Abstract
We revisit the following version of the Gapped String Indexing problem, where the goal is to preprocess a text to enable efficient reporting of all occurrences of a gapped pattern in T. An occurrence of P in T is defined as a pair where substrings and match and , respectively, with a gap lying within the interval . This problem has significant applications in computational biology and text mining. A hardness result on this problem suggests that any index with polylogarithmic query time must occupy near quadratic space. In a recent study [STACS 2024], Bille et al. presented a sub-quadratic space index using space , where is a parameter fixed at the time of index construction. Its query time is , which is sub-linear per occurrence when . We show how to achieve a gap-sensitive query time of using the same space, where denotes the number of occurrences with gap g. This is faster when there are many occurrences with small gaps.
1. Introduction
Let be a string (called the text) over a polynomially sized alphabet and be a gapped pattern, where and are strings and is an integer interval called the gap range. An occurrence of P in T is represented as a pair such that , with gap . Locating occurrences of gapped patterns has numerous applications in computational biology [1,2,3,4,5,6,7] and text mining [8,9,10,11]. The algorithmic variant of the gapped pattern matching problem is well studied [12,13] and can be solved in time ( suppresses polylogarithmic factors, in particular, for any constant k.) [3,6,14,15,16].
This work focuses on an indexing version of the problem where the text T is known during preprocessing. The gapped pattern is provided as a query. Formally, we consider Problem 1.
Problem 1
(Gapped String Indexing).
- Preprocess: A text .
- Query: Given a gapped pattern , report all occurrences of P in T.
In recent work, Bille et al. [17] showed that for all , an index for Problem 1 can be constructed occupying or space, and answering queries time. Our main result is an index that requires similar space but achieves query times parameterized by the gaps present in the occurrences. This is stated formally in Theorem 1 below. In particular, our result improves the case where gaps for most occurrences are small.
Theorem 1.
For all , there exists an index for Gapped String Indexing that occupies space and answers queries in time
where is the number of occurrences with gap g.
Our main technique revolves around solving the following bounded variant of Gapped String Indexing, which may be of independent interest.
Problem 2
(Bounded Gapped String Indexing).
- Preprocess: A text and integer G.
- Query: Given a gapped pattern where , report all occurrences of P in T.
For Problem 2, we provide a solution with space and query complexity stated in Theorem 2.
Theorem 2.
For every , there exists an index for Bounded Gapped String Indexing that occupies space and answers queries in time
where is the size of the output.
To prove Theorem 2, we build on previous results for the Gapped Set Intersection Problem (defined formally in Section 2), developing techniques for the bounded-gap case. We utilize a blocking technique based on binary trees, which when combined with a generalized form of the Kraft–McMillan inequality, allows us to achieve an improved query time (Lemma 9). Our preprocessing techniques differ from those of Bille et al. [17] in that we rely on the bounded nature of the gaps to perform the proposed blocking technique. Indeed, Theorem 2 is accomplished through extra preprocessing (blocking and building data structures for blocks) that is possible only when an upper bound is known in advance. As described in Section 3.6, Theorem 1 is then achieved by applying Theorem 2 for different ranges of G.
1.1. Previous Work
A long line of research has contributed to the current results on Gapped String Indexing. Many of these place some restrictions on the problem. The earliest results are by Peterlongo et al. [18], and are for the heavily restricted version where lengths , , and gap are known for preprocessing. Given these restrictions, their solution uses space and achieves an optimal query time of .
In a slightly generalized variant where only the gap length is given at preprocessing, Iliopoulos and Rahman [19] present an index using space , where is an arbitrarily small constant, with query time . For the same case where g is known in advance, Bille and Gørtz [17] introduced an improved approach, achieving optimal query time with space. In the case where only an upper bound G is given on , the problem can be reduced to 3D range searching with an index using space and query time [20]. Conditioned on the Strong Set-Disjointness Conjecture [21], Bille et al. [22] also demonstrated that any solution with query time must use space. Recently, Ganguly et al. [23] proposed a variant (called Bounded Ratio Gapped String Indexing) where the gap satisfies . Here, represents a gap ratio fixed at index construction time. Under this relaxed constraint, an index can be constructed occupying space and having query time.
The framework employed by Bille et al. [17] utilizes the results for 3-SUM Indexing by Golovnev [24] (see also [25]). In 3-SUM indexing, one needs to preprocess two sets of integers, and , so that given a query integer c, one can efficiently determine if there exists and such that . The reduction from Gapped String Indexing to 3-SUM Indexing, through a series of intermediate problems, forms the basis of both [17] and this work. Leveraging one of these intermediate problems (between Gapped Indexing and 3-SUM Indexing) alleviates some steps for this work relative to Bille et al.’s results [17].
1.2. Notation and Technical Preliminaries
We use to refer to the set and to the set . For an array of integers A, we use to denote the subarray and to denote .
For a string T, we use to refer to the symbol in T, to denote the substring , and the substring . We call a substring of the form a suffix of T and a substring of the form a prefix of T. We use to denote the reverse of the string T. The suffix tree [26] of a string is a compact tree of all suffixes with leaves in corresponding lexicographic order. The suffix array, denoted , is defined such that is the suffix when all suffixes are sorted in lexicographic order. For a pattern P, its suffix range is the maximal range such that for all , has prefix P. The suffix range exists if P occurs in T; otherwise, the suffix range is empty.
For convenience, we assume that all strings are over a polynomially-sized integer alphabet so that the suffix tree and suffix array can be constructed in linear time [27]. Given the suffix tree and suffix array, the suffix range of a string can be found in time.
2. A Preliminary Solution
Before introducing our main solution, we first present a preliminary approach. While this solution does not achieve the query efficiency required to prove Theorem 1, it serves as a valuable foundation for our solution. As an initial step, we introduce the following two problem formulations from [17].
Problem 3
(Gapped Set Intersection (with Reporting)).
- Preprocess: A collection of subsets , ⋯, of total size over integer universe .
- Query: Given , report if there exists (report all, resp.) where there exists such that .
We also define the bounded version of Problem 3, analogous to Problem 2. Specifically, the bounded problem formulation provides a collection of subsets , ⋯, , and an integer G for preprocessing. A query consists of the tuple with the additional guarantee that . We will utilize the following results from Bille et al. [17].
Lemma 1
(Index for Gapped Set Intersection (Theorems 5 and 6 from [17])). For every , there is a data structure for the Gapped Set Intersection that occupies space and answers existential queries in time and reporting queries in time , where is the size of the output.
Lemma 2 allows us to focus on Problem 3 for the majority of the remaining work.
Lemma 2
(Reduction to Bounded Gapped Set Intersection (Adapted from [17]). Assume there is a data structure for the Bounded Gapped Set Intersection (with Reporting) using space that answers existential queries in time and reporting queries in time . Then, there exists a data structure for Bounded Gapped String Indexing using space. It can answer existential queries in time and reporting queries in time , where n is the length of the input text and is the size of the output.
For completeness, we sketch an adaptation of the proof from [17].
Proof.
We begin with the array such that , where is the suffix array of T. We also define the array , where , and is the suffix array of the reverse string . Both arrays are decomposed into subsets corresponding to dyadic intervals of the form , where and . These subsets serve as the input to the Gapped Set Intersection instance. Notably, the sum of the subset cardinalities is .
Given a query , we decompose the suffix range for into a collection of dyadic-sized subarrays of , corresponding to precomputed subsets. We denote this collection by . Similarly, we decompose the suffix range of into a collection of dyadic intervals corresponding to precomputed subsets of denoted by . We then perform queries (For notational brevity here, we abuse notation slightly. The actual query would be on the indices corresponding to subsets A and B.) for all and . It is important to note that the bounds and remain unchanged throughout the reduction. □
We next present a preliminary solution for the Bounded Gapped Set Intersection with space and reporting time .
2.1. The Data Structure
Let be an array containing the elements of in sorted order where we now define . We subdivide A into overlapping blocks of size , with each consecutive block overlapping by G elements. Formally, for , we define block . See Figure 1.

Figure 1.
A preliminary blocking scheme for and .
Given a query where , consider the following: if and there exists an such that , then, since , a and b must lie in the same block. Consequently, we construct the data structure from Lemma 1 for each block, , using the subsets , ⋯, , excluding empty subsets. Thus, the number of subsets in a given block may be fewer than k. We store in sorted order, for each block , the original subset indices i for each non-empty subset . We also store the associated t value in the same sorted order.
2.2. Querying
Given query , we iterate through . For block , we first determine if is empty. This can be accomplished using binary search over the stored indices for , which were described above. If , we are finished for as there are no solutions in this block. Otherwise, we obtain such that . Similarly, we perform a binary search for j over the indices for . If , we are finished for . Otherwise, we obtain such that . We then make the query to the data structure for block and store the reported solutions.
After all blocks are processed, a final sort of the stored solutions is performed and the duplicates are removed. We then output the resulting list of occurrences.
2.3. Analysis
The above approach requires space per block, resulting in an overall space requirement of . Reporting all occurrences within a single block takes , where represents the number of occurrences in . The total time across all blocks is . Note that each solution occurs in at most two blocks, so the final sorting and duplicate removal step does not change the asymptotic query time.
Applying the reduction in Lemma 2, this yields a solution for Bounded Gapped String Indexing with Reporting that requires space and achieves a query time of . However, this does not provide the desired query time complexity, particularly for large values of G. We now present an improved solution.
3. An Improved Solution
The basis of the improved solution is to carefully decompose the array A (defined in Section 2) to avoid having to check every block for occurrences as was done in the preliminary solution. We rely heavily on techniques from [28] and its extensions in [22,29].
3.1. The Data Structure
We will construct a tree data structure over the array A. Each node in the tree will have an associated subarray of A. We construct the tree structure over the array A recursively as follows: The tree’s root is associated with the entire array . We designate the root’s midpoint as . The root node is given a middle child, that is a leaf representing the subarray . We then recursively create two child subtrees: the left child subtree corresponds to , and the right child subtree corresponds to . If at any point the size of a subarray is at most , we treat the node as a leaf node. See Figure 2. For each node in the tree, we create the Gapped Set Intersection Data Structure from Lemma 1. For each leaf node, we create the Gapped Set Intersection Data Structure outlined in Lemma 1. See Algorithm 1 for pseudocode. Like in Section 2, these data structures are constructed over the non-empty subsets of each block, and we maintain the mapping from the query i and j to the corresponding non-empty subset if it exists. These details are omitted from the pseudocode.
| Algorithm 1 Construction Algorithm |
| 1: procedure Construct() |
| 2: if then |
| 3: create_internal_node() |
| 4: |
| 5: create_leaf_node() |
| 6: Construct() |
| 7: Construct() |
| 8: else |
| 9: create_leaf_node() |
| 10: end if |
| 11: return v |
| 12: end procedure |
| 13: procedure Construct(A, G) |
| 14: Construct() |
| 15: end procedure |
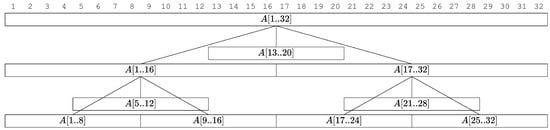
Figure 2.
Tree structure constructed by Algorithm 1 with and .
In terms of notation, we call the leaves created in Line 5 of Algorithm 1 middle children leaves. For a node v with associated subarray , we call the midpoint of v.
3.2. Querying
To query the tree structure, we begin at the root. We first use the data structure from Lemma 1 to check whether any occurrence is contained in the current nodes’ associated subarray. If the current node is a leaf and it contains an occurrence, we report all occurrences using the data structure from Lemma 1. If the current node is not a leaf and contains an occurrence, we recursively search all of its children. This is shown in pseudocode in Algorithm 2.
| Algorithm 2 Query Algorithm |
| 1: procedure Search() |
| 2: if not v.contains_occurrence() then |
| 3: return |
| 4: else if v.contains_occurrence() and v is leaf then |
| 5: v.report_all_occurrences() |
| 6: else if v.contains_occurrence() and v is internal node then |
| 7: Search() |
| 8: Search() |
| 9: Search() |
| 10: end if |
| 11: end procedure |
| 12: procedure Query() |
| 13: Search() |
| 14: end procedure |
3.3. Correctness
The key observation from Figure 2 is that the union of the leaf nodes in the tree constructed by Algorithm 1 resembles the blocking scheme described in Section 2 (see Figure 1 for comparison). Based on this observation, we now formalize the following key lemmas.
Lemma 3.
Every subarray of size at most G is contained in the subarray of some leaf.
Proof.
For the sake of contradiction, assume that subarray is not contained in the subarray of any leaf. Let v be the node of maximum height that contains . Let l and r denote the bounds for the subarray for node v, and let .
Since is not contained in any leaf, it must be that is not fully contained within the range . Therefore, either starts in the range or ends in the range . In the former case, since , we have that must be contained within the subarray of left_child, which contradicts the assumption that v is the highest node containing . In the latter case, since , we have that must be contained within the subarray of v.right_child, which again contradicts the assumption that v is the highest node containing . □
The correctness of the above query procedure then follows from the fact that every block of size at most G is contained within the subarray of some leaf node v. All ancestors u of leaf v will report that they contain an occurrence, allowing the DFS traversal to continue until leaf v is reached and its occurrences are reported.
3.4. Space Analysis
First, considering only the Gapped Set Intersection Data Structure from Lemma 1 on non-leaf nodes, this requires space logarithmic factors from
Since , we have , and the geometric series converges to a constant. Hence, ignoring leaf nodes, the space is .
Next, we include the leaves. We first show that the number of leaves is .
Lemma 4.
Every non-middle child leaf’s associated subarray has size at least G.
Proof.
Suppose, for the sake of contradiction, there exists a leaf u that has a subarray size less than G. Let v be the parent of u with range l to r and midpoint m. If u is a left child, it has a subarray size
The above implies
which leads to . However, this implies , so v had a subarray size of at most . In such a case, our algorithm would not recursively create a left child for v, a contradiction.
Similarly, if u is a right child with subarray size less than G, it has size
meaning . This implies . Hence, . Again, we conclude v has a subarray size small enough that our algorithm would not recursively create a right child for v, a contradiction. □
Lemma 5.
The number of leaves in the tree structure created by Algorithm 1 is .
Proof.
Since all leaves created as non-middle children have disjoint associated subarrays, their union represents a set of cardinality N, and (by Lemma 4) each represents a disjoint subset of size at least G. Therefore, there are at most non-middle child leaves. Next, because the tree (still excluding middle children) is a binary tree, the total number of internal nodes is also . Furthermore, including the middle child leaves at most doubles the total number of nodes in the tree. □
Because each leaf contains a subarray of size , the space for the data structures for each leaf is . Combined with Lemma 5, the total space for leaves is , which, since , is also .
3.5. Query Time Analysis
We now analyze the run time of Algorithm 2. Our first step can be seen as a modification of the Kraft–McMillan inequality.
Lemma 6.
For a rooted binary tree , let denote the height of node v in . Then
Proof.
We use induction on the tree height. The base case holds with a single node having height 0 since . For an arbitrary tree with nodes, let the left subtree of the root be with relative height function , and the right child of the root, , with relative height function . Then,
□
Lemma 7.
Let s be the number of leaves for which we have to run the “report_all_occurrences” subroutine in Algorithm 2. Let be the set of nodes on whichSearchis executed in Algorithm 2. Then, .
Proof.
The height of the tree constructed by Algorithm 1 is . Each root-to-v path for all v where “report_all_occurrences” is called contributes at most calls of Search. Thus, the contribution overall of these paths is . What remains to be counted are nodes where Search is called and “contains_occurrence” reports there are no occurrences. For these, observe that each node on the root-to-v path for all v where “report_all_occurrences” is called has at most two children where this can be the case. Thus, including these nodes at most triples the total number of nodes on which Search is called. □
We take s and as defined in Lemma 7. First, we consider the time used by calls to “contains_occurrence”. Because the size of a subarray for a node v at height is , by Lemma 1, a call to “contains_occurrence” for a node v at height requires time . The combined time used for “contains_occurrence” calls is polylogarithmic factors from
We next apply Hölder’s inequality to obtain the bound
Applying Lemma 6 and 7, we can further bound this as
Substituting into Equation (1), we obtain that the time used for “contains_occurrence” calls is .
Next, we consider the time used by calls to “report_all_occurrences”. By Lemma 1, each leaf v on which “report_all_occurrences” is called takes time , where denotes the number of occurrences contained in the subarray for node v. To bound the time complexity, we should bound the number of times an occurrence can be reported over all blocks. To this end, we prove Lemma 8.
Lemma 8.
Every subarray of size has a non-empty intersection with leaf’s subarrays.
Proof.
we have , where the last inequality follows from Line 6 in Algorithm 1. Hence, .
Consider first the tree structure without any middle child leaves. In this case, the leaves are all disjoint and, by Lemma 4, have subarray size at least G. Hence, at most two non-middle children leaves have non-empty intersections with .
Now, we incorporate the middle children. We consider the middle children leaves as being ordered according to their midpoint.
- Claim: The difference between consecutive midpoints of middle children leaves is at least G. To see this, consider a middle child of u with midpoint and a middle child of v with midpoint immediately preceding in the order. If v is in the left subtree of u, since
If, on the other hand, v is not in the left subtree of u, then since the middle child of v is ordered before the middle child of u, v cannot be in the right subtree of u. If v is the parent of u, then by a similar argument,
and we have , where the last inequality follows from Line 7 in Algorithm 1. Hence, . In any other remaining cases, u and v must share either some lowest common ancestor or an intermediate vertex on the path from v to u. Call this vertex w. Observe that the middle child of w sits in the ordering between the middle children of u and v, a contradiction that makes the last remaining cases impossible.
Applying the above claim, we first observe that the range of possible midpoints of middle children leaves that can have a non-empty intersection with is . Therefore, an upper bound on the number of middle children leaves that can have a non-empty intersection with is given by
Hence, at most three middle children leaves have intersections with . Combined with at most two non-middle children leaves intersecting , we arrive at the desired result. □
As a result of Lemma 8, each occurrence is reported at most times, and the removal of potential duplicates does not affect the total asymptotic time complexity. Over all “report_all_occurrences” calls, the time is .
Summing the time for calls to “contains_occurrence” and “report_all_occurrences” we obtain a total time of . Because each leaf on which “report_all_occurrences” is called contains at least one occurrence, and by Lemma 8, each occurrence is contained in leaves, we have . Furthermore, we have . This gives us the following lemma.
Lemma 9.
For every , there is a data structure for the Bounded Gapped Set Intersection with Reporting that occupies space and answers queries in time
where is the size of the output.
Combining Lemma 9 with the reduction used in Lemma 2, we obtain the result in Theorem 2, which is an space index for Bounded Gapped String Indexing with query time.
3.6. Obtaining Theorem 1
We now apply Theorem 2 to obtain Theorem 1. For convenience, we assume that n is a power of two (if not, we can pad T with extra symbols # not in T’s alphabet until its length is a power of two. We can accomplish this while at most doubling its length). We construct the data structure from Lemma 9 for all G in 1, 2, 4, 8, , n. The space required across all data structures is polylogarithmic factors from
To answer a query , we split into logarithmically many ranges , , , , where in the case , no split is performed. The query is given to the data structure for . By Theorem 2, it reports occurrences in time , where is the number of occurrences with a gap in the range . Continuing in this fashion for each split, the overall complexity is polylogarithmic factors from
where is the number of occurrences with a gap in range . The last equality holds since .
We observe that for a given range , , where is the number of occurrences with gap exactly g. Furthermore, for all . Hence,
We conclude that
giving us an overall query time complexity of
as desired. This completes the proof of Theorem 1.
4. Conclusions
We have presented an index for Gapped String Indexing with a reporting time parameterized by the gap lengths of the occurrences. Potential directions for further development include the following:
- Establishing matching conditional lower bounds based on the Strong Set-Disjointness Conjecture or other conjectures used in fine-grained complexity.
- Extensions to the multi-gap case: That is, preprocess a text to answer queries of the form . It is not immediate how to adapt Problem 3-based techniques to this setting.
- Extensions to the bounded ratio gapped setting of Ganguly et al. [23].
We acknowledge the likelihood that the gap-sensitive approach proposed here may have a worse query time compared to the prior approach by Bille et al. [17] for large gap values due to polylogarithmic factors. In such instances, it may be advantageous to consider some form of a meta-algorithm that employs our gap-sensitive approach for small gaps and the algorithm of Bille et al. [17] for larger gaps. We leave this as a possible direction for future research.
Author Contributions
Conceptualization, M.H.H., D.G. and S.V.T.; methodology, M.H.H., D.G. and S.V.T.; validation, M.H.H., D.G. and S.V.T.; formal analysis, M.H.H., D.G. and S.V.T.; investigation, M.H.H., D.G. and S.V.T.; writing—original draft preparation, M.H.H., D.G. and S.V.T.; writing—review and editing, M.H.H., D.G. and S.V.T.; supervision, D.G. and S.V.T.; project administration, D.G. and S.V.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by U.S. National Science Foundation (NSF) award number CCF-2315822.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
This research is supported in part by the U.S. National Science Foundation (NSF) award CCF-2315822.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bucher, P.; Bairoch, A. A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; pp. 53–61. [Google Scholar]
- Hofmann, K.; Bucher, P.; Falquet, L.; Bairoch, A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999, 27, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, K.; Grabowski, S. Efficient algorithms for pattern matching with general gaps, character classes, and transposition invariance. Inf. Retr. 2008, 11, 335–357. [Google Scholar] [CrossRef]
- MYERS, E.W. Approximate Matching of Network Expressions with Spacers. J. Comput. Biol. 1996, 3, 33–51. [Google Scholar] [CrossRef] [PubMed]
- Mehldau, G.; Myers, G. A system for pattern matching applications on biosequences. Bioinformatics 1993, 9, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Navarro, G.; Raffinot, M. Fast and Simple Character Classes and Bounded Gaps Pattern Matching, with Applications to Protein Searching. J. Comput. Biol. 2003, 10, 903–923. [Google Scholar] [CrossRef] [PubMed]
- Pissis, S.P. MoTeX-II: Structured MoTif eXtraction from large-scale datasets. BMC Bioinform. 2014, 15, 235. [Google Scholar] [CrossRef] [PubMed]
- Miner, G.; Delen, D.; Elder, J.; Fast, A.; Hill, T.; Nisbet, R.A. Practical Text Mining and Statistical Analysis for Non-Structured Text Data Applications; Academic Press: Boston, MA, USA, 2012. [Google Scholar]
- Kroeger, P.R. Analyzing Grammar: An Introduction; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Manning, C.D.; Schütze, H. Foundations of Statistical Natural Language Processing; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Willkomm, J.; Schäler, M.; Böhm, K. Accurate Cardinality Estimation of Co-occurring Words Using Suffix Trees. In Proceedings of the Database Systems for Advanced Applications: 26th International Conference, DASFAA 2021, Taipei, Taiwan, April 11–14 2021; Proceedings, Part II 26. Jensen, C.S., Lim, E.P., Yang, D.N., Lee, W.C., Tseng, V.S., Kalogeraki, V., Huang, J.W., Shen, C.Y., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 721–737. [Google Scholar]
- Gusfield, D. Algorithms on Strings, Trees, and Sequences: Computer Science and Computational Biology; Cambridge University Press: Cambridge, UK, 1997. [Google Scholar]
- Crochemore, M.; Hancart, C.; Lecroq, T. Algorithms on Strings; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Bille, P.; Gørtz, I.L.; Vildhøj, H.W.; Wind, D.K. String matching with variable length gaps. Theor. Comput. Sci. 2012, 443, 25–34. [Google Scholar] [CrossRef]
- Bille, P.; Thorup, M. Regular Expression Matching with Multi-Strings and Intervals. In Proceedings of the Twenty-First Annual ACM-SIAM Symposium on Discrete Algorithms, SODA 2010, Austin, TX, USA, 17–19 January 2010; Charikar, M., Ed.; SIAM: Philadelphia, PA, USA, 2010; pp. 1297–1308. [Google Scholar] [CrossRef]
- Morgante, M.; Policriti, A.; Vitacolonna, N.; Zuccolo, A. Structured Motifs Search. J. Comput. Biol. 2005, 12, 1065–1082. [Google Scholar] [CrossRef] [PubMed]
- Bille, P.; Gørtz, I.L.; Lewenstein, M.; Pissis, S.P.; Rotenberg, E.; Steiner, T.A. Gapped String Indexing in Subquadratic Space and Sublinear Query Time. In Proceedings of the 41st International Symposium on Theoretical Aspects of Computer Science, STACS 2024, Clermont-Ferrand, France, 12–14 March 2024; Beyersdorff, O., Kanté, M.M., Kupferman, O., Lokshtanov, D., Eds.; Schloss Dagstuhl-Leibniz-Zentrum für Informatik: Dagstuhl, Germany, 2024; Volume 289, pp. 16:1–16:21. [Google Scholar] [CrossRef]
- Peterlongo, P.; Allali, J.; Sagot, M. Indexing Gapped-Factors Using a Tree. Int. J. Found. Comput. Sci. 2008, 19, 71–87. [Google Scholar] [CrossRef]
- Iliopoulos, C.S.; Rahman, M.S. Indexing Factors with Gaps. Algorithmica 2009, 55, 60–70. [Google Scholar] [CrossRef]
- Lewenstein, M. Indexing with Gaps. In Proceedings of the String Processing and Information Retrieval, 18th International Symposium, SPIRE 2011, Pisa, Italy, 17–21 October 2011; Grossi, R., Sebastiani, F., Silvestri, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 7024, pp. 135–143. [Google Scholar] [CrossRef]
- Goldstein, I.; Lewenstein, M.; Porat, E. On the Hardness of Set Disjointness and Set Intersection with Bounded Universe. In Proceedings of the 30th International Symposium on Algorithms and Computation, ISAAC 2019, Shanghai, China, 8–11 December 2019; Lu, P., Zhang, G., Eds.; Schloss Dagstuhl-Leibniz-Zentrum für Informatik: Dagstuhl, Germany, 2019; Volume 149, pp. 7:1–7:22. [Google Scholar] [CrossRef]
- Bille, P.; Gørtz, I.L.; Pedersen, M.R.; Steiner, T.A. Gapped Indexing for Consecutive Occurrences. Algorithmica 2023, 85, 879–901. [Google Scholar] [CrossRef]
- Ganguly, A.; Gibney, D.; MacNichol, P.; Thankachan, S.V. Bounded Ratio Gapped String Indexing. In Proceedings of the SPIRE 2024, Puerto Vallarta, Mexico, 23–25 September 2024. [Google Scholar]
- Golovnev, A.; Guo, S.; Horel, T.; Park, S.; Vaikuntanathan, V. Data structures meet cryptography: 3SUM with preprocessing. In Proceedings of the 52nd Annual ACM SIGACT Symposium on Theory of Computing, STOC 2020, Chicago, IL, USA, 22–26 June 2020; Makarychev, K., Makarychev, Y., Tulsiani, M., Kamath, G., Chuzhoy, J., Eds.; ACM: New York, NY, USA, 2020; pp. 294–307. [Google Scholar] [CrossRef]
- Kopelowitz, T.; Porat, E. The Strong 3SUM-INDEXING Conjecture is False. arXiv 2019, arXiv:1907.11206. [Google Scholar]
- Weiner, P. Linear Pattern Matching Algorithms. In Proceedings of the 14th Annual Symposium on Switching and Automata Theory, Iowa City, IA, USA, 15–17 October 1973; IEEE Computer Society: New York, NY, USA, 1973; pp. 1–11. [Google Scholar] [CrossRef]
- Farach, M. Optimal Suffix Tree Construction with Large Alphabets. In Proceedings of the 38th Annual Symposium on Foundations of Computer Science, FOCS ’97, Miami Beach, FL, USA, 19–22 October 1997; IEEE Computer Society: New York, NY, USA, 1997; pp. 137–143. [Google Scholar] [CrossRef]
- Cohen, H.; Porat, E. Fast set intersection and two-patterns matching. Theor. Comput. Sci. 2010, 411, 3795–3800. [Google Scholar] [CrossRef]
- Hon, W.; Shah, R.; Thankachan, S.V.; Vitter, J.S. String Retrieval for Multi-pattern Queries. In Proceedings of the String Processing and Information Retrieval—17th International Symposium, SPIRE 2010, Los Cabos, Mexico, 11–13 October 2010; Chávez, E., Lonardi, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 6393, pp. 55–66. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).