Abstract
Brain MRI segmentation plays a crucial role in neuroimaging studies and clinical trials by enabling the precise localization and quantification of brain tissues and structures. The advent of deep learning has transformed the field, offering accurate and fast tools for MRI segmentation. Nevertheless, several challenges limit the widespread applicability of these methods in practice. In this systematic review, we provide a comprehensive analysis of developments in deep learning-based segmentation of brain MRI in adults, segmenting the brain into tissues, structures, and regions of interest. We explore the key model factors influencing segmentation performance, including architectural design, choice of input size and model dimensionality, and generalization strategies. Furthermore, we address validation practices, which are particularly important given the scarcity of manual annotations, and identify the limitations of current methodologies. We present an extensive compilation of existing segmentation works and highlight the emerging trends and key results. Finally, we discuss the challenges and potential future directions in the field.
1. Introduction
Magnetic Resonance Imaging (MRI) is a non-invasive imaging modality that enables the detailed visualization of anatomical structures, pathological changes, and functional properties of tissues. MRI has become a principal biomarker in the diagnosis and treatment of neurological disorders. Accurate segmentation of brain MRI images is a critical step in many clinical and research applications, including tumor detection and the study and treatment of neurodegenerative diseases such as Alzheimer’s disease (AD) and multiple sclerosis (MS). Brain segmentation involves partitioning brain MRI into different tissues such as gray matter (GM), white matter (WM), cerebrospinal fluid (CSF), anatomical structures or regions of interest, and pathological regions like lesions. The outputs are used for diagnosis, identification of pathology, and morphometric estimation of brain structures, and are provided as input to downstream neuroimaging steps such as volume estimation and reconstruction of cortical geometry.
While manual segmentation by medical experts is typically considered the gold-standard benchmark for brain segmentation, it too can be error-prone and operator-dependent, and is tedious and time-consuming [1]. Numerous automatic tools have therefore been developed for the task. Traditional approaches are typically based on a series of image transformation steps, such as registration, intensity thresholding, edge detection, clustering, and region growing. Simpler traditional approaches typically struggle with segmenting complex and heterogeneous structures of varying contrasts that can be found in the brain. The more sophisticated approaches, such as atlas-based methods, may lack robustness and suffer from long runtimes, especially when including non-rigid registration. This limits their scalability to large studies and clinical trials, which include thousands of scans or require immediate results.
In recent years, the advent of deep learning techniques has revolutionized the field of medical image analysis [2]. Convolutional Neural Networks (CNNs), a class of deep neural networks designed for image processing, demonstrated remarkable success and achieved state-of-the-art performance in medical image segmentation. The application of deep learning methods holds the potential to improve the accuracy, robustness, and automation of brain MRI segmentation. This reduces the burden on medical experts and enhances the reliability and reproducibility of diagnostic procedures. By providing accurate and scalable segmentation, deep learning models can facilitate large-scale neurological studies and support improved diagnosis, treatment planning, and monitoring of neurological diseases, ultimately contributing to improved patient outcomes. Many methods, however, face important challenges that limit their practical deployment.
Several reviews have addressed segmentation in specific contexts such as tumors [3], lesions [4], or fetal populations [5], but none have focused on brain segmentation in adults. The most closely related recent review by Wu et al. [6] surveyed segmentation of brain tissues (GM, WM, and CSF) across different age groups, including 35 studies on adults. However, several important gaps remain unaddressed. Our review builds on the existing literature in four key ways: (1) we cover segmentation into anatomical structures and regions of interest, which is crucial for many neuroimaging applications and presents distinct challenges from tissue segmentation, and we include 66 publications on this task; (2) we include an additional 54 publications on tissue segmentation, extending the scope of Wu et al.’s work; (3) we systematically analyze architectural choices, model dimensionalities (2D, 2.5D, 3D) and input strategies (patch-based vs. full slice/volume), and generalization techniques; and (4) we assess commonly utilized approaches for validation and identify key methodological limitations. Together, these contributions provide a more comprehensive and deeper overview of the current landscape of deep learning-based brain segmentation in adult populations.
2. Methods
This review is registered on the Open Science Framework (OSF; https://osf.io/k879r, accessed on 8 October 2025) and follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines where applicable (see Supplementary Materials S1 for the PRISMA checklist).
We outline our systematic literature search strategy in Section 2.1. We describe brain MRI segmentation and its applications in Section 2.2 and the traditional approaches in Section 2.3 in order to highlight the context of deep learning methods. We discuss deep learning approaches in Section 2.4 and focus on key aspects regarding model architecture in Section 2.5, patch-based and whole-image-based models in Section 2.6, model dimensionality in Section 2.7, and generalization in Section 2.8.
2.1. Literature Search
We conducted a search on PubMed (https://pubmed.ncbi.nlm.nih.gov/, accessed on 23 May 2025) to identify publications on brain segmentation of MRI using deep learning models. The query used was
(brain[MeSH Terms]) AND (magnetic resonance imaging[MeSH Terms]) AND (segmentation[Title/Abstract] OR segmentations[Title/Abstract]) AND (“neural network”[Title/Abstract] OR “neural networks”[Title/Abstract] OR “deep learning”[Title/Abstract] OR transformer[Title/Abstract])
The search was limited to publications from 1 January 2015 onward to exclude studies preceding the influential U-Net [7] segmentation model, and was last conducted on 23 May 2025. The query returned 660 works, from which we excluded exclusively pediatric, ex vivo, and non-human studies, structure segmentation with a low (<7) number of labels, and tumor and lesion segmentation; only publications written in English were considered. This resulted in 131 publications. A subsequent manual review to ensure the eligibility criteria and direct relevance to deep learning brain MRI segmentation yielded 88 publications. For comprehensive coverage, we supplemented these results with forward and backward citation tracking, giving a total of 120 publications. Figure 1 illustrates the steps followed during the screening process to identify eligible publications.
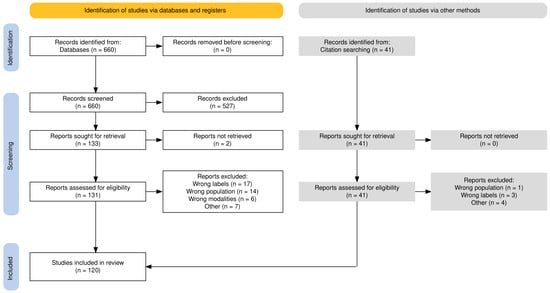
Figure 1.
Flow diagram of the literature screening and selection process.
2.2. Definition and Applications
Brain segmentation partitions the brain into brain tissues, anatomical structures, and regions of interest. Specifically, tissue segmentation classifies each brain voxel as GM, WM, or CSF, and may identify additional elements such as the scalp and skull [8], while structure segmentation assigns voxels to one of several structures or regions. In the literature, whole brain segmentation refers to structure segmentation of the entire brain. Tissue and structure segmentation differ in applications and computational requirements, with whole brain segmentation typically being the most challenging task due to a large number of output labels. Some models segment the brain into both tissues and structures simultaneously [9]. Although structure segmentation outputs can often be combined to produce tissue segmentation, this approach may yield inferior results compared to dedicated tissue segmentation [10] and requires greater computational resources.
Brain segmentation is a critical step in neuroimaging pipelines, providing detailed anatomical delineation necessary for structure visualization, morphometric or volumetric quantification, and downstream computational analyses. It supports large-scale studies examining brain development, structural variability among populations, and pathology associated with neurodegenerative, psychiatric, and neurodevelopmental disorders. As neuroimaging datasets increasingly comprise thousands of scans [11], the demand for automated, scalable, and robust segmentation methods has intensified. This requirement is also present in clinical trials, where high-throughput and accurate volume estimation from MRI is essential for reproducibility and statistical power. In clinical settings, brain segmentation further facilitates surgical planning and image-guided interventions. Segmentation results are also used as inputs to further steps in neuroimaging workflows and for aligning PET and CT images for better anatomical localization.
Segmentation-derived volume and thickness measurements play a crucial role in the assessment and follow-up of neurodegenerative disorders. Healthy aging is characterized by slow GM and WM atrophy, cortical thinning, and expansion of the ventricular system and subarachnoid CSF. Estimates of greater and localized atrophy, especially in the hippocampus, amygdala, entorhinal cortex, and medial temporal lobe [12], are associated with mild cognitive impairment. Such patterns have prominently been studied in AD, which is further distinguished by atrophy of the posterior cingulate and the parietal lobes. Tracking AD progression and treatment effects is additionally motivated by increased atrophy and ventricular enlargement found in recent trials of anti-amyloid drugs [13]. In MS, deep GM atrophy and damage show high clinical relevance in predicting physical disability and cognitive impairment [14]. WM lesions, visible as T1 hypointensities and more prominently as T2 and FLAIR hyperintensities, also correlate with disability in MS. Some whole brain segmentation methods therefore include segmentation of WM lesions, while WM tissue masks can improve separate lesion segmentation [15]. In Huntington’s disease, the primary image marker of progression is atrophy of the caudate and putamen [16]. Volumetry also aids in distinguishing Parkinson’s disease (PD) from Parkinson-plus syndromes, which manifest with additional clinical and structural features. For example, midbrain atrophy is seen in progressive supranuclear palsy (PSP), but is typically absent in PD, while atrophy of the pons, cerebellum, and putamen is more indicative of multiple system atrophy (MSA) [17].
Brain segmentation is also utilized in psychological and neurological conditions, where subcortical volumes differentiate major depressive, bipolar, and remitted major depressive disorder [18], while subcortical enlargement has been observed in schizophrenia [19]. Structural brain changes have also been investigated in relation to conditions such as atrial fibrillation [20], migraine [21], and even hospitalization [22]. Segmentation supports magnetic resonance spectroscopy by enabling metabolite quantification by tissue type [23]. In transcranial electrical stimulation, accurate segmentation of sulci and gyri is essential for precise electric field modeling [24]. Furthermore, segmentation aids in defining functional region boundaries for functional MRI (fMRI), offering applications even when structural MRI reveals no abnormalities, such as in mild traumatic brain injury [25]. Delineation of the cortex, WM, and the meninges is also performed as the first step in cortical reconstruction pipelines [26]. The applications to various domains and settings increase the need for accurate, robust, generalizable, and efficient brain MRI segmentation.
2.3. Traditional Segmentation Approaches
Traditional brain segmentation algorithms are typically multi-stage pipelines that consist of several processing steps [27]. Intensity-based approaches use thresholding, region growing, clustering, and classification, and are suitable for the simpler task of tissue segmentation. Whole brain segmentation is typically performed by the more intensive atlas-based methods. These methods begin with preprocessing steps, such as intensity normalization, bias field correction, and skull stripping, followed by non-rigid registration to an atlas in a standard space. Anatomical labels of the atlas are then propagated onto the subject image. This approach underlies established and publicly available neuroimaging toolkits such as FreeSurfer [28], SPM [29], and CAT [30], which are frequently utilized in studies and clinical trials. However, such methods have very long runtimes due to non-rigid registration: for example, CAT12 and FreeSurfer may require approximately 1 h and 8 h, respectively, to segment a single scan. Furthermore, they are sensitive to registration quality, with high intersubject variability of brain anatomy further limiting accurate atlas matching. Multi-atlas label fusion methods therefore extend the paradigm by using labels from multiple annotated scans to produce the final predictions on the input volume. While such methods produce very accurate segmentations, they require multiple non-linear registrations and are exceptionally computationally intensive, requiring even up to 147 h for a single segmentation [31]. Long runtimes limit the scalability of atlas-based methods for large-scale studies and trials, where throughput is essential to processing thousands of scans. The methods also leave room for improvements in accuracy and reliability. In contrast, deep learning models are a scalable alternative, achieving state-of-the-art accuracy while drastically reducing inference time.
2.4. Deep Learning Segmentation
The success of deep learning models in segmentation tasks has been driven by hardware advances, the availability of annotated datasets, and improvements in deep learning architectures. Since their adaptation to brain MRI segmentation, the models have achieved state-of-the-art accuracy while significantly accelerating segmentation; accurate methods perform segmentation in seconds on GPUs. Such models typically avoid the complex pipelines of traditional methods and do not rely on registration or hand-crafted features; instead, they learn directly from annotated data in a supervised manner. Their performance, therefore, highly depends on the quality and diversity of training data, as well as the model architecture and training strategy.
Deep learning has been extensively applied to tumor [3] and lesion [4] segmentation. The progress has been somewhat slower in brain segmentation, partly due to the longstanding availability of established traditional methods, which have remained widely used in both research and clinical contexts. More critically, there is a notable scarcity of comprehensive, manually labeled ground truth datasets for the entire brain, hindering supervised training and robust evaluation. In comparison, there is a larger and more diverse volume of manually annotated data available for tumor and lesion segmentation. Additionally, brain segmentation encompasses a broad anatomical scope, requiring models to generalize across a wide range of brain structures with varying size, shape, and intensity profiles. Whole brain segmentation, in particular, demands great computational resources due to the large numbers of target labels.
A central design consideration in deep learning-based segmentation is the choice of model architecture. Most brain segmentation models have adapted architectures originally developed for other domains, evolving from early convolutional classification networks applied in a sliding-window manner [32] to fully convolutional networks [33], U-Net variants [7], and, more recently, transformer-based [34] and state space models [35]. Due to the large GPU memory requirements of training on 3D images, initial approaches were commonly patch-based and subdivided the input MRI or slice into smaller regions to reduce memory demands and increase training efficiency. Although the increase in computational resources allowed for many current models to be trained on full volumes, several studies have reported improved results when training on large patches instead [36]. Another important factor is model dimensionality: 2D methods operate on individual slices or thin blocks of slices, 3D methods process full volumes or sub-volumes, while 2.5D ensemble models trained on orthogonal 2D slices. The choice of the segmentation input (patches vs. full slices or volumes) and model dimensionality reflects a trade-off between the amount of utilized spatial context and the computational cost of training and inference. We discuss the segmentation input in Section 2.6 and model dimensionality in Section 2.7. Crucially for practical adoption, segmentation methods must generalize across highly diverse imaging parameters and populations. We discuss generalization strategies in Section 2.8.
We list all structure and tissue segmentation methods in Table 1 and Table 2, respectively, summarizing key properties such as input modalities, model dimensionality, input size, and model architecture. For consistency, we report the highest Dice similarity coefficient (DSC)—the most commonly available performance measure—as an indicator of segmentation accuracy. Additionally, structure segmentation results on the Mindboggle101 dataset [37] are presented in Table 4.
2.5. Segmentation Architectures
Deep learning brain MRI segmentation has closely followed advancements in deep learning for general medical image segmentation. There, early efforts processed images patch-by-patch and classified the center voxel of each patch using a convolutional classification model in a sliding-window fashion [32]. Such approaches struggle to incorporate global context, as each patch is processed in isolation and is computationally expensive due to redundant computations. The introduction of Fully Convolutional Networks (FCNs) [33] addressed these issues by processing images of arbitrary size and generating correspondingly-sized output, enabling end-to-end learning. FCNs replaced the fully-connected layers in classification models with convolutional ones to produce dense, pixel-wise predictions. FCNs utilize transposed convolutions to restore resolution after downsampling and combine coarser predictions from deep layers with fine-grained predictions from shallow layers using skip connections. Compared to sliding-window models, FCNs improved accuracy and efficiency by utilizing global context and eliminating redundant computations [33].
Building on FCNs, “U-shaped” models such as the U-Net [7] introduced a symmetric encoder–decoder architecture with skip connections that combine activations from deep and shallow layers. The encoder and decoder are composed of multiple blocks, each block consisting of a series of convolutional layers, non-linearities such as the ReLU, and normalization layers. The latter include batch, group [36], instance [38,39,40], and weight [41] normalization layers in brain segmentation. The encoder progressively downsamples activations via max pooling or strided convolutions, while the decoder upsamples activations towards the original resolution using transposed convolutions [7] or pooling indices taken from the encoder [42]. Compared to FCNs, the U-Net architecture has greater representational power and greater ability to combine high-level features and fine details.
U-Net achieved notable success in various segmentation tasks and inspired numerous modifications which continue to achieve state-of-the-art results in medical image segmentation. In brain segmentation, it remains the backbone architecture of the majority of models, with notable architectural refinements incorporated over time. These include residual blocks in the encoder and decoder [24,36,43,44], dense [8,45,46] or competitive dense [26,47,48] blocks, the UNet++ architecture [49] with nested dense skip pathways, multiscale convolutional features [38,50], and multiple paths capturing details at different scales [51,52]. Multiple U-shaped models utilize pooling indices instead of or in conjunction with skip connections [26,42,47,48,53,54,55], with the aim of greater preservation of fine details and smaller structures. Works have also focused on reducing model sizes [10,44,50,56]. Some architectures have utilized dilated convolutions as additions or alternatives to U-Net [57,58,59,60,61,62].
A special class of architectural refinements to U-Net models are attention-gating mechanisms, which have been increasingly adopted to improve discrimination and localization capabilities. In brain segmentation, such mechanisms include Squeeze-and-Excitation blocks [63], which weigh feature channels according to their global importance, 3D spatial attention mechanisms [64], global attention modules [54,55], which integrate local features with their respective global dependencies, cross-attention for capturing dependencies between dual-modality features [65], and split-attention for weighing groups of channels [49]. Works have also utilized scale attention mechanisms [38,46]. While models incorporating attention modules demonstrate performance gains compared to baseline, other works report no improvement when incorporating attention [36,48,66,67]. These differences highlight scenario dependence and a need for more extensive validation.
Initially developed for sequence-to-sequence language processing, transformers have demonstrated success in vision tasks and have been widely adopted in segmentation [34]. Transformers utilize self-attention mechanisms, which allow for each input element to directly interact with and aggregate information from all other input elements based on learned pairwise relationships. This enables better capturing of long-range dependencies and global context than in conventional CNNs, which have limited receptive fields in shallow layers due to fixed-sized kernels, yielding longer chains between distant inputs. However, transformer-based models have higher computational requirements—quadratic with regards to input size—and require more training data, hindering straightforward application to brain segmentation. There, CNN-Transformer hybrid models have utilized self-attention in bottleneck blocks between the encoders and decoders of U-Net backbones, including full-volume models for tissue segmentation [68] and, partly due to large GPU memory requirements, patch-based models for subcortical [69] and whole brain [70] segmentation. Yu et al. [71] designed a hierarchical transformer encoder block and utilized it for whole brain segmentation combined with intracranial and posterior fossa segmentation. Transformers have also been combined with generative adversarial networks in order to more easily segment MS lesions alongside healthy tissues [72]. Interestingly, Rao et al. [68] obtained marginal improvements over a residual U-Net on datasets used in training, but noticeably better generalization to unseen sites.
Most recently, Mamba-based models have been adopted to medical image segmentation. Mamba utilizes a state space model with a selective scan mechanism and a hardware-aware algorithm which allow for the model to scale linearly with input size. Wei et al. [73] found that the U-Mamba architecture outperforms nnU-Net and the transformer-based SwinUNETR model in segmenting 122 GM structures. Cao et al. [74] used a hybrid CNN-Mamba U-Net with state space model blocks as the bottleneck for subcortical segmentation and reported improvements compared to state-of-the-art models, including the transformer-based TABSurfer [69].
Despite architectural improvements compared to the U-Net baseline, the influential nnU-Net works [75,76] have questioned the significance of the reported results in 2D and 3D medical image segmentation. In various settings, they reported minimal or non-existent improvements over optimized U-Net models and hence suggested an ongoing innovation bias. The works emphasized the need for stricter validation standards and presented the self-adapting nnU-Net framework designed for the rigorous validation of new methods.
We report the model architecture for each of the brain segmentation models in Table 1 and Table 2. We show the progression of architectures over time in Figure 2.
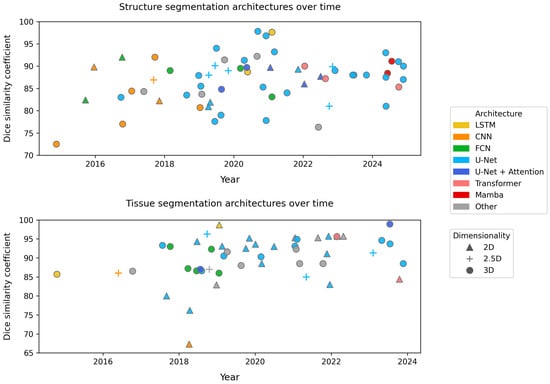
Figure 2.
Temporal evolution of structural (top) and tissue (bottom) brain MRI segmentation. Each point represents a publication, positioned by year and highest reported Dice similarity coefficient.
2.6. Patch-Based and Whole-Image Based Models
An important distinction in brain MRI segmentation is between patch-based models and those operating on whole volumes or slices. Patch-based models approaches subdivide the MRI data into smaller, manageable regions, which can simplify learning by emphasizing localized features and substantially reduce memory requirements. The reduction is particularly advantageous for models processing high-resolution (HiRes) scans, multi-modal inputs, or performing structural segmentation involving a large number of output labels. Patch-based models have demonstrated robustness in handling HiRes data and in mitigating the effects of noise and variability in smaller regions. Some works have shown that the overlapping of patches is crucial for good prediction accuracy at border voxels [36,77], as it allows for the aggregation of predictions across wider spatial context. Moreover, overlapping patches also increase the number of effective training samples for certain models, which can improve generalization. On the other hand, Yamanakkanavar, Lee et al. [78,79] report improved performance when using non-overlapping patches in a 2D setting. Despite some of the advantages, patch-based models nevertheless face limitations in capturing global context and integrating information across larger brain structures, with the use of overlapping also introducing redundant computation. Furthermore, artifacts on patch borders may be visible, even when overlapping is used [80].
With the growth of computational resources, patch sizes progressed from small local ones—e.g., 32 × 32 [81]—to those encompassing large portions of the brain, e.g., 128 × 128 × 128 or single-hemisphere volumes [82]. Several early works have utilized patches of different sizes [51,83,84,85] either across separate network pathways or by combining small local patches with more global downsampled ones. In structural segmentation, adaptive patch sizing based on the target region of interest has also been explored for enabling more efficient training and testing [86].
In contrast, many models segment the full MRI volume or slice in a single forward pass. By leveraging the full spatial context of the brain, these models can capture long-range dependencies and improve segmentation consistency across large or spatially distant structures. Such segmentation also avoids the computational redundancy associated with overlapping patches and generally results in faster inference times. However, full-volume models in particular require substantially more training memory, which can limit their feasibility for large models or HiRes inputs, especially in structural segmentation tasks. Even with the large memory capacities of modern GPUs, such models are typically limited to training on small batch sizes, which may compromise training stability; group normalization layers [36,87] can be used to mitigate this issue. Despite these challenges, many recent methods adopt full-volume inputs specifically to benefit from global image context. A direct structural 3D segmentation comparison by Roy et al. [36], however, demonstrated better performance when using large overlapping subvolumes, suggesting that full-volume inputs do not universally guarantee improved accuracy. The input configuration used in each brain segmentation model reviewed in this work is reported in Table 1 and Table 2.

Table 1.
Summary of brain MRI structure segmentation methods, including input modalities, number of anatomical labels, dimensionality, input type (patch, full slice/volume), backbone architecture, implementation availability (Avail.), and highest reported mean Dice similarity coefficient (DSC) across the labels.
Table 1.
Summary of brain MRI structure segmentation methods, including input modalities, number of anatomical labels, dimensionality, input type (patch, full slice/volume), backbone architecture, implementation availability (Avail.), and highest reported mean Dice similarity coefficient (DSC) across the labels.
| Method | Modalities | Labels | Dim. | Input | Arch. | Avail. | DSC (%) |
|---|---|---|---|---|---|---|---|
| Brébisson et al. [83] | T1 | 133 | 2.5D and 3D | Patch | CNN | No | 72.5 |
| Shakeri et al. [88] | T1 | 8 subcortical | 2D | Full | FCN | Yes | 82.4 |
| Moeskops et al. [84] | T1, T2 | 8 | 2D | Patch | CNN | No | 89.8 |
| Milletari et al. [89] | QSM | 26 subcortical | 2D, 2.5D, or 3D | Patch | CNN | No | 77 |
| Bao et al. [90] | T1 | 7 subcortical | 2D | Patch | CNN | No | 82.22 |
| Moeskops et al. [58] | T1 | 7 | 2D | Patch | FCN | No | 92 |
| Dolz et al. [91] | T1 | 8 subcortical | 3D | Patch | FCN | Yes | 89 |
| Kushibar et al. [81] | T1 | 14 subcortical | 2.5D | Patch | CNN | Yes | 86.9 |
| Mehta et al. [85] | T1 | 32-134 | 2D and 3D | Patch | CNN | No | 84.4 |
| Mehta et al. [92] | T1 | 7 subcortical | 2D and 3D | Patch | U-Net | No | 83 |
| Wachinger et al. [93] | T1 | 25 | 3D | Patch | CNN | Yes | 92 |
| Li et al. [57] | T1 | 155 | 3D | Patch | Dilated | Yes | 84.3 |
| Karani et al. [94] | T1, T2 | 7 subcortical | / | / | U-Net | No | 89.3 |
| Roy et al. [47] | T1 | 27 | 2.5D | Full | U-Net | Yes | 90.1 |
| Roy et al. [63] | T1 | 27 | / | / | U-Net + Attention | No | 86.2 |
| Li et al. [59] | T1, T1-IR, FLAIR | 10 | 2D | Full | U-Net | No | 80.9 |
| Kaku et al. [95] | T1 | 102 | 2D | Full | U-Net | Yes | 81.9 |
| Huo et al. [96] | T1 | 133 | 3D | Patch | U-Net | Yes | 77.6 |
| Jog et al. [97] | T1, T2 | 9, 12 | 3D | Patch | U-Net | Yes | 94 |
| Novosad et al. [98] | T1 | 8 and 12 subcortical | 3D | Patch | FCN | Yes | 89.5 |
| Novosad et al. [99] | T1 | 12 subcortical | 3D | Patch | CNN | Yes | 80.7 |
| Sun et al. [64] | T1, T1-IR, FLAIR | Tissue, 25 | 3D | Patch | U-Net + Attention | No | 84.8 |
| Dai et al. [100] | T1 | 15-138 | 3D | Patch | U-Net | No | 87.9 |
| Roy et al. [101] | T1 | 33 | 2.5D | Full | U-Net | Yes | 88 |
| Luna et al. [102] | T1, T1-IR, FLAIR | 8 | 3D | Patch | U-Net | No | 85.5 |
| McClure et al. [61] | T1 | 50 | 3D | Patch | Dilated | Yes | 83.7 |
| Dalca et al. [103] | T1, PD | 12 | 3D | Full | U-Net | Yes | 83.5 |
| Coupe et al. [104] | T1 | 133 | 3D | Patch | U-Net | Yes | 79 |
| Ramzan et al. [62] | T1, T1-IR, FLAIR | Tissues, 8 | 3D | Patch | Dilated | No | 91.4 |
| Henschel et al. [26] | T1 | 95 | 2.5D | Full | U-Net | Yes | 89 |
| Bontempi et al. [80] | T1 | 8 | 3D | Full | U-Net | Yes | 91.3 |
| Liu et al. [105] | T1 | 14 subcortical | 3D | Patch | U-Net + LSTM | No | 88.7 |
| Lee et al. [49] | T1 | 33, 100+ | 3D | Patch | U-Net + Attention | No | 89.7 |
| Zopes et al. [106] | T1, T2, DWI, CT | 27 | 3D | Patch | U-Net | No | 85.3 |
| Li et al. [16] | T1 | 8 subcortical | 3D | Patch | C-LSTM | No | 97.6 |
| Li et al. [107] | T1 | 8 subcortical | 3D | Patch | U-Net | No | 96.8 |
| Svanera et al. [108] | T1 | 8 | 3D | Full | U-Net | Yes | 97.8 |
| Li et al. [109] | T1 | 133 | 2D | Full | U-Net + Attention | Yes | 89.7 |
| Greve et al. [110] | T1 | 12 subcortical | 3D | Full | U-Net | Yes | 77.8 |
| Li et al. [40] | T1 | 54 | 3D | Full | FCN | Yes | 83.1 |
| Meyer et al. [41] | T1 | 8 | 3D | Patch | U-Net | Yes | 93.2 |
| Wu et al. [86] | T1 | 14, 54 | 3D | Patch | M-FCN | No | 92.2 |
| Nejad et al. [111] | T1 | 12 | 2D | Patch | U-Net | Yes | 89.3 |
| Liu et al. [112] | T1 | 5, 7 | 3D | Full | CLMorph | No | 76.3 |
| Ghazi et al. [113] | T1 | 133 | 2.5D | Full | U-Net | Yes | 81 |
| Henschel et al. [48] | T1 | 95 | 2.5D | Full | U-Net | Yes | 89.9 |
| Laiton-Bonadiez et al. [70] | T1 | 37 | 3D | Patch | Transformer | No | 90 |
| Wei et al. [114] | T1 | 136 | 2D | Full | U-Net + Attention | Yes | 86 |
| Yee et al. [82] | T1 | 102 | 3D | Patch | U-Net | No | 84 |
| Baniasadi et al. [115] | T1 | 30 subcortical | 3D | Patch | U-Net | Yes | 89 |
| Billot et al. [116] | T1, T2, PD, DBS | 110 | 3D | Full | U-Net | Yes | 88 |
| Billot et al. [117] | T1, T2, DBS, FLAIR, PD, CT | 33 | 3D | Full | U-Net | Yes | 88 |
| Cao et al. [69] | T1 | 31 subcortical | 3D | Patch | Transformer | No | 87.2 |
| Li et al. [46] | T1 | 28, 139 | 2D | Full | U-Net + Attention | Yes | 87.7 |
| Moon et al. [118] | T1 | 109 | 3D | / | U-Net | No | / |
| Cao et al. [74] | T1 | 31 subcortical | 3D | Patch | Mamba | Yes | 88.4 |
| Diaz et al. [119] | T1, T2, FLAIR | 7 | 3D | Patch | U-Net | Yes | 88 |
| Kujawa et al. [120] | T1 | 108 | 3D | Patch | U-Net | No | 87.5 |
| Lorzel et al. [25] | T1 | 58 | 3D | Patch | U-Net | No | 81 |
| Svanera et al. [52] | T1 | 7 | 3D | Full | LOD-brain | Yes | 93 |
| Goto et al. [121] | T1 | 107 | 3D | Full | U-Net | No | / |
| Le Bot et al. [122] | FLAIR | 133 | 3D | Patch | U-Net | No | 91 |
| Li et al. [65] | T1, PET | 45 | 3D | Full | Transformer | No | 85.3 |
| Li et al. [17] | T1 | 12 | 3D | Patch | U-Net | Yes | 90 |
| Puzio et al. [123] | T1, T2 | 38 | 3D | Patch | U-Net | No | 87 |
| Wei et al. [73] | T1 | 122 GM | 3D | Patch | Mamba | No | 91.1 |
2.7. Model Dimensionality
Brain segmentation models can be categorized into 2D, 2.5D, and 3D, depending on the dimensionality of their input and processing. Early deep segmentation models were developed for 2D images, and thus splitting MRI volumes into 2D slices along the axial, coronal, or sagittal plane allowed for easy application of the models onto brain MRI [84,88,90]. Compared to 3D approaches, this strategy reduces memory requirements and, for some models, increases the number of effective training samples per volume. However, 2D models lack access to inter-slice context and volumetric spatial dependencies, which are critical for accurate segmentation. Although most of the recent slice-based models operate on thin blocks of adjacent slices [46,114,124] in order to capture the important local 3D information, they remain limited in their ability to leverage broader volumetric context.
Three-dimensional segmentation models directly segment subvolumes or entire image volumes, and are equivalent to standard two-dimensional networks with an added spatial dimension. These approaches are able to utilize a wide, three-dimensional image context. However, a significant drawback of 3D models is the memory requirements due to large data volumes, which scale rapidly with input resolution, network depth, and the number of output labels. Large parameter counts and potentially smaller numbers of training samples per volume additionally lead to generalization problems. Computational requirements grow especially large in structural segmentation involving numerous target regions; for example, a 3D U-Net segmenting a full 256 × 256 × 256 volume into 79 structures may exceed 100 GB of memory with computation sharded across multiple GPUs [36]. Techniques have therefore been developed to reduce the requirements by grouping and merging labels before inference and recovering the labels at inference time [120]. This reduces the practical feasibility of training large 3D networks, especially if operating on full volume inputs, and restricts batch sizes. In our review, we found that most recent works utilize 3D models with full volumes or large patches as inputs.
In contrast, 2.5D models represent a hybrid solution that ensembles models segmenting orthogonal 2D slices or blocks of adjacent slices, thus combining some of the advantages of 2D and 3D segmentation methods. Typically, 2.5D models first generate slice segmentations for the axial, coronal, and sagittal planes separately. The final classification for each voxel is then derived by combining the predictions from the three slices encompassing the voxel. This approach provides a compromise between computational efficiency and use of spatial context by requiring significantly less computation compared to 3D approaches while still incorporating the orthogonal 3D context. Crucially, 2.5D models often deliver the same levels of segmentation accuracy as 3D models [26,36,47,48].
Early studies comparing 2D, 2.5D, and 3D models reported no consistent ranking in terms of performance metrics [77,89]. However, qualitative differences were noted—for example, Bernal et al. [77] observed that 3D models produced smoother predictions across slices than 2D ones, as the latter processed slices individually. However, this property also reduced the sensitivity of 2D models to variations in voxel spacing. More recent comparisons have consistently shown that 3D models outperform 2D models in terms of accuracy [125,126,127]. The relationship between 2.5D and 3D models remains less clear. A direct comparison in [36] reported that 2.5D models outperformed full-volume 3D models, but underperformed relative to 3D models trained on large overlapping patches. We note that the 2.5D models in the comparison by Avesta et al. [126] on three subcortical structures were not based on orthogonal slices, but instead used stacks of five adjacent slices from a single plane.
Models are not limited to one-dimensionality and can combine inputs of different spatial properties [83,85,92]. We note that manual segmentation by medical experts may follow different strategies, analyzing slices from a single views, using orthogonal slices, or utilizing 3D context, which are important, as manual segmentation is often used as a ground truth benchmark. In Table 1 and Table 2, we specify the dimensionality of each model.
Higher-dimensional models capture richer spatial information, but require greater computational resources and, in some cases, can still be more efficient by reducing redundant context aggregation. Efficiency is also shaped by factors such as model size, architectural complexity, and hyperparameter choices, making comparisons across studies difficult. A major limitation is the inconsistent reporting of computational complexity, which highlights the need for a more systematic evaluation in future work.

Table 2.
Summary of brain MRI tissue segmentation methods, including input modalities, dimensionality, input type (patch, full slice/volume), backbone architecture, implementation availability (Avail.), and highest reported mean Dice similarity coefficient (DSC) across the tissues.
Table 2.
Summary of brain MRI tissue segmentation methods, including input modalities, dimensionality, input type (patch, full slice/volume), backbone architecture, implementation availability (Avail.), and highest reported mean Dice similarity coefficient (DSC) across the tissues.
| Method | Modality | Dim. | Input | Arch. | Avail. | DSC (%) |
|---|---|---|---|---|---|---|
| Stollenga et al. [128] | T1, T1-IR, T2-FLAIR | 3D | Patch | PyraMiD-LSTM | No | 85.7 |
| Nguyen et al. [129] | T1 | 2.5D | Patch | CNN | No | 86 |
| Fedorov et al. [130] | T1 | 3D | Patch | Dilated | No | 86.5 |
| Chen et al. [43] | T1, T1-IR, T2-FLAIR | 3D | Patch | FCN | No | 86.6 |
| Khagi et al. [131] | T1 | 2D | Full | SegNet | No | 76.2 |
| Rajchl et al. [132] | T1 | 3D | Patch | FCN | Yes | 93 |
| Kumar et al. [53] | T1 | 2D | Patch | SegNet | No | 80 |
| Gottapu et al. [133] | T1 | 2D | Patch | CNN | No | 67.3 |
| Mahbod et al. [134] | T1, T1-IR, FLAIR | / | / | ANN | No | 85.3 |
| Chen et al. [135] | T1, T1-IR, T2-FLAIR | 2D | Patch | OctopusNet | No | 82.9 |
| Kong et al. [136] | T1 | 2D | Patch | CNN | No | / |
| Bernal et al. [77] | T1, and T1 and T2 | 2D, 3D | Patch | FCN, U-Net | Yes | 92.9 |
| Dolz et al. [45] | T1, T1-IR, T2-FLAIR | 3D | Patch | FCN | Yes | 87.2 |
| Gabr et al. [137] | T1, T2, T2-FLAIR, PD | 2D | Full | U-Net | Yes | 93 |
| Ito et al. [138] | T1 | 3D | Patch | FCN | No | 86 |
| Yogananda et al. [139] | T1 | 3D | Patch | U-Net | No | 86.6 |
| Mujica-Vargas et al. [140] | T1, T2, T2-FLAIR | 2D | Full | U-Net | No | 93.1 |
| Wang et al. [141] | T1 | 3D | Patch | U-Net | No | 90.4 |
| Xie et al. [142] | T1, T2, PD | 2D | Full | LSTM | No | 98.7 |
| Yan et al. [143] | T1 | 3D | Full | GCN | No | 91.6 |
| Li et al. [60] | T1, T1-IR, T2-FLAIR | 2.5D | Full | Dilated | Yes | 87 |
| Wei et al. [51] | T1 | 2.5D | Patch | U-Net | No | 96.3 |
| Sun et al. [64] | T1, T1-IR and T2-FLAIR | 3D | Patch | U-Net + Attention | No | 87 |
| Ramzan et al. [62] | T1, T1-IR, T2-FLAIR | 3D | Patch | Dilated | No | 88 |
| Lee et al. [79] | T1 | 2D | Patch | U-Net | Yes | 93.6 |
| Mostapha et al. [39] | T1 | 3D | Full | U-Net | No | 90.3 |
| Narayana et al. [144] | T1, T2, T2-FLAIR, PD | 2D | Full | U-Net | Yes | 92.5 |
| Yamanakkanavar et al. [78] | T1 | 2D | Patch | U-Net | No | 95.2 |
| Sendra-Balcells et al. [24] | T1 | 2D | Full | U-Net | No | 88.5 |
| Dayananda et al. [54] | T1 | 2D | Patch | Squeeze U-Net | No | 95.3 |
| Basnet et al. [10] | T1, T2 | 3D | Patch | U-Net | Yes | 93.1 |
| Long et al. [145] | T1, T1-IR, T2-FLAIR | 3D | Patch | MSCD-UNet | No | 88.5 |
| Woo et al. [125] | T1 | 3D | Full | U-Net | No | 94.9 |
| Yamanakkanavar et al. [50] | T1 | 2D | Patch | U-Net | No | 95.7 |
| Zhang et al. [146] | Diffusion, T1, T2 | 2.5D | Full | U-Net | No | 85 |
| Zhang et al. [147] | T1 | 3D | Full | GCN | No | 92.3 |
| Wei et al. [66] | T1 | 3D | Patch | Nes-Net | No | 88.5 |
| Niu et al. [67] | T1 | 2D | Full | U-Net | Yes | 91.1 |
| Goyal et al. [148] | T1 | 2D | Patch | SegNet | No | 83 |
| Prajapati et al. [124] | T1 | 2D | Full | U-Net | No | 95.7 |
| Rao et al. [68] | T1 | 3D | Full | Transformer | Yes | 95.6 |
| Yamanakkanavar et al. [50] | T1 | 2D | Patch | Squeeze U-Net | No | 96 |
| Yamanakkanavar et al. [149] | T1 | 2D | Patch | MF2-Net | No | 95.3 |
| Dayananda et al. [44] | T1 | 2D | Patch | Squeeze U-Net | No | 95.7 |
| Clerigues et al. [150] | T1, T2-FLAIR | 3D | Patch | U-Net | Yes | 94.6 |
| Guven et al. [151] | T1 | 2D | Full | GAN | No | / |
| Gi et al. [8] | T2 | 2.5D, 3D | Patch, full | U-Net | No | 91.3 |
| Oh et al. [152] | T1 | 3D | Full | U-Net | Yes | 88.5 |
| Simarro et al. [9] | T1 | 3D | Patch | U-Net | Yes | / |
| Hossain et al. [127] | T1 | 3D | Patch | U-Net | Yes | 93.7 |
| Liu et al. [38] | T1 | 3D | Patch | U-Net + Attention | Yes | 98.9 |
| Mohammadi et al. [72] | T1, T2 | 2D | Full | Transformer | No | 84.4 |
2.8. Generalization Strategies
A crucial consideration in deep learning is generalization: the ability to perform reliably on domains beyond those used in training. Brain MRI data are highly heterogeneous in imaging parameters (scanner vendor, modality, sequence type, field strength, contrast, resolution) and populations (age, condition, demographics). A segmentation model must generalize effectively in order to handle diverse datasets, multi-center studies, and longitudinal trials where brain structure changes due to age or disease; conventional methods have shown significant robustness in such settings. Furthermore, in clinical scenarios physicians may prefer sparse sets of 2D slices instead of the isotropic 3D volumes common in research studies. Clinical adoption, therefore, requires adaptation to variability in in-plane resolution, slice orientation, and slice spacing, and to larger partial voluming. As deep learning models are highly sensitive to training data characteristics, many domain adaptation strategies have been proposed for brain MRI, especially in structure segmentation. These include data augmentation, domain randomization, and the use of large and diverse training sets.
Data augmentation is widely used to improve generalization. Svanera et al. [52] systematically evaluate geometrical transformations, six noise distortions, and ghosting and MR field in homogeneity artifacts as augmentations. They found that flips and grid transformations, noise distortions, and artifact introductions improve robustness, while rotations and translations do not. Meyer et al. [41] propose a Gaussian Mixture Model-based data augmentation that modifies tissue intensities to vary contrast while preserving anatomy. The method improves generalization to unseen scanners, even with multi-scanner training data, with further work needed to assess performance without bias-field correction and to adapt it to MS lesions without available masks.
SynthSeg [117] trains entirely on synthetic data sampled from a generative model conditioned on segmentation maps. By fully randomizing contrast, resolution, orientation, and slice spacing, the model learns domain-independent features. SynthSeg achieves remarkable generalizability across diverse sequences and modalities (including FLAIR and CT), anisotropic low-resolution (up to 7.0 mm) MRI, and atrophy [116], without any retraining or fine-tuning, and outperforms supervised and domain-adaptation methods in experiments. Its ability to handle sparse 2D acquisitions makes it particularly relevant for clinical workflows. On FLAIR, the specialized FLAIRBrainSeg [122] supervised learning method was shown to outperform such modality-agnostic approaches.
FastSurferVINN [48] achieves resolution-independence by integrating resolution normalization directly into the network. Leveraging diverse training data, the model combines the abundance of 1.0 mm scans with the fine detail of HiRes data (0.7–0.9 mm). This not only enables generalization to resolutions within the training range, but also to those outside it (from 0.7 mm to 1.6 mm), while outperforming fixed-resolution models. The authors further demonstrated that scaling the training set from 120 to ∼1300 scans significantly improves performance across all resolutions, even under severe resolution imbalance. Similarly, Simarro et al. [9] showed that training on diverse age groups (2–81 years) yields a single model, with performance on par with age-specific models, facilitating consistent segmentation methodology in developmental studies.
A powerful generalization strategy is to leverage large and diverse MRI datasets. In the simpler task of brain extraction, Fletcher et al. [153] systematically showed that training data diversity is the most critical factor, followed by dataset size, with network complexity playing a lesser role. Their model achieved “production-level” performance on diverse data when trained on 8 cohorts and thousands of scans. For brain segmentation, Svanera et al. [52] similarly demonstrated the power of diverse training data, while observing a performance plateau across sites with 32–70 training datasets and a slight performance drop on individual sites. More volumes per dataset improved accuracy for both seen and unseen sites, with diminishing returns after ∼1000 scans (further extensively augmented). Their model showed excellent generalizability and outperformed QuickNat, FastSurfer, and SynthSeg on motion artifacts, and several established traditional methods on single-hemisphere subjects.
Several tissue segmentation studies report good performance on internal datasets using scarce training data. Narayana et al. [144] systematically showed good segmentation with just 10 training scans, although segmenting T2 MS lesions required hundreds. Importantly, domain-adaptation may benefit from fine-tuning: Roy et al. [47] fine-tuned on 28 manually segmented scans, Karani et al. [94] on 4 scans using domain-specific batch normalization, and Sun et al. [64] bridged labeling differences using a single image.
3. Results
We present the validation techniques found in brain MRI segmentation studies and discuss their limitations in Section 3.1. We discuss the results obtained by specific studies in Section 3.2.
3.1. Validation Strategies
Robust validation is essential for evaluating model accuracy, generalizability, and potential for clinical application. Nevertheless, validation remains a fundamental challenge in brain MRI segmentation, in large part due to the scarcity of manually annotated datasets.
The most common approach to validation in the works in our review is the direct comparison to ground truth using the DSC and Hausdorff distance [154]; the 95th percentile Hausdorff distance and the average Hausdorff distance [154] are frequently utilized to reduce sensitivity to outliers. Related metrics include the volumetric similarity coefficient [154], absolute volumetric difference [127], average surface distance [48], and average symmetric surface distance [69]. While the DSC was reported in all ground truth comparisons, we note that it is highly sensitive to small voxel-level differences, limiting its relevance for small subcortical structures, and does not account for the spatial distribution of errors; Taha et al. provide a discussion of the metrics [154].
The golden ground truth in direct comparison involves segmentations produced manually by medical experts. For tissue segmentation, the most commonly utilized publicly available datasets are MRBrainS13 [155] (20 healthy subjects), [155] (23 subjects with pathologies), and IBSR 18 [156] (18 healthy subjects), which also include segmentations into 8, 9, and 32 structures, respectively. In structure segmentation, high-quality manual datasets additionally include Mindboggle101 [37] (101 healthy subjects, 95 structures) and MICCAI 2012 (MALC) [157] (30 healthy subjects), 20 repeat (20 healthy subjects, two scans each), ADNI30 (15 healthy subjects and 14 AD patients), and OASIS30 (30 healthy subjects), which includes 132 structures [158].
Despite their importance, manual labels are in general susceptible to inter- and intra-rater variability issues and errors, and leave out many pathological populations. Notably, it is known that IBSR 18 tissue labels contain sulcal CSF voxels misclassified as GM [159], while Svanera et al. [52] completely excluded the manual structure annotations of MALC and IBSR due to quality concerns and replaced them with FreeSurfer outputs, retaining only Mindboggle-101 labels. Similarly, Kaku et al. [95] report artifacts of MALC labels in saggital and axial views, as the manual segmentation was performed in the coronal plane, and use Mindboggle101 instead. On the other hand, Cao et al. [69] report rough contours in Mindboggle-101. Some works utilize their own proprietary manual annotations, often to tailor validation to specific populations [9], resolutions [48], and field strengths [66,108]. However, such approaches limit reproducibility and comparability across studies. Therefore, a critical need remains for diverse, high-quality manual labels, particularly for diverse populations and acquisition parameters.
Numerous studies include ’silver’ ground truth for validation. These are segmentations obtained by using established tools such as FSL FAST [38,62,132,150,160], MRIAP [144,161], SPM [132,139], and FreeSurfer [24,28,125] for tissue segmentation and, most commonly, FreeSurfer for structure and ROI segmentation; tools such as ANTS [73,104,152,162], GIF [57,163], NLSS [96,164], icobrain [9,41,165], and SPM [121] are also used to produce structure annotations. Rajchl et al. [132] use outputs from multiple tools for training and validation. However, the ground truth obtained from established methods includes systematic errors and shows variability across software versions and even platforms, significantly limiting their reliability [117] and reducing the significance of direct comparisons [130]. Billot et al. [117], therefore, only considers the DSC w.r.t FreeSurfer segmentations valid only up to a specific threshold. Notably, many deep learning methods outperform these well-established tools in terms of accuracy against manual labels, test–retest reliability, sensitivity to clinical effects, and qualitative analysis [26,47,61,66,69,80], making silver-standard-based validation increasingly insufficient for meaningful comparison. Worryingly, numerous works completely rely on silver ground truth and do not include comparisons to manual data or perform additional tests.
To supplement direct comparison to ground truth labels, several works perform additional statistical tests. Test–retest reliability is estimated by computing the similarity of segmentations across two or more scans taken in a short time-frame. It is commonly utilized to assess stability in a scenario where anatomy is expected to remain stable. Certain studies quantify reliability directly on segmentations using metrics such as the DSC and Hausdorff distance [98,104], with Kondrateva et al. [166] stressing the importance of using multiple metrics to capture different aspects of segmentation quality. On the other hand, other works perform volume estimation instead and utilize the intra-class correlation coefficient, absolute volume difference, and percent relative difference metrics [26,110,121]. Puzio et al. [123] motivate the latter approach by the high sensitivity of the DSC to minor variations along structure boundaries, which may lack clinical relevance. Furthermore, Worth et al. [158] have shown the test–retest DSC of expert manual segmentations to be limited; Novosad et al. [98] achieve a highly similar coefficient (86.6%) to that reported in Worth et al. on several subcortical structures.
Several works perform sensitivity analyses to estimate the capability to detect known variations in brain morphology between diagnostic or treatment groups. For example, group differences between controls, individuals with mild cognitive impairment, and AD patients are estimated utilizing GM atrophy, volume loss in subcortical structures (namely the hippocampus, amygdala, and thalamus), ventricular enlargement, and thinning of cortical regions of interest. Sensitivity is evaluated by computing effect sizes [47] or fitting linear models and reporting p-values [26]. In MS, gray matter volumes have been used to estimate the sample sizes needed to detect treatment effects [167].
An important property of models is their generalizability to different imaging sites, populations, and acquisition protocols, especially in ensuring their practical utility in real-world clinical scenarios and large-scale multi-center studies. Many works therefore evaluate their models on diverse datasets, incorporating scans from multiple sites, scanners, and subject groups. Importantly, to assess robustness to domain shifts, models are trained on data with limited diversity and tested on that with characteristics not encountered during training, such as unseen scanner types [41,115], acquisition protocols [38,50], resolutions [48], demographics, or clinical conditions [26,47,107]. Robustness is also evaluated on simulated artifacts [52], unseen MRI modalities or CT [117], and single-hemisphere brains [52]. Furthermore, test–retest analysis on scans acquired by different scanners can be used to evaluate a model’s applicability across various scanning conditions [123].
Quantitative analyses are criticized by human experts, such as some physicians and neuroscientists, as they do not necessarily account for the severity of each segmentation error [154]. Kaku et al. [95] therefore measure segmentation agreement between their model and a group of medical experts, while Bontempi, Svanera et al. [80,108] include a “Turing test” in which they survey a group of experts to choose the best segmentation results among the competing methods.
3.2. Results
Validation practices in brain MRI segmentation exhibit substantial variability in terms of methods, datasets, and metrics, with comparison across works further complicated by the use of different training approaches and validation datasets.
The highest DSC reported by structure and tissue segmentation studies are summarized in Table 1 and Table 2, respectively. For each segmentation type, box plots of the DSC by input size and by dimensionality are shown in Figure 3, and the mean differences and p-values (Mann–Whitney U test) between methods are summarized in Table 3. The boxplots provide a descriptive overview of performance trends, with the lack of statistical significance (all p-values > 0.34) expected due to the limited number of studies.
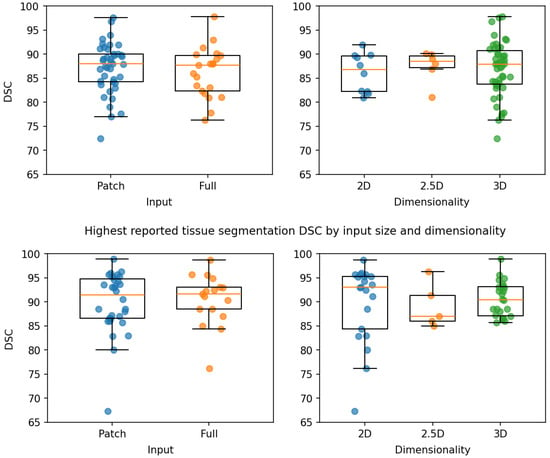
Figure 3.
Boxplots of the highest reported mean structure (top) and tissue (bottom) Dice similarity coefficient (DSC) by input size (left) and dimensionality (right).

Table 3.
Comparison of segmentation methods across input size and dimensionality for structure and tissue segmentation. Mean indicates the difference between the mean DSC of method groups 1 and 2.
In tissue segmentation, most studies utilized the manually annotated IBSR18, MRBrainS13, and MRBrainS18 datasets and the simulated BrainWeb dataset [168] for ground truth reference. Structural segmentation studies demonstrated a much greater diversity in both validation strategies and test datasets, with considerable variation in the number and definitions of anatomical labels. To provide a limited but meaningful comparison, we report the performance results for the methods evaluated on the Mindboggle101 dataset [37], the manually segmented dataset with the largest number of scans and with few reported quality concerns. The dataset consists of 101 scans with manually corrected labels based on the DKT cortical labeling protocol [37], including 33 subcortical and 62 cortical labels. T1 scans of healthy adults (age: 19–61) were collected specifically for the dataset or from a diverse set of publicly available studies, with field strengths ranging from 1.5 T to 7 T. In Table 4, we show the DSC reported by all nine structure segmentation publications that validate on Mindboggle101. For each work, we report the models evaluated on the dataset, whether the dataset was utilized in training, the number of output structure labels, and the DSC, which is the most commonly used metric among these works.

Table 4.
Summary of studies employing Mindboggle101 for validation, including evaluated methods, use of Mindboggle101 in training, number of anatomical labels, and reported Dice similarity coefficient (DSC).
The models in Table 4 segment the brain into different anatomical labels. The highest reported DSC, 96.5%, was achieved by FastSurfer, as reported by Svanera et al. [52], who grouped the finer-grained DKT labels into seven broader classes. The authors of FastSurfer [26] obtained DSCs of ∼80% for both cortical and subcortical segmentation, with 95 labels in total, while the FastSurferVINN model obtains higher performance [48]. Cao et al. report a noticeably lower score of 75.8% for FastSurferVINN on the same subcortical labels, but with the largest structure (cortical white matter) excluded; FreeSurfer, the most frequently utilized traditional method, achieves a DSC of 74%. Some studies in Table 4 incorporated Mindboggle101 scans into training, generally reporting higher DSC, with Kaku et al. [95] demonstrating a large improvement of approximately 7% following fine-tuning.
The results in Table 4 are strongly influenced by the choice of output classes and training strategies. We note that, apart from the DSC, the set of reported evaluation metrics varies considerably across the studies. Together, these factors highlight the difficulty of comparisons across works, even when the models are validated on the same dataset.
4. Discussion
Deep learning methods have emerged as powerful tools for brain MRI segmentation, offering advantages in computational efficiency, robustness, and accuracy compared to conventional approaches. Nevertheless, deep learning models remain underutilized in neurological studies and clinical trials, which still feature traditional approaches such as atlas-based methods. This is in part due to a much larger volume of studies validating conventional methods, and is also due to greater robustness across diverse image conditions and populations and the easier interpretability of such methods. With the growing availability of publicly available MRI datasets and the use of conventional methods to obtain silver-truth training data, deep learning models have started to show very high consistency across sites [52].
In our review, we observed a steady increase in publications on deep learning-based brain MRI segmentation, reflecting a growing interest in the field. Early models were constrained by limited GPU memory, often relying on 2D architectures that processed individual slices or small blocks of slices. While computationally feasible at the time, such methods are unable to capture full 3D anatomical context. In structure segmentation, we found that models have primarily shifted toward 3D processing. Furthermore, 2.5D architectures are also utilized and offer a compromise between computational feasibility and context utilization, with studies reporting state-of-the-art results from such models in whole brain segmentation [48]. Our survey also revealed a clear progression from small patch-based inputs to very large patches or entire volumes. Some studies reported improved accuracy when using very large patches instead of full volumes [36], suggesting that the strategy may confer benefits beyond computational feasibility.
Architectural developments in brain MRI segmentation have closely paralleled progress in the broader medical image segmentation. Early approaches were based on sliding-window classification and fully convolutional networks. The U-Net architecture has since become the dominant backbone and remains central to most modern brain MRI segmentation methods. Numerous additions and alternatives to the architecture have been proposed, with the most recent being the transformer and state space models. However, the benefit of many modifications over optimized U-Net models remains uncertain, with performance gains often dependent on specific datasets and evaluation conditions. As in the broader medical segmentation landscape [76], there is a need for greater validation and comparisons of different models.
Several works have focused on improving the generalization capabilities of brain MRI segmentation models. Strategies such as data augmentation, domain randomization, training on large and diverse datasets, and fine-tuning on limited data have produced robust performance across sites, protocols, and populations. A consistent set of findings emerge across studies: diverse training cohorts are essential for broad generalization, do not reduce performance on individual datasets, and may even enhance it; increasing the number of training scans improves performance, but with diminishing returns; and dataset size imbalances do not necessarily disadvantage smaller cohorts. Moreover, targeted fine-tuning may be effective with a small number of scans. Together, these insights highlight the power of dataset diversity and scale, complemented when needed by small-scale fine-tuning.
Validation strategies varied widely across studies, but showed important limitations. Manually labeled datasets remain small, biased toward healthy populations, and often contain segmentation errors, underscoring the need for high-quality annotations across diverse cohorts and increasingly common MRI modalities such as submillimeter imaging. Furthermore, many structure segmentation works rely on imperfect silver-standard references rather than manual labels. Statistical measures such as test–retest reliability, group differences, and longitudinal sensitivity are sometimes used to supplement evaluation, while qualitative assessment by independent experts remains rare. Standardized frameworks like nnU-Net [75,76], which provide rigorous validation, have been used in some studies but remain underutilized. Their adoption is strongly recommended, as they improve reproducibility and comparisons across works, helping mitigate evaluation bias.
The diversity of validation techniques complicates cross-study comparisons. As shown in Table 4, comparison across structure segmentation studies remains challenging even when the same test sets are utilized, owing to differences in training strategies, output labels, and evaluation metrics. There is therefore a clear need to unify validation approaches for method comparison.
Despite substantial progress in accuracy and efficiency, the practical adoption of deep learning-based brain MRI segmentation remains limited. Many methods are not trained or evaluated for robustness across diverse acquisition parameters and populations, a key concern for widespread applicability. Only a few works explicitly address the increasingly used submillimeter acquisitions [48] or the anisotropic low-resolution scans [116] widely used by clinicians. Models are typically tied to a fixed set of labels, offering less flexibility than established methods such as SPM and CAT, which can register to multiple atlases. Independent large-scale validation studies remain scarce, and many methods lack thorough documentation or even public availability. Like conventional tools that have been refined for decades, deep learning models would benefit from continuous development to keep pace with evolving user needs and MRI advances. Together, these issues highlight the gap between promising research results and routine, reliable deployment. Tools like FastSurfer [48], SynthSeg+ [116], and LOD-Brain [52] have taken strong steps in this direction, providing extensive documentation and validation across diverse datasets. FastSurfer has been continuously updated, introducing resolution-independence and subfield segmentation. SynthSeg+ offers high performance on both GPUs and CPUs, automatic quality control, and support for clinical scans, and has been incorporated into the FreeSurfer toolbox.
Future work in brain MRI segmentation should leverage advancements in MRI. The increasing availability of ultra-high-field 7T MRI, offering superior tissue contrast for subcortical delineation, and submillimeter imaging, which can capture fine cortical folding patterns, remains underexploited, despite its potential to enhance segmentation accuracy. In addition, current efforts toward improving generalization should be extended into robust pipelines capable of handling scans acquired under diverse conditions, spanning different scanners, populations, age groups, and imaging modalities. Longitudinal brain segmentation, which utilizes multiple scans of the same patient, is used by conventional methods to reduce sensitivity to noise and better monitor changes due to age, disease, and treatment. This strategy, however, remains very underexplored in deep learning literature. Critically, efforts to strengthen validation are essential. Larger manually annotated datasets, derived from diverse cohorts and imaging settings, are needed alongside the unification of validation protocols. Expanded benchmarking and comparative analyses will be crucial to rigorously assess model performance, disentangle the influence of architecture from training configurations, and ultimately support the reliable deployment of deep learning-based segmentation in clinical trials, neuroimaging studies, and clinical practice. Broader adoption in practice will also benefit from more comprehensive documentation, compatibility with existing neuroimaging pipelines and downstream tasks, and the public availability of implementations.
5. Conclusions
Deep learning has markedly advanced brain MRI segmentation, enabling fast, accurate, and scalable analysis. Models have evolved from slice- and patch-based CNNs to 3D and 2.5D U-Nets and emerging transformer-based architectures. Recent generalization techniques have enabled remarkable robustness across diverse data. Despite state-of-the-art accuracy, deep learning methods remain underutilized in large studies and clinical practice. Key challenges include the limited application of generalization strategies, lack of independent validation, difficult cross-study comparability, and a scarcity of high-quality manual segmentations. Moreover, most methods lack thorough documentation, regular updates, or integration into broader neuroimaging tools. Future work should focus on expanding diverse, high-quality datasets, unifying validation protocols, leveraging MRI advancements, and developing robust models that generalize across populations and scanners. Addressing these issues will facilitate broader adoption in research and clinical practice, supporting more reliable neuroimaging studies and improved patient outcomes.
Supplementary Materials
The PRISMA 2020 Checklist can be downloaded at: https://www.mdpi.com/article/10.3390/a18100636/s1.
Author Contributions
Conceptualization, N.Š. and P.R.; methodology, N.Š. and P.R.; investigation, N.Š.; writing—original draft preparation, N.Š.; writing—review and editing, N.Š. and P.R.; supervision, P.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Slovenian Research and Innovation Agency, Research Program P2-0250.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yepes-Calderon, F.; McComb, J.G. Eliminating the need for manual segmentation to determine size and volume from MRI. A proof of concept on segmenting the lateral ventricles. PLoS ONE 2023, 18, e0285414. [Google Scholar] [CrossRef]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A Review of Deep Learning in Medical Imaging: Imaging Traits, Technology Trends, Case Studies with Progress Highlights, and Future Promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef] [PubMed]
- Magadza, T.; Viriri, S. Deep Learning for Brain Tumor Segmentation: A Survey of State-of-the-Art. J. Imaging 2021, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Gu, L.; Liu, Z.; Zhao, S. Review of Deep Learning Approaches for the Segmentation of Multiple Sclerosis Lesions on Brain MRI. Front. Neuroinform. 2020, 14, 610967. [Google Scholar] [CrossRef] [PubMed]
- Ciceri, T.; Squarcina, L.; Giubergia, A.; Bertoldo, A.; Brambilla, P.; Peruzzo, D. Review on deep learning fetal brain segmentation from Magnetic Resonance images. Artif. Intell. Med. 2023, 143, 102608. [Google Scholar] [CrossRef]
- Wu, L.; Wang, S.; Liu, J.; Hou, L.; Li, N.; Su, F.; Yang, X.; Lu, W.; Qiu, J.; Zhang, M.; et al. A survey of MRI-based brain tissue segmentation using deep learning. Complex Intell. Syst. 2025, 11. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Gi, Y.; Oh, G.; Jo, Y.; Lim, H.; Ko, Y.; Hong, J.; Lee, E.; Park, S.; Kwak, T.; Kim, S.; et al. Study of multistep Dense U-Net-based automatic segmentation for head MRI scans. Med. Phys. 2024, 51, 2230–2238. [Google Scholar] [CrossRef]
- Simarro, J.; Meyer, M.I.; Van Eyndhoven, S.; Phan, T.V.; Billiet, T.; Sima, D.M.; Ortibus, E. A deep learning model for brain segmentation across pediatric and adult populations. Sci. Rep. 2024, 14, 11735. [Google Scholar] [CrossRef]
- Basnet, R.; Ahmad, M.O.; Swamy, M. A deep dense residual network with reduced parameters for volumetric brain tissue segmentation from MR images. Biomed. Signal Process. Control 2021, 70, 103063. [Google Scholar] [CrossRef]
- Bethlehem, R.A.I.; Seidlitz, J.; White, S.R.; Vogel, J.W.; Anderson, K.M.; Adamson, C.; Adler, S.; Alexopoulos, G.S.; Anagnostou, E.; Areces-Gonzalez, A.; et al. Brain charts for the human lifespan. Nature 2022, 604, 525–533. [Google Scholar] [CrossRef]
- Singh, V.; Chertkow, H.; Lerch, J.P.; Evans, A.C.; Dorr, A.E.; Kabani, N.J. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer’s disease. Brain 2006, 129, 2885–2893. [Google Scholar] [CrossRef]
- Alves, F.; Kalinowski, P.; Ayton, S. Accelerated Brain Volume Loss Caused by Anti-β-Amyloid Drugs: A Systematic Review and Meta-analysis. Neurology 2023, 100, e2114–e2124. [Google Scholar] [CrossRef]
- Chu, R.; Kim, G.; Tauhid, S.; Khalid, F.; Healy, B.C.; Bakshi, R. Whole brain and deep gray matter atrophy detection over 5 years with 3T MRI in multiple sclerosis using a variety of automated segmentation pipelines. PLoS ONE 2018, 13, e0206939. [Google Scholar] [CrossRef] [PubMed]
- Uhr, V.; Diaz, I.; Rummel, C.; McKinley, R. Exploring Robustness of Cortical Morphometry in the presence of white matter lesions, using Diffusion Models for Lesion Filling. arXiv 2025. [Google Scholar] [CrossRef]
- Li, H.; Zhang, H.; Johnson, H.; Long, J.D.; Paulsen, J.S.; Oguz, I. Longitudinal subcortical segmentation with deep learning. In Proceedings of the Medical Imaging 2021: Image Processing, Online, 15–19 February 2021; p. 43. [Google Scholar] [CrossRef]
- Li, M.; Magnússon, M.; Kristjánsdóttir, I.; Lund, S.H.; Van Eimeren, T.; Ellingsen, L.M. Region-based U-nets for fast, accurate, and scalable deep brain segmentation: Application to Parkinson Plus Syndromes. NeuroImage Clin. 2025, 47, 103807. [Google Scholar] [CrossRef] [PubMed]
- Sacchet, M.D.; Livermore, E.E.; Iglesias, J.E.; Glover, G.H.; Gotlib, I.H. Subcortical volumes differentiate Major Depressive Disorder, Bipolar Disorder, and remitted Major Depressive Disorder. J. Psychiatr. Res. 2015, 68, 91–98. [Google Scholar] [CrossRef]
- Hokama, H.; Shenton, M.E.; Nestor, P.G.; Kikinis, R.; Levitt, J.J.; Metcalf, D.; Wible, C.G.; O’Donnella, B.F.; Jolesz, F.A.; McCarley, R.W. Caudate, putamen, and globus pallidus volume in schizophrenia: A quantitative MRI study. Psychiatry Res. Neuroimaging 1995, 61, 209–229. [Google Scholar] [CrossRef]
- Moazzami, K.; Shao, I.Y.; Chen, L.Y.; Lutsey, P.L.; Jack, C.R.; Mosley, T.; Joyner, D.A.; Gottesman, R.; Alonso, A. Atrial Fibrillation, Brain Volumes, and Subclinical Cerebrovascular Disease (from the Atherosclerosis Risk in Communities Neurocognitive Study [ARIC-NCS]). Am. J. Cardiol. 2020, 125, 222–228. [Google Scholar] [CrossRef]
- Yu, S.Y.; Chen, X.Y.; Chen, Z.Y.; Dong, Z.; Liu, M.Q. Regional volume changes of the brain in migraine chronification. Neural Regen. Res. 2020, 15, 1701. [Google Scholar] [CrossRef]
- Walker, K.A.; Gottesman, R.F.; Wu, A.; Knopman, D.S.; Mosley, T.H.; Alonso, A.; Kucharska-Newton, A.; Brown, C.H. Association of Hospitalization, Critical Illness, and Infection with Brain Structure in Older Adults. J. Am. Geriatr. Soc. 2018, 66, 1919–1926. [Google Scholar] [CrossRef]
- Auer, D.P.; Wilke, M.; Grabner, A.; Heidenreich, J.O.; Bronisch, T.; Wetter, T.C. Reduced NAA in the thalamus and altered membrane and glial metabolism in schizophrenic patients detected by 1H-MRS and tissue segmentation. Schizophr. Res. 2001, 52, 87–99. [Google Scholar] [CrossRef]
- Sendra-Balcells, C.; Salvador, R.; Pedro, J.B.; Biagi, M.C.; Aubinet, C.; Manor, B.; Thibaut, A.; Laureys, S.; Lekadir, K.; Ruffini, G. Convolutional neural network MRI segmentation for fast and robust optimization of transcranial electrical current stimulation of the human brain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lorzel, H.M.; Allen, M.D. Development of the next-generation functional neuro-cognitive imaging protocol—Part 1: A 3D sliding-window convolutional neural net for automated brain parcellation. NeuroImage 2024, 286, 120505. [Google Scholar] [CrossRef] [PubMed]
- Henschel, L.; Conjeti, S.; Estrada, S.; Diers, K.; Fischl, B.; Reuter, M. FastSurfer—A fast and accurate deep learning based neuroimaging pipeline. NeuroImage 2020, 219, 117012. [Google Scholar] [CrossRef] [PubMed]
- Despotović, I.; Goossens, B.; Philips, W. MRI Segmentation of the Human Brain: Challenges, Methods, and Applications. Comput. Math. Methods Med. 2015, 2015, 450341. [Google Scholar] [CrossRef] [PubMed]
- Fischl, B. FreeSurfer. NeuroImage 2012, 62, 774–781. [Google Scholar] [CrossRef]
- Friston, K.J. (Ed.) Statistical Parametric Mapping: The Analysis of Funtional Brain Images, 1st ed.; Elsevier: Amsterdam, The Netherlands; Academic Press: Boston, MA, USA, 2007. [Google Scholar]
- Gaser, C.; Dahnke, R.; Thompson, P.M.; Kurth, F.; Luders, E.; Alzheimer’s Disease Neuroimaging Initiative. CAT: A computational Anatomy Toolbox for the Analysis of Structural MRI Data. GigaScience 2024, 13, giae049. [Google Scholar] [CrossRef]
- Puonti, O.; Iglesias, J.E.; Van Leemput, K. Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. NeuroImage 2016, 143, 235–249. [Google Scholar] [CrossRef]
- Ciresan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Deep Neural Networks Segment Neuronal Membranes in Electron Microscopy Images. In Proceedings of the Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar] [CrossRef]
- Li, X.; Ding, H.; Yuan, H.; Zhang, W.; Pang, J.; Cheng, G.; Chen, K.; Liu, Z.; Loy, C.C. Transformer-Based Visual Segmentation: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10138–10163. [Google Scholar] [CrossRef]
- Ma, J.; Li, F.; Wang, B. U-Mamba: Enhancing Long-range Dependency for Biomedical Image Segmentation. arXiv 2024. [Google Scholar] [CrossRef]
- Roy, S.; Kügler, D.; Reuter, M. Are 2.5D approaches superior to 3D deep networks in whole brain segmentation? In Proceedings of the International Conference on Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022. [Google Scholar]
- Klein, A.; Tourville, J. 101 Labeled Brain Images and a Consistent Human Cortical Labeling Protocol. Front. Neurosci. 2012, 6, 171. [Google Scholar] [CrossRef]
- Liu, Y.; Song, C.; Ning, X.; Gao, Y.; Wang, D. nnSegNeXt: A 3D Convolutional Network for Brain Tissue Segmentation Based on Quality Evaluation. Bioengineering 2024, 11, 575. [Google Scholar] [CrossRef]
- Mostapha, M.; Mailhe, B.; Chen, X.; Ceccaldi, P.; Yoo, Y.; Nadar, M. Braided Networks for Scan-Aware MRI Brain Tissue Segmentation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 136–139. [Google Scholar] [CrossRef]
- Li, Y.; Cui, J.; Sheng, Y.; Liang, X.; Wang, J.; Chang, E.I.C.; Xu, Y. Whole brain segmentation with full volume neural network. Comput. Med. Imaging Graph. 2021, 93, 101991. [Google Scholar] [CrossRef]
- Meyer, M.I.; De La Rosa, E.; Pedrosa De Barros, N.; Paolella, R.; Van Leemput, K.; Sima, D.M. A Contrast Augmentation Approach to Improve Multi-Scanner Generalization in MRI. Front. Neurosci. 2021, 15, 708196. [Google Scholar] [CrossRef] [PubMed]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Dou, Q.; Yu, L.; Qin, J.; Heng, P.A. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 2018, 170, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Dayananda, C.; Choi, J.Y.; Lee, B. A Squeeze U-SegNet Architecture Based on Residual Convolution for Brain MRI Segmentation. IEEE Access 2022, 10, 52804–52817. [Google Scholar] [CrossRef]
- Dolz, J.; Gopinath, K.; Yuan, J.; Lombaert, H.; Desrosiers, C.; Ben Ayed, I. HyperDense-Net: A Hyper-Densely Connected CNN for Multi-Modal Image Segmentation. IEEE Trans. Med. Imaging 2019, 38, 1116–1126. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, C.; Zhang, Y.; Wang, X.; Ma, X.; Zhang, H.; Wu, S. CAN: Context-assisted full Attention Network for brain tissue segmentation. Med. Image Anal. 2023, 85, 102710. [Google Scholar] [CrossRef]
- Guha Roy, A.; Conjeti, S.; Navab, N.; Wachinger, C. QuickNAT: A fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage 2019, 186, 713–727. [Google Scholar] [CrossRef]
- Estrada, S.; Kügler, D.; Bahrami, E.; Xu, P.; Mousa, D.; Breteler, M.M.; Aziz, N.A.; Reuter, M. FastSurfer-HypVINN: Automated sub-segmentation of the hypothalamus and adjacent structures on high-resolutional brain MRI. Imaging Neurosci. 2023, 1, 1–32. [Google Scholar] [CrossRef]
- Lee, M.; Kim, J.; Ey Kim, R.; Kim, H.G.; Oh, S.W.; Lee, M.K.; Wang, S.M.; Kim, N.Y.; Kang, D.W.; Rieu, Z.; et al. Split-Attention U-Net: A Fully Convolutional Network for Robust Multi-Label Segmentation from Brain MRI. Brain Sci. 2020, 10, 974. [Google Scholar] [CrossRef] [PubMed]
- Yamanakkanavar, N.; Choi, J.Y.; Lee, B. SM-SegNet: A Lightweight Squeeze M-SegNet for Tissue Segmentation in Brain MRI Scans. Sensors 2022, 22, 5148. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Xia, Y.; Zhang, Y. M3Net: A multi-model, multi-size, and multi-view deep neural network for brain magnetic resonance image segmentation. Pattern Recognit. 2019, 91, 366–378. [Google Scholar] [CrossRef]
- Svanera, M.; Savardi, M.; Signoroni, A.; Benini, S.; Muckli, L. Fighting the scanner effect in brain MRI segmentation with a progressive level-of-detail network trained on multi-site data. Med. Image Anal. 2024, 93, 103090. [Google Scholar] [CrossRef]
- Kumar, P.; Nagar, P.; Arora, C.; Gupta, A. U-Segnet: Fully Convolutional Neural Network Based Automated Brain Tissue Segmentation Tool. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 3503–3507. [Google Scholar] [CrossRef]
- Dayananda, C.; Choi, J.Y.; Lee, B. Multi-Scale Squeeze U-SegNet with Multi Global Attention for Brain MRI Segmentation. Sensors 2021, 21, 3363. [Google Scholar] [CrossRef]
- Yamanakkanavar, N.; Lee, B. A novel M-SegNet with global attention CNN architecture for automatic segmentation of brain MRI. Comput. Biol. Med. 2021, 136, 104761. [Google Scholar] [CrossRef]
- Paschali, M.; Gasperini, S.; Roy, A.G.; Fang, M.Y.S.; Navab, N. 3DQ: Compact Quantized Neural Networks for Volumetric Whole Brain Segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11766, pp. 438–446. [Google Scholar] [CrossRef]
- Li, W.; Wang, G.; Fidon, L.; Ourselin, S.; Cardoso, M.J.; Vercauteren, T. On the Compactness, Efficiency, and Representation of 3D Convolutional Networks: Brain Parcellation as a Pretext Task. In Information Processing in Medical Imaging; Niethammer, M., Styner, M., Aylward, S., Zhu, H., Oguz, I., Yap, P.T., Shen, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 10265, pp. 348–360. [Google Scholar] [CrossRef]
- Moeskops, P.; Veta, M.; Lafarge, M.W.; Eppenhof, K.A.J.; Pluim, J.P.W. Adversarial Training and Dilated Convolutions for Brain MRI Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Cardoso, M.J., Arbel, T., Carneiro, G., Syeda-Mahmood, T., Tavares, J.M.R., Moradi, M., Bradley, A., Greenspan, H., Papa, J.P., Madabhushi, A., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 10553, pp. 56–64. [Google Scholar] [CrossRef]
- Li, H.; Zhygallo, A.; Menze, B. Automatic Brain Structures Segmentation Using Deep Residual Dilated U-Net. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., Van Walsum, T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11383, pp. 385–393. [Google Scholar] [CrossRef]
- Li, J.; Yu, Z.L.; Gu, Z.; Liu, H.; Li, Y. MMAN: Multi-modality aggregation network for brain segmentation from MR images. Neurocomputing 2019, 358, 10–19. [Google Scholar] [CrossRef]
- McClure, P.; Rho, N.; Lee, J.A.; Kaczmarzyk, J.R.; Zheng, C.Y.; Ghosh, S.S.; Nielson, D.M.; Thomas, A.G.; Bandettini, P.; Pereira, F. Knowing What You Know in Brain Segmentation Using Bayesian Deep Neural Networks. Front. Neuroinform. 2019, 13, 67. [Google Scholar] [CrossRef]
- Ramzan, F.; Khan, M.U.G.; Iqbal, S.; Saba, T.; Rehman, A. Volumetric Segmentation of Brain Regions From MRI Scans Using 3D Convolutional Neural Networks. IEEE Access 2020, 8, 103697–103709. [Google Scholar] [CrossRef]
- Roy, A.G.; Navab, N.; Wachinger, C. Concurrent Spatial and Channel ‘Squeeze & Excitation’ in Fully Convolutional Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11070, pp. 421–429. [Google Scholar] [CrossRef]
- Sun, L.; Ma, W.; Ding, X.; Huang, Y.; Liang, D.; Paisley, J. A 3D Spatially Weighted Network for Segmentation of Brain Tissue From MRI. IEEE Trans. Med. Imaging 2020, 39, 898–909. [Google Scholar] [CrossRef]
- Li, W.; Huang, Z.; Zhang, Q.; Zhang, N.; Zhao, W.; Wu, Y.; Yuan, J.; Yang, Y.; Zhang, Y.; Yang, Y.; et al. Accurate Whole-Brain Segmentation for Bimodal PET/MR Images via a Cross-Attention Mechanism. IEEE Trans. Radiat. Plasma Med. Sci. 2025, 9, 47–56. [Google Scholar] [CrossRef]
- Wei, J.; Wu, Z.; Wang, L.; Bui, T.D.; Qu, L.; Yap, P.T.; Xia, Y.; Li, G.; Shen, D. A cascaded nested network for 3T brain MR image segmentation guided by 7T labeling. Pattern Recognit. 2022, 124, 108420. [Google Scholar] [CrossRef] [PubMed]
- Niu, K.; Guo, Z.; Peng, X.; Pei, S. P-ResUnet: Segmentation of brain tissue with Purified Residual Unet. Comput. Biol. Med. 2022, 151, 106294. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.M.; Wan, Z.; Arabshahi, S.; Ma, D.J.; Lee, P.Y.; Tian, Y.; Zhang, X.; Laine, A.F.; Guo, J. Improving across-dataset brain tissue segmentation for MRI imaging using transformer. Front. Neuroimaging 2022, 1, 1023481. [Google Scholar] [CrossRef]
- Cao, A.; Rao, V.M.; Liu, K.; Liu, X.; Laine, A.F.; Guo, J. TABSurfer: A Hybrid Deep Learning Architecture for Subcortical Segmentation. arXiv 2023. [Google Scholar] [CrossRef]
- Laiton-Bonadiez, C.; Sanchez-Torres, G.; Branch-Bedoya, J. Deep 3D Neural Network for Brain Structures Segmentation Using Self-Attention Modules in MRI Images. Sensors 2022, 22, 2559. [Google Scholar] [CrossRef]
- Yu, X.; Tang, Y.; Yang, Q.; Lee, H.H.; Bao, S.; Huo, Y.; Landman, B.A. Enhancing hierarchical transformers for whole brain segmentation with intracranial measurements integration. In Proceedings of the Medical Imaging 2024: Clinical and Biomedical Imaging, San Diego, CA, USA, 18–22 February 2024; p. 18. [Google Scholar] [CrossRef]
- Mohammadi, Z.; Aghaei, A.; Moghaddam, M.E. CycleFormer: Brain tissue segmentation in the presence of Multiple Sclerosis lesions and Intensity Non-Uniformity artifact. Biomed. Signal Process. Control 2024, 93, 106153. [Google Scholar] [CrossRef]
- Wei, Y.; Jagtap, J.M.; Singh, Y.; Khosravi, B.; Cai, J.; Gunter, J.L.; Erickson, B.J. Comprehensive Segmentation of Gray Matter Structures on T1-Weighted Brain MRI: A Comparative Study of Convolutional Neural Network, Convolutional Neural Network Hybrid-Transformer or -Mamba Architectures. Am. J. Neuroradiol. 2025, 46, 742–749. [Google Scholar] [CrossRef]
- Cao, A.; Li, Z.; Jomsky, J.; Laine, A.F.; Guo, J. MedSegMamba: 3D CNN-Mamba Hybrid Architecture for Brain Segmentation. arXiv 2024. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Isensee, F.; Wald, T.; Ulrich, C.; Baumgartner, M.; Roy, S.; Maier-Hein, K.; Jaeger, P.F. nnU-Net Revisited: A Call for Rigorous Validation in 3D Medical Image Segmentation. arXiv 2024. [Google Scholar] [CrossRef]
- Bernal, J.; Kushibar, K.; Cabezas, M.; Valverde, S.; Oliver, A.; Llado, X. Quantitative Analysis of Patch-Based Fully Convolutional Neural Networks for Tissue Segmentation on Brain Magnetic Resonance Imaging. IEEE Access 2019, 7, 89986–90002. [Google Scholar] [CrossRef]
- Yamanakkanavar, N.; Lee, B. Using a Patch-Wise M-Net Convolutional Neural Network for Tissue Segmentation in Brain MRI Images. IEEE Access 2020, 8, 120946–120958. [Google Scholar] [CrossRef]
- Lee, B.; Yamanakkanavar, N.; Choi, J.Y. Automatic segmentation of brain MRI using a novel patch-wise U-net deep architecture. PLoS ONE 2020, 15, e0236493. [Google Scholar] [CrossRef]
- Bontempi, D.; Benini, S.; Signoroni, A.; Svanera, M.; Muckli, L. CEREBRUM: A fast and fully-volumetric Convolutional Encoder-decodeR for weakly-supervised sEgmentation of BRain strUctures from out-of-the-scanner MRI. Med. Image Anal. 2020, 62, 101688. [Google Scholar] [CrossRef]
- Kushibar, K.; Valverde, S.; González-Villà, S.; Bernal, J.; Cabezas, M.; Oliver, A.; Lladó, X. Automated sub-cortical brain structure segmentation combining spatial and deep convolutional features. Med. Image Anal. 2018, 48, 177–186. [Google Scholar] [CrossRef]
- Yee, E.; Ma, D.; Popuri, K.; Chen, S.; Lee, H.; Chow, V.; Ma, C.; Wang, L.; Beg, M.F. 3D hemisphere-based convolutional neural network for whole-brain MRI segmentation. Comput. Med. Imaging Graph. 2022, 95, 102000. [Google Scholar] [CrossRef]
- De Brebisson, A.; Montana, G. Deep neural networks for anatomical brain segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; pp. 20–28. [Google Scholar] [CrossRef]
- Moeskops, P.; Viergever, M.A.; Mendrik, A.M.; De Vries, L.S.; Benders, M.J.N.L.; Isgum, I. Automatic Segmentation of MR Brain Images With a Convolutional Neural Network. IEEE Trans. Med. Imaging 2016, 35, 1252–1261. [Google Scholar] [CrossRef]
- Mehta, R.; Majumdar, A.; Sivaswamy, J. BrainSegNet: A convolutional neural network architecture for automated segmentation of human brain structures. J. Med. Imaging 2017, 4, 024003. [Google Scholar] [CrossRef]
- Wu, J.; Tang, X. Brain segmentation based on multi-atlas and diffeomorphism guided 3D fully convolutional network ensembles. Pattern Recognit. 2021, 115, 107904. [Google Scholar] [CrossRef]
- Wu, Y.; He, K. Group Normalization. Int. J. Comput. Vis. 2020, 128, 742–755. [Google Scholar] [CrossRef]
- Shakeri, M.; Tsogkas, S.; Ferrante, E.; Lippe, S.; Kadoury, S.; Paragios, N.; Kokkinos, I. Sub-cortical brain structure segmentation using F-CNN’S. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 269–272. [Google Scholar] [CrossRef]
- Milletari, F.; Ahmadi, S.A.; Kroll, C.; Plate, A.; Rozanski, V.; Maiostre, J.; Levin, J.; Dietrich, O.; Ertl-Wagner, B.; Bötzel, K.; et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image Underst. 2017, 164, 92–102. [Google Scholar] [CrossRef]
- Bao, S.; Chung, A.C.S. Multi-scale structured CNN with label consistency for brain MR image segmentation. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2018, 6, 113–117. [Google Scholar] [CrossRef]
- Dolz, J.; Desrosiers, C.; Ben Ayed, I. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. NeuroImage 2018, 170, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.; Sivaswamy, J. M-net: A Convolutional Neural Network for deep brain structure segmentation. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 437–440. [Google Scholar] [CrossRef]
- Wachinger, C.; Reuter, M.; Klein, T. DeepNAT: Deep convolutional neural network for segmenting neuroanatomy. NeuroImage 2018, 170, 434–445. [Google Scholar] [CrossRef]
- Karani, N.; Chaitanya, K.; Baumgartner, C.; Konukoglu, E. A Lifelong Learning Approach to Brain MR Segmentation Across Scanners and Protocols. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 11070, pp. 476–484. [Google Scholar] [CrossRef]
- Kaku, A.; Hegde, C.V.; Huang, J.; Chung, S.; Wang, X.; Young, M.; Radmanesh, A.; Lui, Y.W.; Razavian, N. DARTS: DenseUnet-based Automatic Rapid Tool for brain Segmentation. arXiv 2019, arXiv:1911.05567. [Google Scholar] [CrossRef]
- Huo, Y.; Xu, Z.; Xiong, Y.; Aboud, K.; Parvathaneni, P.; Bao, S.; Bermudez, C.; Resnick, S.M.; Cutting, L.E.; Landman, B.A. 3D whole brain segmentation using spatially localized atlas network tiles. NeuroImage 2019, 194, 105–119. [Google Scholar] [CrossRef]
- Jog, A.; Hoopes, A.; Greve, D.N.; Van Leemput, K.; Fischl, B. PSACNN: Pulse sequence adaptive fast whole brain segmentation. NeuroImage 2019, 199, 553–569. [Google Scholar] [CrossRef]
- Novosad, P.; Fonov, V.; Collins, D.L.; Alzheimer’s Disease Neuroimaging Initiative. Accurate and robust segmentation of neuroanatomy in T1-weighted MRI by combining spatial priors with deep convolutional neural networks. Hum. Brain Mapp. 2020, 41, 309–327. [Google Scholar] [CrossRef]
- Novosad, P.; Fonov, V.; Collins, D.L. Unsupervised domain adaptation for the automated segmentation of neuroanatomy in MRI: A deep learning approach. bioRxiv 2019. [Google Scholar] [CrossRef]
- Dai, C.; Mo, Y.; Angelini, E.; Guo, Y.; Bai, W. Transfer Learning from Partial Annotations for Whole Brain Segmentation. In Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data; Wang, Q., Milletari, F., Nguyen, H.V., Albarqouni, S., Cardoso, M.J., Rieke, N., Xu, Z., Kamnitsas, K., Patel, V., Roysam, B., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11795, pp. 199–206. [Google Scholar] [CrossRef]
- Roy, A.G.; Conjeti, S.; Navab, N.; Wachinger, C. Bayesian QuickNAT: Model uncertainty in deep whole-brain segmentation for structure-wise quality control. NeuroImage 2019, 195, 11–22. [Google Scholar] [CrossRef]
- Luna, M.; Park, S.H. 3D Patchwise U-Net with Transition Layers for MR Brain Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., Van Walsum, T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11383, pp. 394–403. [Google Scholar] [CrossRef]
- Dalca, A.V.; Yu, E.; Golland, P.; Fischl, B.; Sabuncu, M.R.; Eugenio Iglesias, J. Unsupervised Deep Learning for Bayesian Brain MRI Segmentation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11766, pp. 356–365. [Google Scholar] [CrossRef]
- Coupé, P.; Mansencal, B.; Clément, M.; Giraud, R.; Denis De Senneville, B.; Ta, V.T.; Lepetit, V.; Manjon, J.V. AssemblyNet: A large ensemble of CNNs for 3D whole brain MRI segmentation. NeuroImage 2020, 219, 117026. [Google Scholar] [CrossRef]
- Liu, L.; Hu, X.; Zhu, L.; Fu, C.W.; Qin, J.; Heng, P.A. ψ-Net: Stacking Densely Convolutional LSTMs for Sub-Cortical Brain Structure Segmentation. IEEE Trans. Med. Imaging 2020, 39, 2806–2817. [Google Scholar] [CrossRef] [PubMed]
- Zopes, J.; Platscher, M.; Paganucci, S.; Federau, C. Multi-Modal Segmentation of 3D Brain Scans Using Neural Networks. Front. Neurol. 2021, 12, 653375. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, H.; Johnson, H.; Long, J.D.; Paulsen, J.S.; Oguz, I. MRI subcortical segmentation in neurodegeneration with cascaded 3D CNNs. In Proceedings of the Medical Imaging 2021: Image Processing, Online, 15–19 February 2021; p. 25. [Google Scholar] [CrossRef]
- Svanera, M.; Benini, S.; Bontempi, D.; Muckli, L. CEREBRUM-7T: Fast and Fully Volumetric Brain Segmentation of 7 Tesla MR Volumes. Hum. Brain Mapp. 2021, 42, 5563–5580. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, H.; Fan, Y. ACEnet: Anatomical context-encoding network for neuroanatomy segmentation. Med. Image Anal. 2021, 70, 101991. [Google Scholar] [CrossRef]
- Greve, D.N.; Billot, B.; Cordero, D.; Hoopes, A.; Hoffmann, M.; Dalca, A.V.; Fischl, B.; Iglesias, J.E.; Augustinack, J.C. A deep learning toolbox for automatic segmentation of subcortical limbic structures from MRI images. NeuroImage 2021, 244, 118610. [Google Scholar] [CrossRef]
- Nejad, A.; Masoudnia, S.; Nazem-Zadeh, M.R. A Fast and Memory-Efficient Brain MRI Segmentation Framework for Clinical Applications. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 2140–2143. [Google Scholar] [CrossRef]
- Liu, L.; Aviles-Rivero, A.I.; Schönlieb, C.B. Contrastive Registration for Unsupervised Medical Image Segmentation. arXiv 2022, arXiv:2011.08894. [Google Scholar] [CrossRef]
- Ghazi, M.M.; Nielsen, M. FAST-AID Brain: Fast and Accurate Segmentation Tool Using Artificial Intelligence Developed for Brain. In Image Analysis; Petersen, J., Dahl, V.A., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2025; Volume 15725, pp. 161–176. [Google Scholar] [CrossRef]
- Wei, C.; Yang, Y.; Guo, X.; Ye, C.; Lv, H.; Xiang, Y.; Ma, T. MRF-Net: A multi-branch residual fusion network for fast and accurate whole-brain MRI segmentation. Front. Neurosci. 2022, 16, 940381. [Google Scholar] [CrossRef]
- Baniasadi, M.; Petersen, M.V.; Gonçalves, J.; Horn, A.; Vlasov, V.; Hertel, F.; Husch, A. DBSegment: Fast and robust segmentation of deep brain structures considering domain generalization. Hum. Brain Mapp. 2023, 44, 762–778. [Google Scholar] [CrossRef] [PubMed]
- Billot, B.; Magdamo, C.; Cheng, Y.; Arnold, S.E.; Das, S.; Iglesias, J.E. Robust machine learning segmentation for large-scale analysis of heterogeneous clinical brain MRI datasets. Proc. Natl. Acad. Sci. USA 2023, 120, e2216399120. [Google Scholar] [CrossRef] [PubMed]
- Billot, B.; Greve, D.N.; Puonti, O.; Thielscher, A.; Van Leemput, K.; Fischl, B.; Dalca, A.V.; Iglesias, J.E. SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. Med. Image Anal. 2023, 86, 102789. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.M.; Lee, Y.Y.; Hyeong, K.E.; Yoon, W.; Baek, B.H.; Heo, S.H.; Shin, S.S.; Kim, S.K. Development and validation of deep learning-based automatic brain segmentation for East Asians: A comparison with Freesurfer. Front. Neurosci. 2023, 17, 1157738. [Google Scholar] [CrossRef]
- Diaz, I.; Geiger, M.; McKinley, R.I. Leveraging SO(3)-steerable convolutions for pose-robust semantic segmentation in 3D medical data. Mach. Learn. Biomed. Imaging 2024, 2, 834–855. [Google Scholar] [CrossRef]
- Kujawa, A.; Dorent, R.; Ourselin, S.; Vercauteren, T. Label Merge-and-Split: A Graph-Colouring Approach for Memory-Efficient Brain Parcellation. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2024; Volume 15009, pp. 350–360. [Google Scholar] [CrossRef]
- Goto, M.; Kamagata, K.; Andica, C.; Takabayashi, K.; Uchida, W.; Goto, T.; Yuzawa, T.; Kitamura, Y.; Hatano, T.; Hattori, N.; et al. Deep Learning-based Hierarchical Brain Segmentation with Preliminary Analysis of the Repeatability and Reproducibility. Magn. Reson. Med. Sci. 2025, 24. [Google Scholar] [CrossRef]
- Bot, E.L.; Giraud, R.; Mansencal, B.; Tourdias, T.; Manjon, J.V.; Coupé, P. FLAIRBrainSeg: Fine-grained brain segmentation using FLAIR MRI only. arXiv 2025. [Google Scholar] [CrossRef]
- Puzio, T.; Matera, K.; Karwowski, J.; Piwnik, J.; Białkowski, S.; Podyma, M.; Dunikowski, K.; Siger, M.; Stasiołek, M.; Grzelak, P.; et al. Deep learning-based automatic segmentation of brain structures on MRI: A test-retest reproducibility analysis. Comput. Struct. Biotechnol. J. 2025, 28, 128–140. [Google Scholar] [CrossRef]
- Prajapati, R.; Kwon, G.R. SIP-UNet: Sequential Inputs Parallel UNet Architecture for Segmentation of Brain Tissues from Magnetic Resonance Images. Mathematics 2022, 10, 2755. [Google Scholar] [CrossRef]
- Woo, B.; Lee, M. Comparison of tissue segmentation performance between 2D U-Net and 3D U-Net on brain MR Images. In Proceedings of the 2021 International Conference on Electronics, Information, and Communication (ICEIC), Jeju, Republic of Korea, 31 January–3 February 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Avesta, A.; Hossain, S.; Lin, M.; Aboian, M.; Krumholz, H.M.; Aneja, S. Comparing 3D, 2.5D, and 2D Approaches to Brain Image Auto-Segmentation. Bioengineering 2023, 10, 181. [Google Scholar] [CrossRef]
- Hossain, M.I.; Amin, M.Z.; Anyimadu, D.T.; Suleiman, T.A. Comparative Study of Probabilistic Atlas and Deep Learning Approaches for Automatic Brain Tissue Segmentation from MRI Using N4 Bias Field Correction and Anisotropic Diffusion Pre-processing Techniques. arXiv 2024. [Google Scholar] [CrossRef]
- Stollenga, M.F.; Byeon, W.; Liwicki, M.; Schmidhuber, J. Parallel Multi-Dimensional LSTM, With Application to Fast Biomedical Volumetric Image Segmentation. In Proceedings of the Neural Information Processing Systems, Istanbul, Turkey, 9–12 November 2015. [Google Scholar]
- Nguyen, D.M.H.; Vu, H.T.; Ung, H.Q.; Nguyen, B.T. 3D-Brain Segmentation Using Deep Neural Network and Gaussian Mixture Model. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; pp. 815–824. [Google Scholar] [CrossRef]
- Fedorov, A.; Johnson, J.; Damaraju, E.; Ozerin, A.; Calhoun, V.; Plis, S. End-to-end learning of brain tissue segmentation from imperfect labeling. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 3785–3792. [Google Scholar] [CrossRef]
- Khagi, B.; Kwon, G.R. Pixel-Label-Based Segmentation of Cross-Sectional Brain MRI Using Simplified SegNet Architecture-Based CNN. J. Healthc. Eng. 2018, 2018, 3640705. [Google Scholar] [CrossRef]
- Rajchl, M.; Pawlowski, N.; Rueckert, D.; Matthews, P.M.; Glocker, B. NeuroNet: Fast and Robust Reproduction of Multiple Brain Image Segmentation Pipelines. arXiv 2018, arXiv:1806.04224. [Google Scholar] [CrossRef]
- Gottapu, R.D.; Dagli, C.H. DenseNet for Anatomical Brain Segmentation. Procedia Comput. Sci. 2018, 140, 179–185. [Google Scholar] [CrossRef]
- Mahbod, A.; Chowdhury, M.; Smedby, O.; Wang, C. Automatic brain segmentation using artificial neural networks with shape context. Pattern Recognit. Lett. 2018, 101, 74–79. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Wei, D.; Li, Y.; Zheng, Y. OctopusNet: A Deep Learning Segmentation Network for Multi-modal Medical Images. In Multiscale Multimodal Medical Imaging; Li, Q., Leahy, R., Dong, B., Li, X., Eds.; Springer International Publishing: Cham, Switzerland, 2020; Volume 11977, pp. 17–25. [Google Scholar] [CrossRef]
- Kong, Z.; Li, T.; Luo, J.; Xu, S. Automatic Tissue Image Segmentation Based on Image Processing and Deep Learning. J. Healthc. Eng. 2019, 2019, 2912458. [Google Scholar] [CrossRef]
- Gabr, R.E.; Coronado, I.; Robinson, M.; Sujit, S.J.; Datta, S.; Sun, X.; Allen, W.J.; Lublin, F.D.; Wolinsky, J.S.; Narayana, P.A. Brain and lesion segmentation in multiple sclerosis using fully convolutional neural networks: A large-scale study. Mult. Scler. J. 2020, 26, 1217–1226. [Google Scholar] [CrossRef]
- Ito, R.; Nakae, K.; Hata, J.; Okano, H.; Ishii, S. Semi-supervised deep learning of brain tissue segmentation. Neural Netw. 2019, 116, 25–34. [Google Scholar] [CrossRef]
- Yogananda, C.G.B.; Wagner, B.C.; Murugesan, G.K.; Madhuranthakam, A.; Maldjian, J.A. A Deep Learning Pipeline for Automatic Skull Stripping and Brain Segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 727–731. [Google Scholar] [CrossRef]
- Mújica-Vargas, D.; Martínez, A.; Matuz-Cruz, M.; Luna-Alvarez, A.; Morales-Xicohtencatl, M. Non-parametric Brain Tissues Segmentation via a Parallel Architecture of CNNs. In Pattern Recognition; Carrasco-Ochoa, J.A., Martínez-Trinidad, J.F., Olvera-López, J.A., Salas, J., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 11524, pp. 216–226. [Google Scholar] [CrossRef]
- Wang, L.; Xie, C.; Zeng, N. RP-Net: A 3D Convolutional Neural Network for Brain Segmentation From Magnetic Resonance Imaging. IEEE Access 2019, 7, 39670–39679. [Google Scholar] [CrossRef]
- Xie, K.; Wen, Y. LSTM-MA: A LSTM Method with Multi-Modality and Adjacency Constraint for Brain Image Segmentation. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 240–244. [Google Scholar] [CrossRef]
- Yan, Z.; Youyong, K.; Jiasong, W.; Coatrieux, G.; Huazhong, S. Brain Tissue Segmentation based on Graph Convolutional Networks. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 1470–1474. [Google Scholar] [CrossRef]
- Narayana, P.A.; Coronado, I.; Sujit, S.J.; Wolinsky, J.S.; Lublin, F.D.; Gabr, R.E. Deep-Learning-Based Neural Tissue Segmentation of MRI in Multiple Sclerosis: Effect of Training Set Size. J. Magn. Reson. Imaging 2020, 51, 1487–1496. [Google Scholar] [CrossRef]
- Long, J.S.; Ma, G.Z.; Song, E.M.; Jin, R.C. Learning U-Net Based Multi-Scale Features in Encoding-Decoding for MR Image Brain Tissue Segmentation. Sensors 2021, 21, 3232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Breger, A.; Kevin Cho, K.I.; Ning, L.; Westin, C.F.; O’Donnell, L.J.; Pasternak, O. Deep Learning Based Segmentation of Brain Tissue from Diffusion MRI. NeuroImage 2021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Kong, Y.; Wu, J.; Yang, J.; Shu, H.; Coatrieux, G. GSCFN: A graph self-construction and fusion network for semi-supervised brain tissue segmentation in MRI. Neurocomputing 2021, 455, 23–37. [Google Scholar] [CrossRef]
- Goyal, P. Shallow SegNet with bilinear interpolation and weighted cross-entropy loss for Semantic segmentation of brain tissue. In Proceedings of the 2022 IEEE International Conference on Signal Processing, Informatics, Communication and Energy Systems (SPICES), Thiruvananthapuram, India, 10–12 March 2022; pp. 361–365. [Google Scholar] [CrossRef]
- Yamanakkanavar, N.; Lee, B. MF2-Net: A multipath feature fusion network for medical image segmentation. Eng. Appl. Artif. Intell. 2022, 114, 105004. [Google Scholar] [CrossRef]
- Clèrigues, A.; Valverde, S.; Salvi, J.; Oliver, A.; Lladó, X. Minimizing the effect of white matter lesions on deep learning based tissue segmentation for brain volumetry. Comput. Med. Imaging Graph. 2023, 103, 102157. [Google Scholar] [CrossRef]
- Altun Güven, S.; Talu, M.F. Brain MRI high resolution image creation and segmentation with the new GAN method. Biomed. Signal Process. Control 2023, 80, 104246. [Google Scholar] [CrossRef]
- Oh, K.; Lee, J.; Heo, D.W.; Shen, D.; Suk, H.I. Transferring Ultrahigh-Field Representations for Intensity-Guided Brain Segmentation of Low-Field Magnetic Resonance Imaging. arXiv 2024. [Google Scholar] [CrossRef]
- Fletcher, E.; DeCarli, C.; Fan, A.P.; Knaack, A. Convolutional Neural Net Learning Can Achieve Production-Level Brain Segmentation in Structural Magnetic Resonance Imaging. Front. Neurosci. 2021, 15, 683426. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Mendrik, A.M.; Vincken, K.L.; Kuijf, H.J.; Breeuwer, M.; Bouvy, W.H.; De Bresser, J.; Alansary, A.; De Bruijne, M.; Carass, A.; El-Baz, A.; et al. MRBrainS Challenge: Online Evaluation Framework for Brain Image Segmentation in 3T MRI Scans. Comput. Intell. Neurosci. 2015, 2015, 813696. [Google Scholar] [CrossRef]
- Rohlfing, T. Image Similarity and Tissue Overlaps as Surrogates for Image Registration Accuracy: Widely Used but Unreliable. IEEE Trans. Med. Imaging 2012, 31, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Landman, B.A.; Warfield, S.K. (Eds.) MICCAI 2012 Workshop on Multi-Atlas Labeling; CreateSpace Independent Publishing Platform: Scotts Valley, CA, USA, 2012. [Google Scholar]
- Worth, A.; Tourville, J. Acceptable values of similarity coefficients in neuroanatomical labeling in MRI. In Proceedings of the Society for Neuroscience Annual Meeting, Chicago, IL, USA, 17–21 October 2015. Program No. 829.21. [Google Scholar]
- Valverde, S.; Oliver, A.; Cabezas, M.; Roura, E.; Lladó, X. Comparison of 10 brain tissue segmentation methods using revisited IBSR annotations. J. Magn. Reson. Imaging 2015, 41, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. FSL. NeuroImage 2012, 62, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Narayana, P.A. A comprehensive approach to the segmentation of multichannel three-dimensional MR brain images in multiple sclerosis. NeuroImage Clin. 2013, 2, 184–196. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 2011, 54, 2033–2044. [Google Scholar] [CrossRef]
- Cardoso, M.J.; Modat, M.; Wolz, R.; Melbourne, A.; Cash, D.; Rueckert, D.; Ourselin, S. Geodesic Information Flows: Spatially-Variant Graphs and Their Application to Segmentation and Fusion. IEEE Trans. Med. Imaging 2015, 34, 1976–1988. [Google Scholar] [CrossRef]
- Asman, A.J.; Landman, B.A. Hierarchical performance estimation in the statistical label fusion framework. Med. Image Anal. 2014, 18, 1070–1081. [Google Scholar] [CrossRef]
- Struyfs, H.; Sima, D.M.; Wittens, M.; Ribbens, A.; Pedrosa De Barros, N.; Phan, T.V.; Ferraz Meyer, M.I.; Claes, L.; Niemantsverdriet, E.; Engelborghs, S.; et al. Automated MRI volumetry as a diagnostic tool for Alzheimer’s disease: Validation of icobrain dm. NeuroImage Clin. 2020, 26, 102243. [Google Scholar] [CrossRef]
- Kondrateva, E.; Barg, S.; Vasiliev, M. Benchmarking the Reproducibility of Brain MRI Segmentation Across Scanners and Time. arXiv 2025, arXiv:2504.15931. [Google Scholar] [CrossRef]
- Battaglini, M.; Jenkinson, M.; De Stefano, N.; Alzheimer’s Disease Neuroimaging Initiative. SIENA-XL for improving the assessment of gray and white matter volume changes on brain MRI. Hum. Brain Mapp. 2018, 39, 1063–1077. [Google Scholar] [CrossRef]
- Cocosco, C.A.; Kollokian, V.; Kwan, R.K.S.; Evans, A.C. BrainWeb: Online Interface to a 3D MRI Simulated Brain Database. NeuroImage 1997, 5, 425. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).