Abstract
Background: Sepsis is one of the major causes of in-hospital death, and is frequent in patients presenting to the emergency department (ED). Early identification of high-risk septic patients is critical. Machine learning (ML) techniques have been proposed for identification and prognostication of ED septic patients, but these models often lack pre-hospital data and lack validation against early sepsis identification scores (such as qSOFA) and scores for critically ill patients (SOFA, APACHE II). Methods We conducted an electronic health record (EHR) study to test whether interpretable and scalable ML models predict mortality in septic ED patients and compared their performance with clinical scores. Consecutive adult septic patients admitted to ED over 18 months were included. We built ML models, ranging from a simple-classifier model, to unbalanced and balanced logistic regression, and random forest, and compared their performance to qSOFA, SOFA, and APACHE II scores. Results: We included 425 sepsis patients after screening 38,500 EHR for sepsis criteria. Overall mortality was 15.2% and peaked in patients coming from retirement homes (38%). Random forest, like balanced (0.811) and unbalanced logistic regression (0.863), identified patients at risk of mortality (0.813). All ML models outperformed qSOFA, APACHE II, and SOFA scores. Age, mean arterial pressure, and serum sodium were major mortality predictors. Conclusions: We confirmed that random forest models outperform previous models, including qSOFA, SOFA, and APACHE II, in identifying septic patients at higher mortality risk, while maintaining good interpretability. Machine learning models may gain further adoption in the future with increasing diffusion and granularity of EHR data, yielding the advantage of increased scalability compared to standard statistical techniques.
1. Introduction
Sepsis is a life-threatening organ dysfunction due to a dysregulated host response to infection [1], and is a significant public health challenge and economic burden in the industrialized world [2].
The incidence of sepsis spans between 650,000 and 750,000 new cases per year in the United States, where it is the 10th highest cause of death [3,4]. Sepsis numbers are rising as the incidence of sepsis increases with age, and the proportion of elderly patients accessing the emergency department increases [5].
Early care of sepsis and septic shock is essential for the patient’s prognosis, and include early diagnosis, resuscitation, and prompt antibiotic treatment, which is often performed in the emergency department (ED) [6,7,8]. Postponed diagnosis of sepsis increases morbidity and mortality [9].
In the emergency department (ED) population, predictive scores have been extensively studied to identify sepsis and stratify mortality in septic patients [5,10]. Most published models adopt traditional statistical techniques for mortality prediction and identification of septic patients [10,11,12,13,14]. Recently, machine learning has been proposed in this field, primarily for early identification of septic patients [15,16]. By discovering predictive patterns in the data beyond linear relations, machine learning models can naturally handle the abundance and complexity of digital patient data, allowing for precise forecasts of which patients will develop sepsis. Although a few recent ML studies also focused on mortality prediction [17], none of them integrated information from patient provenience and validated it against well-known critically ill scores such as APACHE II, qSOFA, and SOFA [18,19]. Therefore, the aim of the current study was to develop an interpretable and scalable ML model for identification of high-risk patients based on clinical, laboratory, and pre-hospital data, and to compare its performance with qSOFA, APACHE II, and SOFA scores.
2. Materials and Methods
We conducted an electronic health record study. We extracted de-identified data from hospital electronic health records of all patients admitted to a tertiary clinical center, over 18 months. Study received ethical approval from Humanitas Research Hospital independent Ethical Committee. We identified septic patients according to Angus criteria and Agency for Healthcare Research and Quality (AHRQ) criteria, two tools frequently adopted for studies using EHR and administrative data [20,21,22,23]. Accordingly, International Classification of Diseases, Ninth Edition, Clinical Modification (ICD-9-CM) codes of patients admitted from the ED were extracted from EHR data, and screened for Angus or AHRQ criteria [21,22,23]. In these criteria, there are two ways for a hospitalization to become selected by Angus. The patient is selected when (a) explicit ICD-9 codes for severe sepsis or septic shock are found or (b) there is an ICD-9 diagnosis code for infection. Detailed AHRQ criteria according to ICD9 codes are available on the AHRQ website [23]. Full EHR data of patients identified through these criteria were subsequently re-evaluated in depth by a doctor according to Sepsis Criteria before inclusion in the study [24]. Criteria are reported in Supplemental Material File S1. All patients admitted to the ED were treated according to Surviving Sepsis Campaign guidelines [25]. The site and type of underlying infection were determined by clinical signs and symptoms, medical history, laboratory exams, and biological cultures.
The primary study goal was to assess whether an interpretable and scalable machine learning model can predict mortality in septic patients in the ED better than SOFA, qSOFA, and APACHE II scores, and to compare the model to logistic regression and a dummy classifier. To evaluate the models produced and to assess their performance, we chose to describe and analyze five different parameters: precision, recall, accuracy, receiver operating characteristic (ROC) curve with area under the curve (AUC), and F1-score. Precision is defined as the number of true septic patients divided by the total number of positive predictions; recall quantifies how many true positives were retrieved by our models. F1-score represents a harmonic mean of precision and recall. Accuracy is the number of correctly predicted data across both positive and negative classes, while ROC and its AUC are measures of the ability of a classifier to distinguish between classes. The secondary objective was the descriptive analysis of epidemiological and microbiological characteristics of the study population, specifically on covariates that were available in the EHR (a list of all the covariates can be found in the Supplemental Materials Table S1).
Statistical analysis
Categorical data are described as frequency and percentage. Continuous data are expressed as the median and interquartile range (IQR). We tested the association between independent variable and outcome at univariate analysis, using the chi-squared test for categorical variables or the Wilcoxon rank-sum test as a non-parametric test for continuous variables.
Missing data were explored for visual patterns of missingness as reported in Supplemental Material Figure S1, and considered missing at random (MAR). To account for missing data, we used a chained random forest to perform imputation of missing data using the missRanger package [26], S, which basically imputes each variable using predictions from a random forest that includes all other variables as covariates, and iterating this process multiple times until the process stops improving. Results of the imputation process are reported in Supplemental Material Figure S2.
A baseline reference model was created using a dummy classifier, which is a simple uniform classifier commonly employed as a baseline in ML models [27]. We then developed a logistic regression model with unbalanced classes, and a logistic regression model with classes balanced using the SMOTE technique [28], which is an oversampling technique where synthetic data samples are generated for the class less represented (septic patients in our case). The oversampling technique was performed during the cross-validation phase, after the split into training and test datasets. Finally, we trained a random forest to identify patients at risk of mortality [29]. All models were built on two datasets based on early EHR data, with only one dataset including APACHE-II score, qSOFA score, or SOFA score as predictive features. Each dataset was split in the proportions of 0.7 training set and 0.3 test set, with a 10 k-fold cross validation to improve generalizability and reduce overfitting. Each fit was performed on a training set consisting of 90% of the total training set selected at random, with the remaining 10% used as a hold-out set for validation.
Feature importance was calculated differently depending on the model used: for logistic regression we used the magnitude and the sign of the coefficients of the features given by the trained models, while for random forests we used the Mean Decrease in Impurity (MDI). This method calculates the average decrease in Gini impurity due to the splits in the analyzed feature across all the trees trained in the forest.
Precision, recall, accuracy, F1-score, and ROC analysis with AUC were calculated to compare the performance of all developed models. Subsequently, we compared the performance of developed models with logistic regression models of the APACHE II, qSOFA, and SOFA scores [18,19,30].
3. Results
From a total of 38,500 patients admitted to the emergency department over 18 months, we extracted 640 patients according to Angus and AHRQ sepsis criteria. Finally, 425 individuals were included in the research after a physician reviewed the extracted information to control for sepsis and removed 215 patients who were not verified as sepsis cases. Figure 1 illustrates the study flow chart.
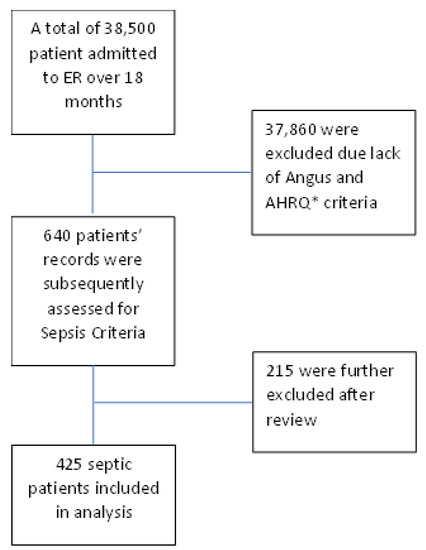
Figure 1.
Study flow chart.
There were 249 males (58.6%) and 176 females (41.4%) in study population. The median age was 77 (IQR 69–83, range 25–97). Hypertension was the most common comorbidity (173 patients, 40.7%), followed by diabetes (105 patients, 24.7%) and cancer (98 patients, 23.1%). Baseline and demographic data are reported in Table 1.

Table 1.
Baseline data and source of sepsis.
Most patients came to ED from home (333 patients, 78.4%), 9.2% (20 patients) from long-term care, and 12.5% (53 patients) from ambulatory clinics or within four weeks from hospital discharge. Mortality was strongly related to patient provenience, with patients from long-term care bearing the highest risk for mortality (48.7%), compared to readmission after hospital discharge (20.7%) or admission from home (10.5%). Overall mortality was 15.3% (65 patients).
The most represented sites of infection were the abdomen (33.2%) and lungs (30.8%). Pneumonia was the most prevalent sepsis site (30.8%) in non-survivors (Table 1). A total of 52 patients (12.2%) presented with septic shock, resulting in higher mortality than sepsis patients (26.2% vs. 9.7%, p = 0.001). Heart rate, mean arterial pressure, and respiratory rate were significantly different between survivors and non-survivors (p = 0.001).
Laboratory values and clinical scores are presented in Table 2.

Table 2.
Laboratory values and clinical scores.
C-reactive protein was higher in non-survivors than survivors (17 [8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25] vs. 13 [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20], p = 0.008), while serum lactate levels were similar. The median APACHE II score was higher in non-survivors than in survivors (15 [11,12,13,14,15,16,17] vs. 12 [8,9,10,11,12,13,14,15,16], p < 0.001). A positive SOFA was more frequent in non-survivors than in survivors (37.5% vs. 12.9%, p = 0.001).
We found no association between the type of pathogen identified by specimen cultures and mortality. The most frequent bacterium identified through blood culture was E. coli (38.1% of cases), followed by E. faecalis (6.2%) and S. aureus (5.1%). Negative blood cultures were present in about 40% of cases (Table 3).

Table 3.
Blood culture and urinary culture microbiological data.
E. coli was the most frequently identified pathogen for urinary cultures (21.7%), followed by K. pneumoniae (5.7%).
The predictive performance of random forest, balanced logistic regression, unbalanced logistic regression, and dummy classifier (reference model) is reported in Table 4.

Table 4.
Performance of random forest, balanced and unbalanced logistic regression, and simple classifier as the baseline, in training and test set.
All ML scores achieved a higher predictive performance after inclusion of SOFA and APACHE-II scored as a feature. Random forest achieved higher discrimination power (AUC 0.863) than balanced (AUC 0.811) or unbalanced (AUC 0.813) logistic regression, but lower overall accuracy (0.76 vs. 0.77 vs. 0.84). Unbalanced logistic regression achieved the highest F1 score, at the expense of a lower precision (0.87). Figure 2 reports feature importance for random forest and logistic regression models.
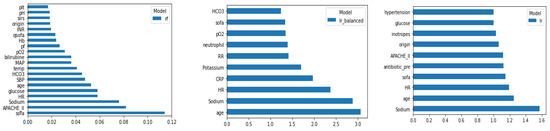
Figure 2.
Feature importance for random forest (left) and odds ratio for balanced (middle) and unbalanced (right) logistic regression model.
APACHE II, qSOFA, and SOFA scores had lower sensitivity and discrimination ability compared to both logistic and random forest models, but were superior compared to the dummy classifier.
4. Discussion
In this study, we confirmed that interpretable and scalable machine learning models may outperform published qSOFA, APACHE II, and SOFA scores in identifying patients at highest risk of mortality. We demonstrated that random forest achieved the highest discriminative performance compared to standard statistical techniques such as balanced and unbalanced logistic regression.
Our study population was composed of elderly and frail patients (median age 77 [IQR 69–83], with a median age in non-survivors of 83 [IQR 75–89]) and several baseline comorbidities. Given the frailty of this population, mortality was high, reaching 15.3% in the whole population, similarly to mortality reported in other studies on septic patients admitted to the ED, resulting in higher sepsis mortality compared to other hospital populations [10,31]. This is due to the specific characteristics of the ED patient population, a mixed group that includes very frail patients from long-term care facilities with a poor physiological reserve and baseline prognosis. In contrast, the cohort of septic hospitalized patients often includes younger patients and surgical patients, which are characterized by lower baseline frailty [10].
The Acute Physiology and Chronic Health Evaluation II (APACHE II) and the Sequential Organ Assessment Score are among the most diffuse predictive scores for mortality, and were used as a reference in this population [18,32]. The qSOFA, an early reduced version of the SOFA score proposed by the Surviving Sepsis Campaign for screening purposes, was also included as a comparison in the model. The SOFA score achieved an intermediate discriminative performance (AUC= 0.712), which was slightly higher than the performance of the APACHE II score (AUC = 0.647). The qSOFA score had an AUC of 0.706. qSOFA, SOFA, and APACHE II were superior to the dummy classifier (a simple model which classifies patients according to chance, and included as a reference only). In contrast, they performed worse than logistic regression, balanced logistic regression, and random forest. Random forest may thus represent a simple solution to approaching data in sepsis ED patients, and outperforms standard statistical scores. Moreover, random forests are more easily scalable than standard statistical techniques, as they do not rely on assumptions of linearity between the log-odds of the outcomes and independent predictors, independency, and multicollinearity, challenges that are common in EHR data, and which are used for standard logistic regression techniques [33]. Random forests may further prove useful in clinical setting, as they may be used in the pre-processing phase for imputation of missing data. We adopted this method in the present study, and it again outperformed standard imputation techniques. Moreover, in clinical situations, random forests retain interpretability, an advantage over more complicated machine learning approaches such as unsupervised or deep learning. On this point, a growing body of research emphasizes the significance of explainable machine learning algorithms applied to the prediction of clinical outcomes [34]. Lemańska-Perek et al. provide a two local-level explanation technique to aid clinicians in the understanding of model prediction for a specific patient with sepsis [35]. Other methods, such as the use of heatmaps to allow interpretability of deep-learning models by highlighting the most influential variables in temporal sequences, have also been proposed [36].
Mortality in patients arriving at the ED from home was significantly lower in univariate analysis than mortality in long-term care patients (10% vs. 48%, p < 0.001). As most of our patients (about 3 out of 4 patients) arrived at the ED from home, they were exposed to common community-acquired pathogens with low antibiotic resistance, compared to frail populations. This, in turn, implied that the ED physician could apply guidelines for empiric antibiotic treatment of community-acquired infections with satisfactory outcomes [37].
Age was significantly associated with mortality in both univariate analysis and machine learning models. Extreme age is a very well-known factor influencing survival in the ED population. The median age in our population is higher than that in previous studies, while mortality is comparable with that of the population [38,39]. Vorker et al., for example, reported a population of ED patients with sepsis with a mean age of 69 and mortality of 23%, compared to the median age of 77 years (IQR 69–83) in our cohort, which reached 15% mortality [38].
Both hyponatremia and hypernatremia have been recognized as independent factors for mortality in ED and critically ill patients [40,41]. In our results, the random forest model and the balanced and unbalanced logistic regression identified levels of natremia as an independent factor for mortality, confirming previous findings.
High heart rate (HR) and elevated C-reactive protein (CPR) levels were also found to be associated with increased mortality, consistent with previous literature. Castilho et al., for example, investigated whether heart rate variability (HRV) was a predictor of mortality in septic patients. They found that several HRV parameters were reduced in non-surviving septic patients [16]. Devran et al., in the same fashion, found that the risk of sepsis-related mortality appears to be increased when the third-day CRP value is greater than 100 mg/dL [17].
A further interesting correlation emerged between blood glucose levels and mortality during sepsis, reflecting the most current literature data. In a retrospective sub-analysis of a multicenter, prospective cohort study, Kushimoto and colleagues investigated the impact of blood glucose abnormalities on outcome and disease severity in patients with severe sepsis. They found that hypoglycemia was related to increased severity and high mortality in septic patients [42].
Gender did not influence survival in this population. Studies on hospitalized patients suggest gender influences mortality, and consider variations in the inflammatory response according to patient gender. Patients coming to ED may be subjected to a shorter ongoing inflammatory process than other hospitalized patients, which often present systemic inflammation related to other conditions such as recent surgery or cancer [43]. The prompt resolution of the infective noxa in the ED could allow a more rapid resolution of the inflammatory process and reduce gender imbalances [44].
Serum lactate levels were only marginally elevated in this population, a finding that may be related to the low proportion of septic shock (12%). Lactate is one of the most significant markers in septic patients, particularly in septic shock [45]. An increase in lactate is classically related to a shift toward anaerobic tissue metabolism in various types of shock, but can also reflect other metabolic and hepatic clearance conditions [45].
Most patients in this cohort had at least one comorbidity. Cardiovascular disease, COPD, and active cancer are all markers of increased mortality in septic patients presenting to the ED [37]. Nonetheless, comorbidities alone are not a good proxy for patient frailty. Accordingly, only unbalanced logistic regression retained hypertension as an independent contributor for mortality, while balanced logistic and random forest included laboratory data and vital parameters as independent predictors of mortality [30,33,37]. The most represented sites of infection were the urinary tract (UTI, 31%) and the abdomen (33%). These data are relevant to give a practical indication for the ED approach: since UTI in the elderly can be asymptomatic, UTI as a sepsis site must always be suspected and excluded. Similarly, we found no association between the type of micro-organism and mortality [37]. E. Coli, a common pathogen in the elderly population, was the most commonly identified bacteria in both urinary and blood samples.
Limitations
This study has the limitation of an observational study. Some data were missing and could not be retrieved, due to the lack of reporting within the electronic health record. We performed analysis and imputation of missing data to reduce the influence of missingness. Data were considered missing at random after visual analysis, while no formal analysis of the type of missingness was performed. Patients included in this study relied on ICD coding for the identification of EHR data, an approach with several limitations, including suboptimal sensitivity and specificity in record retrieval. The total number of patients retrieved in this study is not comparable to that ascribed to big data analysis. However, it is comparable to previous studies on ML applied to ED [46]. In addition, as previously demonstrated by our group, machine learning techniques can be used in settings where big data are not yet available, with performance and reliability comparable to those of standard statistical techniques [47]. Accordingly, machine learning can be a useful resource for patient stratification and classification in an emergency setting and in smaller hospitals. In this work, we opted for the random forest algorithm, which is described as one of the most easily interpretable and scalable models available, and can be further increased by special techniques such as aggressive subsampling. [48]. Due to the small sample size, we were, however, unable to assess the scalability of our model within the present study.
5. Conclusions
Random forest has good predictive performance in identifying septic patients at risk of mortality in the emergency department, outperforming published predictive scores for critically ill patients, and unbalanced logistic regression. Random forests may also be a useful alternative in clinical settings since they retain interpretability, are more scalable than conventional statistical techniques, and are more suitable for nonlinear EHR data because they are dependent on fewer assumptions than logistic regression. Prospective studies on the application of interpretable and scalable machine learning techniques are needed to assess their role as clinical aids for the identification of patients, and to test whether their real-time application can change patient prognosis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/a16020076/s1, Figure S1: Heat maps of missing data by outcome; Figure S2: Results of imputation of missing data by chained random forest; Table S1: Variables included in study; File S1: Sepsis Criteria.
Author Contributions
Conceptualization, M.G., A.V. and M.C.; methodology, M.G. and P.F.C.; formal analysis, M.G. and P.F.C.; investigation, M.G.; resources, S.S., G.C. and A.D.; data curation, A.M., A.D. and R.A.; writing—original draft preparation, M.G., A.D. and A.V.; writing—review and editing, E.C., A.V. and M.C.; supervision, E.C., A.V. and M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are available upon reasonable request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Tsertsvadze, A.; Royle, P.; Seedat, F.; Cooper, J.; Crosby, R.; McCarthy, N. Community-Onset Sepsis and Its Public Health Burden: A Systematic Review. Syst. Rev. 2016, 5, 81. [Google Scholar] [CrossRef]
- Stoller, J.; Halpin, L.; Weis, M.; Aplin, B.; Qu, W.; Georgescu, C.; Nazzal, M. Epidemiology of Severe Sepsis: 2008–2012. J. Crit. Care 2016, 31, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Angus, D.C.; Linde-Zwirble, W.T.; Lidicker, J.; Clermont, G.; Carcillo, J.; Pinsky, M.R. Epidemiology of Severe Sepsis in the United States: Analysis of Incidence, Outcome, and Associated Costs of Care. Crit. Care Med. 2001, 29, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.S.; Mannino, D.M.; Moss, M. The Effect of Age on the Development and Outcome of Adult Sepsis. Crit. Care Med. 2006, 34, 15–21. [Google Scholar] [CrossRef]
- Gaieski, D.F.; Edwards, J.M.; Kallan, M.J.; Carr, B.G. Benchmarking the Incidence and Mortality of Severe Sepsis in the United States. Crit. Care Med. 2013, 41, 1167–1174. [Google Scholar] [CrossRef]
- Nguyen, H.B.; Rivers, E.P.; Abrahamian, F.M.; Moran, G.J.; Abraham, E.; Trzeciak, S.; Huang, D.T.; Osborn, T.; Stevens, D.; Talan, D.A. Severe Sepsis and Septic Shock: Review of the Literature and Emergency Department Management Guidelines. Ann. Emerg. Med. 2006, 48, 54.e1. [Google Scholar] [CrossRef]
- Wang, H.E.; Shapiro, N.I.; Angus, D.C.; Yealy, D.M. National Estimates of Severe Sepsis in United States Emergency Departments. Crit. Care Med. 2007, 35, 1928–1936. [Google Scholar] [CrossRef]
- Peake, S.L.; Delaney, A.; Bailey, M.; Bellomo, R.; Cameron, P.A.; Cooper, D.J.; Higgins, A.M.; Holdgate, A.; Howe, B.D.; SA, W.; et al. Goal-Directed Resuscitation for Patients with Early Septic Shock. New Engl. J. Med. 2014, 371, 1496–1506. [Google Scholar] [CrossRef]
- Freund, Y.; Lemachatti, N.; Krastinova, E.; van Laer, M.; Claessens, Y.E.; Avondo, A.; Occelli, C.; Feral-Pierssens, A.L.; Truchot, J.; Ortega, M.; et al. Prognostic Accuracy of Sepsis-3 Criteria for In-Hospital Mortality Among Patients With Suspected Infection Presenting to the Emergency Department. JAMA 2017, 317, 301–308. [Google Scholar] [CrossRef]
- Askim, Å.; Moser, F.; Gustad, L.T.; Stene, H.; Gundersen, M.; Åsvold, B.O.; Dale, J.; Bjørnsen, L.P.; Damås, J.K.; Solligård, E. Poor Performance of Quick-SOFA (QSOFA) Score in Predicting Severe Sepsis and Mortality—A Prospective Study of Patients Admitted with Infection to the Emergency Department. Scand. J. Trauma Resusc Emerg. Med. 2017, 25, 56. [Google Scholar] [CrossRef] [PubMed]
- Usman, O.A.; Usman, A.A.; Ward, M.A. Comparison of SIRS, QSOFA, and NEWS for the Early Identification of Sepsis in the Emergency Department. Am. J. Emerg Med. 2019, 37, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Holder, A.L.; Gupta, N.; Lulaj, E.; Furgiuele, M.; Hidalgo, I.; Jones, M.P.; Jolly, T.; Gennis, P.; Birnbaum, A. Predictors of Early Progression to Severe Sepsis or Shock among Emergency Department Patients with Nonsevere Sepsis. Int. J. Emerg. Med. 2016, 9, 10. [Google Scholar] [CrossRef]
- Bewersdorf, J.P.; Hautmann, O.; Kofink, D.; Abdul Khalil, A.; Zainal Abidin, I.; Loch, A. The SPEED (Sepsis Patient Evaluation in the Emergency Department) Score: A Risk Stratification and Outcome Prediction Tool. Eur. J. Emerg. Med. 2017, 24, 170. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Jay, M.; Hoffman, J.L.; Calvert, J.; Barton, C.; Shimabukuro, D.; Shieh, L.; Chettipally, U.; Fletcher, G.; Kerem, Y.; et al. Multicentre Validation of a Sepsis Prediction Algorithm Using Only Vital Sign Data in the Emergency Department, General Ward and ICU. BMJ Open 2018, 8, e017833. [Google Scholar] [CrossRef]
- McCoy, A.; Das, R. Reducing Patient Mortality, Length of Stay and Readmissions through Machine Learning-Based Sepsis Prediction in the Emergency Department, Intensive Care Unit and Hospital Floor Units. BMJ Open Qual. 2017, 6, e000158. [Google Scholar] [CrossRef]
- Perng, J.W.; Kao, I.H.; Kung, C.; Hung, S.C.; Lai, Y.H.; Su, C.M. Mortality Prediction of Septic Patients in the Emergency Department Based on Machine Learning. J. Clin. Med. 2019, 8, 1906. [Google Scholar] [CrossRef]
- Knaus, W.A.; Draper, E.A.; Wagner, D.P.; Zimmerman, J.E. APACHE II: A Severity of Disease Classification System. Crit. Care Med. 1985, 13, 818–829. [Google Scholar] [CrossRef]
- Vincent, J.L.; Moreno, R.; Takala, J.; Willatts, S.; de Mendonça, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-Related Organ Failure Assessment) Score to Describe Organ Dysfunction/Failure. On Behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef]
- Iwashyna, T.J.; Odden, A.; Rohde, J.; Bonham, C.; Kuhn, L.; Malani, P.; Chen, L.; Flanders, S. Identifying Patients with Severe Sepsis Using Administrative Claims: Patient-Level Validation of the Angus Implementation of the International Consensus Conference Definition of Severe Sepsis. Med. Care 2014, 52, 39–43. [Google Scholar] [CrossRef]
- McDonald, K.M.; Romano, P.S.; Geppert, J.; Davies, S.M.; Duncan, B.W.; Shojania, K.G.; Hansen, A. Measures of Patient Safety Based on Hospital Administrative Data—The Patient Safety Indicators. PMC 2002, 370. Report No.: 02-0038. [Google Scholar] [PubMed]
- Angus, D.C.; Seymour, C.W.; Coopersmith, C.M.; Deutschman, C.S.; Klompas, M.; Levy, M.M.; Martin, G.S.; Osborn, T.M.; Rhee, C.; Watson, R.S. A Framework for the Development and Interpretation of Different Sepsis Definitions and Clinical Criteria. Crit. Care Med. 2016, 44, e113–e121. [Google Scholar] [CrossRef] [PubMed]
- AHRQ. AHRQ Quality IndicatorsTM (AHRQ QITM) ICD-10-CM/PCS Specification V2022; AHRQ: Rockville, MD, USA, 2022.
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.-L.; Ramsay, G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003, 31, 1250–1256. [Google Scholar] [CrossRef]
- Dellinger, R.P.; Levy, M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock: 2012. Crit. Care Med. 2013, 41, 580–637. [Google Scholar] [CrossRef] [PubMed]
- Stekhoven, D.J.; Bühlmann, P. MissForest--Non-Parametric Missing Value Imputation for Mixed-Type Data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Hwang, S.Y.; Jo, I.J.; Lee, S.U.; Lee, T.R.; Yoon, H.; Cha, W.C.; Sim, M.S.; Shin, T.G. Low Accuracy of Positive QSOFA Criteria for Predicting 28-Day Mortality in Critically Ill Septic Patients During the Early Period After Emergency Department Presentation. Ann. Emerg Med. 2018, 71, 1–9.e2. [Google Scholar] [CrossRef]
- Gaieski, D.F.; Mikkelsen, M.E.; Band, R.A.; Pines, J.M.; Massone, R.; Furia, F.F.; Shofer, F.S.; Goyal, M. Impact of Time to Antibiotics on Survival in Patients with Severe Sepsis or Septic Shock in Whom Early Goal-Directed Therapy Was Initiated in the Emergency Department. Crit. Care Med. 2010, 38, 1045–1053. [Google Scholar] [CrossRef]
- Vincent, J.L.; De Mendonça, A.; Cantraine, F.; Moreno, R.; Takala, J.; Suter, P.M.; Sprung, C.L.; Colardyn, F.; Blecher, S. Use of the SOFA Score to Assess the Incidence of Organ Dysfunction/Failure in Intensive Care Units: Results of a Multicenter, Prospective Study. Crit Care Med. 1998, 26, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Caruso, P.F.; Cecconi, M. Artificial Intelligence in the Intensive Care Unit. Semin. Respir. Crit. Care Med. 2020, 2020, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Lemańska-Perek, A.; Krzyżanowska-Gołąb, D.; Kobylińska, K.; Biecek, P.; Skalec, T.; Tyszko, M.; Gozdzik, W.; Adamik, B. Explainable Artificial Intelligence Helps in Understanding the Effect of Fibronectin on Survival of Sepsis. Cells 2022, 11, 2433. [Google Scholar] [CrossRef]
- Viton, F.; Elbattah, M.; Guerin, J.L.; Dequen, G. Heatmaps for Visual Explainability of CNN-Based Predictions for Multivariate Time Series with Application to Healthcare. In Proceedings of the 2020 IEEE International Conference on Healthcare Informatics, ICHI 2020, Oldenburg, Germany, 30 November–3 December 2020. [Google Scholar] [CrossRef]
- Drumheller, B.C.; Agarwal, A.; Mikkelsen, M.E.; Sante, S.C.; Weber, A.L.; Goyal, M.; Gaieski, D.F. Risk Factors for Mortality despite Early Protocolized Resuscitation for Severe Sepsis and Septic Shock in the Emergency Department. J. Crit. Care 2016, 31, 13–20. [Google Scholar] [CrossRef]
- Vorwerk, C.; Loryman, B.; Coats, T.J.; Stephenson, J.A.; Gray, L.D.; Reddy, G.; Florence, L.; Butler, N. Prediction of Mortality in Adult Emergency Department Patients with Sepsis. Emerg. Med. J. 2009, 26, 254–258. [Google Scholar] [CrossRef]
- Shapiro, N.I.; Howell, M.D.; Talmor, D.; Donnino, M.; Ngo, L.; Bates, D.W. Mortality in Emergency Department Sepsis (MEDS) Score Predicts 1-Year Mortality. Crit. Care Med. 2007, 35, 192–198. [Google Scholar] [CrossRef]
- Arampatzis, S.; Funk, G.C.; Leichtle, A.B.; Fiedler, G.M.; Schwarz, C.; Zimmermann, H.; Exadaktylos, A.K.; Lindner, G. Impact of Diuretic Therapy-Associated Electrolyte Disorders Present on Admission to the Emergency Department: A Cross-Sectional Analysis. BMC Med. 2013, 11, 83. [Google Scholar] [CrossRef]
- Lindner, G.; Schwarz, C.; Haidinger, M.; Ravioli, S. Hyponatremia in the Emergency Department. Am. J. Emerg Med. 2022, 60, 1–8. [Google Scholar] [CrossRef]
- Kushimoto, S.; Abe, T.; Ogura, H.; Shiraishi, A.; Saitoh, D.; Fujishima, S.; Mayumi, T.; Hifumi, T.; Shiino, Y.; Nakada, T.-A.; et al. Impact of blood glucose abnormalities on outcomes and disease severity in patients with severe sepsis: An analysis from a multicenter, prospective survey of severe sepsis. PLoS ONE 2020, 15, e0229919. [Google Scholar] [CrossRef]
- Kuttab, H.I.; Lykins, J.D.; Hughes, M.D.; Wroblewski, K.; Keast, E.P.; Kukoyi, O.; Kopec, J.A.; Hall, S.; Ward, M.A. Evaluation and Predictors of Fluid Resuscitation in Patients With Severe Sepsis and Septic Shock. Crit. Care Med. 2019, 47, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Robert Boter, N.; Mòdol Deltell, J.M.; Casas Garcia, I.; Rocamora Blanch, G.; Lladós Beltran, G.; Carreres Molas, A. Activation of a Code Sepsis in the Emergency Department Is Associated with a Decrease in Mortality. Med. Clin. 2019, 152, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Messina, A.; Cecconi, M. Lactate in Critically Ill Patients: At the Crossroads Between Perfusion and Metabolism. Annu. Update Intensive Care Emerg. Med. 2019, 2019, 199–211. [Google Scholar] [CrossRef]
- Karlsson, A.; Stassen, W.; Loutfi, A.; Wallgren, U.; Larsson, E.; Kurland, L. Predicting Mortality among Septic Patients Presenting to the Emergency Department-a Cross Sectional Analysis Using Machine Learning. BMC Emerg. Med. 2021, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; Angelotti, G.; Caruso, P.F.; Zanella, A.; Stomeo, N.; Costantini, E.; Protti, A.; Pesenti, A.; Grasselli, G.; Cecconi, M. Outcome Prediction during an ICU Surge Using a Purely Data-Driven Approach: A Supervised Machine Learning Case-Study in Critically Ill Patients from COVID-19 Lombardy Outbreak. Int. J. Med. Inform. 2022, 164, 104807. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Carman, M. Improving Scalability and Performance of Random Forest Based Learning-to-Rank Algorithms by Aggressive Subsampling. In Proceedings of the 12th Australasian Data Mining Conference (AusDM 2014), Brisbane, Australia, 28–29 November 2014. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).