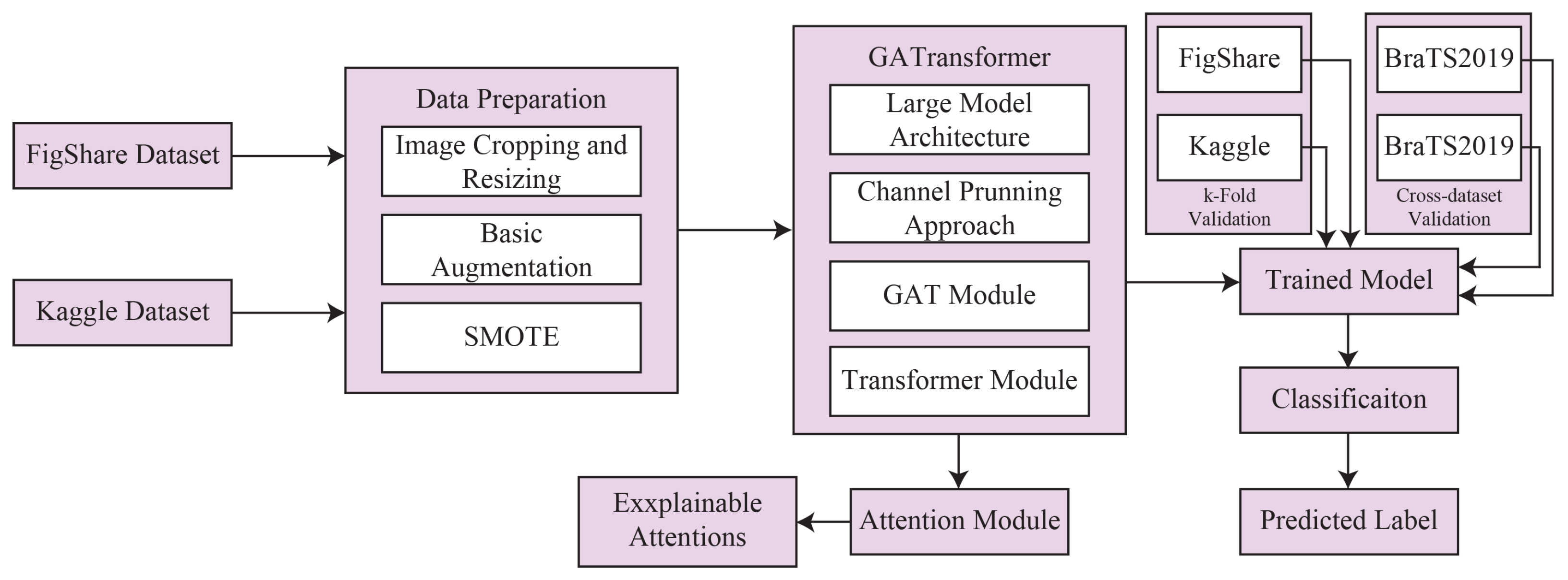
3. Proposed Methodology
3.1. Datasets
3.1.1. Data Preparation
3.1.2. Image Cropping and Resizing
3.1.3. Basic Augmentation
3.1.4. SMOTE
3.2. Proposed GATransformer
3.2.1. Large Model Architecture (LMA)
3.2.2. Channel Pruning Approach
3.2.3. GAT Module
3.2.4. Transformer Module
3.2.5. Classification Module
3.2.6. Attention Module
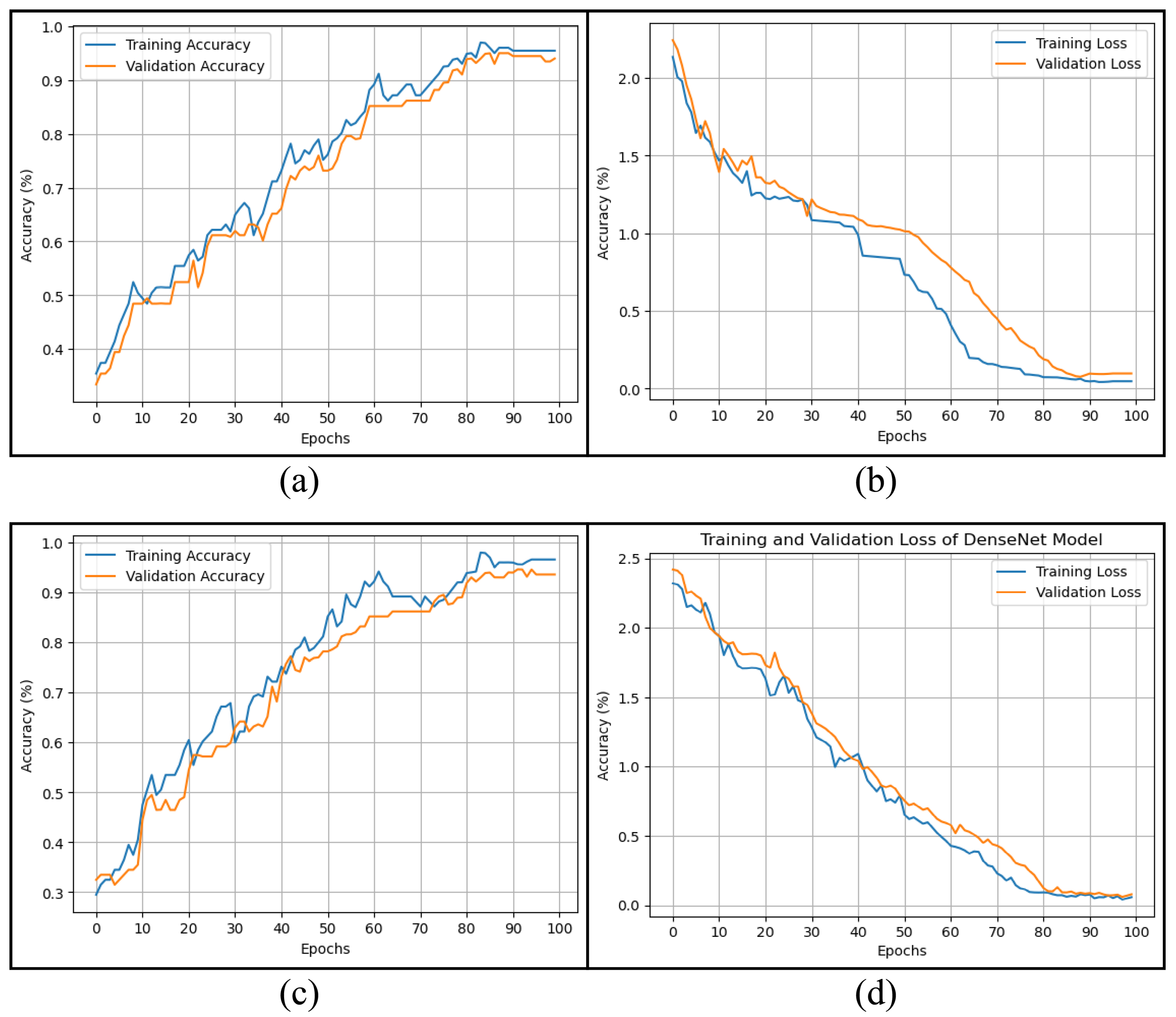
4. Experimental Results
4.1. Experimental Setup
4.2. Performance Metrics
4.3. Classification Analysis
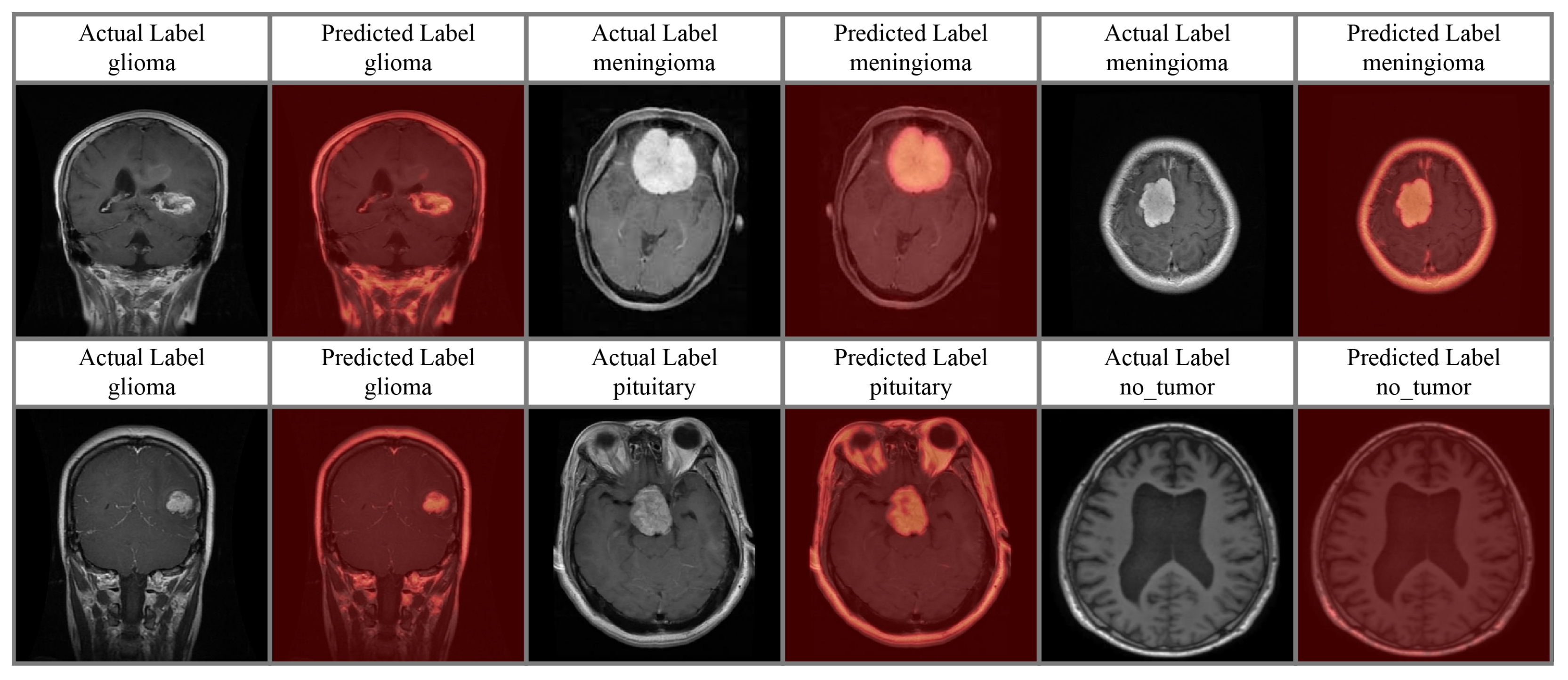
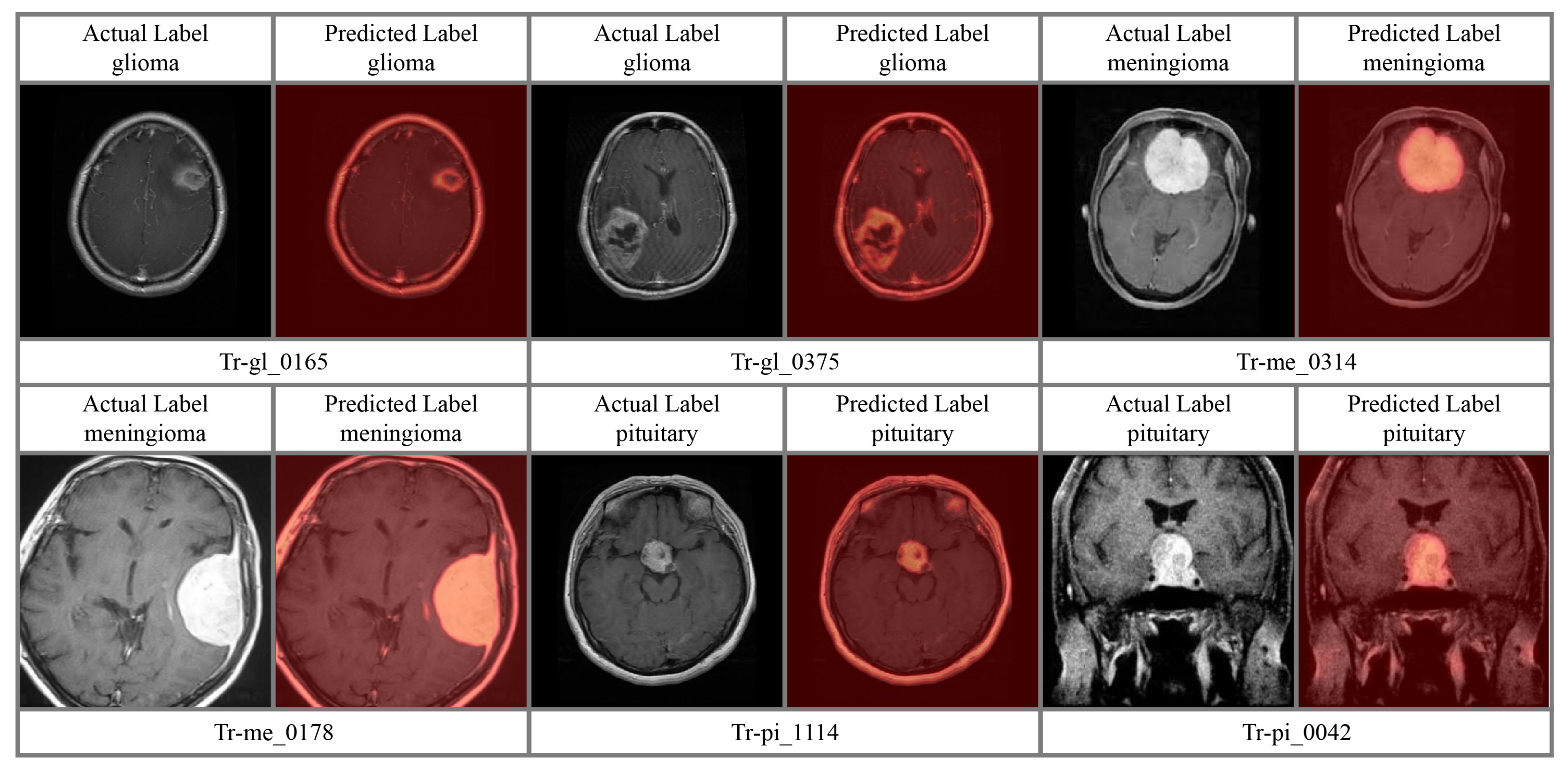
4.4. Attention Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shoeibi, A.; Khodatars, M.; Jafari, M.; Ghassemi, N.; Moridian, P.; Alizadehsani, R.; Ling, S.H.; Khosravi, A.; Alinejad-Rokny, H.; Lam, H.K.; et al. Diagnosis of brain diseases in fusion of neuroimaging modalities using deep learning: A review. Inf. Fusion 2023, 93, 85–117. [Google Scholar] [CrossRef]
- Ranjbarzadeh, R.; Caputo, A.; Tirkolaee, E.B.; Ghoushchi, S.J.; Bendechache, M. Brain tumor segmentation of MRI images: A comprehensive review on the application of artificial intelligence tools. Comput. Biol. Med. 2023, 152, 106405. [Google Scholar] [CrossRef]
- Al Mudawi, N.; Alazeb, A. A model for predicting cervical cancer using machine learning algorithms. Sensors 2022, 22, 4132. [Google Scholar] [CrossRef] [PubMed]
- Biswas, N.; Uddin, K.M.M.; Rikta, S.T.; Dey, S.K. A comparative analysis of machine learning classifiers for stroke prediction: A predictive analytics approach. Healthc. Anal. 2022, 2, 100116. [Google Scholar] [CrossRef]
- Sabu, K.; Ramnath, M.; Choudhary, A.; Raj, G.; Prakash Agrawal, A. A comparison of traditional and ensemble machine learning approaches for parkinson’s disease classification. In Machine Intelligence and Data Science Applications: Proceedings of MIDAS 2021; Springer: Berlin/Heidelberg, Germany, 2022; pp. 25–33. [Google Scholar]
- Ghimire, S.; Nguyen-Huy, T.; Deo, R.C.; Casillas-Perez, D.; Salcedo-Sanz, S. Efficient daily solar radiation prediction with deep learning 4-phase convolutional neural network, dual stage stacked regression and support vector machine CNN-REGST hybrid model. Sustain. Mater. Technol. 2022, 32, e00429. [Google Scholar] [CrossRef]
- Badjie, B.; Deniz Ülker, E. A Deep Transfer Learning Based Architecture for Brain Tumor Classification Using MR Images. Inf. Technol. Control 2022, 51, 332–344. [Google Scholar] [CrossRef]
- Garzón, A.; Kapelan, Z.; Langeveld, J.; Taormina, R. Machine learning-based surrogate modeling for urban water networks: Review and future research directions. Water Resour. Res. 2022, 58, e2021WR031808. [Google Scholar] [CrossRef]
- Mani, R.C.; Kamalakannan, J. The comparative study of CNN models for breast histopathological image classification. In Proceedings of the 2023 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 23–25 January 2023; pp. 1–5. [Google Scholar]
- Tummala, S.; Kim, J.; Kadry, S. BreaST-Net: Multi-class classification of breast cancer from histopathological images using ensemble of swin transformers. Mathematics 2022, 10, 4109. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Q.; Yang, Z.; Li, Y.; Lee, C.H.; Shen, L. Recent progress in transformer-based medical image analysis. Comput. Biol. Med. 2023, 164, 107268. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Sakaguchi, Y.; Kouno, N.; Takasawa, K.; Ishizu, K.; Akagi, Y.; Aoyama, R.; Teraya, N.; Bolatkan, A.; Shinkai, N.; et al. Comparison of Vision Transformers and Convolutional Neural Networks in Medical Image Analysis: A Systematic Review. J. Med. Syst. 2024, 48, 84. [Google Scholar] [CrossRef]
- Mosqueira-Rey, E.; Hernández-Pereira, E.; Alonso-Ríos, D.; Bobes-Bascarán, J.; Fernández-Leal, Á. Human-in-the-loop machine learning: A state of the art. Artif. Intell. Rev. 2023, 56, 3005–3054. [Google Scholar]
- Casini, L.; Marchetti, N.; Montanucci, A.; Orrù, V.; Roccetti, M. A human–AI collaboration workflow for archaeological sites detection. Sci. Rep. 2023, 13, 8699. [Google Scholar] [CrossRef]
- Asiri, A.A.; Shaf, A.; Ali, T.; Shakeel, U.; Irfan, M.; Mehdar, K.M.; Halawani, H.T.; Alghamdi, A.H.; Alshamrani, A.F.A.; Alqhtani, S.M. Exploring the power of deep learning: Fine-tuned vision transformer for accurate and efficient brain tumor detection in MRI scans. Diagnostics 2023, 13, 2094. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Bajwa, U.I.; Mehmood, Y. Multi-class classification of brain tumor types from MR images using EfficientNets. Biomed. Signal Process. Control 2023, 84, 104777. [Google Scholar] [CrossRef]
- Hossain, S.; Chakrabarty, A.; Gadekallu, T.R.; Alazab, M.; Piran, M.J. Vision transformers, ensemble model, and transfer learning leveraging explainable AI for brain tumor detection and classification. IEEE J. Biomed. Health Inform. 2023, 28, 1261–1272. [Google Scholar] [CrossRef]
- Tummala, S.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling. Curr. Oncol. 2022, 29, 7498–7511. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, Y.; Lin, X.; Dong, J.; Cheng, T.; Liang, J. SwinBTS: A method for 3D multimodal brain tumor segmentation using swin transformer. Brain Sci. 2022, 12, 797. [Google Scholar] [CrossRef] [PubMed]
- ZainEldin, H.; Gamel, S.A.; El-Kenawy, E.S.M.; Alharbi, A.H.; Khafaga, D.S.; Ibrahim, A.; Talaat, F.M. Brain tumor detection and classification using deep learning and sine-cosine fitness grey wolf optimization. Bioengineering 2022, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ngo, H.C.; Zhang, Y.; Yusof, N.F.A.; Wang, X. Imaging Segmentation of Brain Tumors Based on the Modified U-net Method. Inf. Technol. Control 2024, 53, 1074–1087. [Google Scholar] [CrossRef]
- Odusami, M.; Maskeliūnas, R.; Damaševičius, R.; Misra, S. Explainable Deep-Learning-Based Diagnosis of Alzheimer’s Disease Using Multimodal Input Fusion of PET and MRI Images. J. Med. Biol. Eng. 2023, 43, 291–302. [Google Scholar] [CrossRef]
- Tehsin, S.; Nasir, I.M.; Damaševičius, R.; Maskeliūnas, R. DaSAM: Disease and Spatial Attention Module-Based Explainable Model for Brain Tumor Detection. Big Data Cogn. Comput. 2024, 8, 97. [Google Scholar] [CrossRef]
- Ullah, M.S.; Khan, M.A.; Albarakati, H.M.; Damaševičius, R.; Alsenan, S. Multimodal brain tumor segmentation and classification from MRI scans based on optimized DeepLabV3+ and interpreted networks information fusion empowered with explainable AI. Comput. Biol. Med. 2024, 182, 109183. [Google Scholar] [CrossRef] [PubMed]
- Nickparvar, M. Brain Tumor MRI Dataset. KAGGLE 2021. [Google Scholar] [CrossRef]
- Cheng, J. Brain magnetic resonance imaging tumor dataset. Figshare MRI Dataset Version 2017, 5. [Google Scholar] [CrossRef]
- Rosebrock, A. Finding Extreme Points in Contours with Open CV. 2016. Available online: https://pyimagesearch.com/2016/04/11/finding-extreme-points-in-contours-with-opencv/ (accessed on 24 October 2024).
- Nitesh, V.C. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. arXiv 2017, arXiv:1710.10903. [Google Scholar]
- Vaswani, A. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Redmon, J. Darknet: Open Source Neural Networks in C. 2013–2016. Available online: http://pjreddie.com/darknet/ (accessed on 24 October 2024).
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Bahdanau, D. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:1409.0473. [Google Scholar]
- Ba, J.L. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Dosovitskiy, A. An image is worth 16 · 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Akhter, A.; Acharjee, U.K.; Talukder, M.A.; Islam, M.M.; Uddin, M.A. A robust hybrid machine learning model for Bengali cyber bullying detection in social media. Nat. Lang. Process. J. 2023, 4, 100027. [Google Scholar] [CrossRef]
- Ahmad, W.; Ali, H.; Shah, Z.; Azmat, S. A new generative adversarial network for medical images super resolution. Sci. Rep. 2022, 12, 9533. [Google Scholar] [CrossRef] [PubMed]











| Dataset | Glioma | Meningioma | Pituitary | No_Tumor | Total |
|---|---|---|---|---|---|
| Kaggle [25] | 1321 | 1339 | 1457 | 1595 | 5712 |
| FigShare [26] | 1426 | 708 | 930 | - | 3064 |
| LMA Combination | Kaggle Dataset | FigShare Dataset | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A (%) | P (%) | R (%) | F1 (%) | RMSE (%) | A (%) | P (%) | R (%) | F1 (%) | RMSE (%) | |
| In + De + E0 | 63.6 | 61.9 | 62.9 | 62.4 | 17.2 | 61.2 | 60.9 | 58.9 | 59.9 | 23.0 |
| In + De + Da | 74.7 | 74.8 | 74.0 | 74.4 | 18.8 | 76.6 | 73.7 | 75.4 | 74.5 | 14.7 |
| In + De + Mo | 85.0 | 84.2 | 83.8 | 84.0 | 6.6 | 85.3 | 83.4 | 82.0 | 82.7 | 9.5 |
| In + E0 + Da | 82.1 | 82.7 | 82.8 | 82.1 | 14.8 | 82.2 | 82.1 | 81.8 | 81.9 | 10.1 |
| In + E0 + Mo | 71.5 | 70.7 | 71.4 | 70.0 | 16.0 | 69.7 | 67.7 | 66.4 | 67.0 | 14.3 |
| In + Da + Mo | 52.8 | 51.4 | 52.4 | 51.9 | 15.6 | 56.9 | 56.0 | 55.1 | 55.5 | 14.1 |
| De + E0 + Da | 72.1 | 73.8 | 73.2 | 72.5 | 11.1 | 77.3 | 74.5 | 76.0 | 75.2 | 13.5 |
| De + E0 + Mo | 75.9 | 75.9 | 74.6 | 76.7 | 18.5 | 79.7 | 78.6 | 77.7 | 78.1 | 16.4 |
| De + Da + Mo | 81.5 | 81.4 | 80.3 | 82.3 | 12.8 | 80.0 | 81.1 | 81.3 | 81.2 | 12.4 |
| E0 + Da + Mo | 70.1 | 70.3 | 71.2 | 69.7 | 23.0 | 68.2 | 65.6 | 67.5 | 66.5 | 15.4 |
| Dataset | Kaggle Dataset | FigShare Dataset | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A (%) | P (%) | R (%) | F1 (%) | RMSE (%) | A (%) | P (%) | R (%) | F1 (%) | RMSE (%) | |
| Original dataset | 85.0 | 84.2 | 83.8 | 84.0 | 6.6 | 85.3 | 83.4 | 82.0 | 82.7 | 9.5 |
| Basic augmentation | 85.0 | 84.2 | 83.8 | 84.0 | 6.6 | 85.3 | 83.4 | 82.0 | 82.7 | 9.5 |
| SMOTE | 87.1 | 86.7 | 86.3 | 86.5 | 5.5 | 87.5 | 86.4 | 86.0 | 86.2 | 8.3 |
| Dataset | Kaggle Dataset | FigShare Dataset | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A (%) | P (%) | R (%) | F1 (%) | RMSE (%) | A (%) | P (%) | R (%) | F1 (%) | RMSE (%) | |
| Original dataset | 92.0 | 91.5 | 91.2 | 91.3 | 8.0 | 92.2 | 91.0 | 90.8 | 90.9 | 7.8 |
| Basic augmentation | 95.0 | 94.6 | 94.4 | 94.5 | 5.5 | 95.3 | 94.7 | 94.2 | 94.4 | 5.3 |
| SMOTE | 99.0 | 98.7 | 98.5 | 98.6 | 2.0 | 99.2 | 98.8 | 98.6 | 98.7 | 1.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tehsin, S.; Nasir, I.M.; Damaševičius, R. GATransformer: A Graph Attention Network-Based Transformer Model to Generate Explainable Attentions for Brain Tumor Detection. Algorithms 2025, 18, 89. https://doi.org/10.3390/a18020089
Tehsin S, Nasir IM, Damaševičius R. GATransformer: A Graph Attention Network-Based Transformer Model to Generate Explainable Attentions for Brain Tumor Detection. Algorithms. 2025; 18(2):89. https://doi.org/10.3390/a18020089
Chicago/Turabian StyleTehsin, Sara, Inzamam Mashood Nasir, and Robertas Damaševičius. 2025. "GATransformer: A Graph Attention Network-Based Transformer Model to Generate Explainable Attentions for Brain Tumor Detection" Algorithms 18, no. 2: 89. https://doi.org/10.3390/a18020089
APA StyleTehsin, S., Nasir, I. M., & Damaševičius, R. (2025). GATransformer: A Graph Attention Network-Based Transformer Model to Generate Explainable Attentions for Brain Tumor Detection. Algorithms, 18(2), 89. https://doi.org/10.3390/a18020089







