1. Introduction
Clustering is an unsupervised learning method that seeks to partition objects in a dataset into several natural groupings called clusters such that objects within a cluster tend to have similar attributes while objects belonging to different clusters have dissimilar attributes. In other words, clustering methods that can produce self-similar groups characterized by intra-group homogeneity and heterogeneity across groups, tend to create good natural partitions of the dataset. This is conceptually different from supervised classification or discriminant analysis where labeled data is used to train a classifier that is eventually used to classify unlabeled data. Clustering is useful in several exploratory data analyses, visualization, decision making, and machine-learning applications such as data mining, image segmentation, and pattern recognition. There are several ways to categorize different clustering methodologies, one of the most common is categorizing clustering methods as hierarchical clustering or partitional clustering methods [
1]. Hierarchical clustering methods organize data into hierarchical structures of partitions starting from singleton clusters (each object in the data is its own cluster) to one cluster over the entire data and every structure in between. Partitional clustering methods, on the other hand, produce a single partition of data for a pre-specified number of clusters based on the optimization of a pre-determined clustering criterion. While defining a suitable proximity measure (or a distance measure) is all that is often required for hierarchical clustering, partitional clustering methodologies require specifying clustering criteria (objective functional), number of clusters sought, and other parameters depending on the type of algorithm being used.
We focus on fuzzy clustering in this paper. Unlike hard clustering where an object belongs to a single cluster, the notion of graded belongingness of objects to all clusters is key to fuzzy clustering [
2]. The most widely used fuzzy clustering algorithm is fuzzy c-means [
3,
4] which is the fuzzy equivalent of the hard
k-means algorithm. It uses a membership function grade to associate each object in the data to clusters. The clusters themselves are defined as cluster centers or cluster prototypes or simply centroids (a centroid is an object instance which is most central to the cluster being described). Objects are assigned to the clusters based on their membership in the cluster. The alternating optimization (AO) algorithm is the most popular implementation scheme for FCM [
5]. The prototypes are initialized, either randomly or procedurally. At each optimization step, the partition memberships and the prototypes are updated, until a pre-defined stopping criterion is met such as when prototypes have stabilized. The number of clusters to be found is defined
a priori. However, FCM is sensitive to the initialization of prototypes and the fact that in many applications it is difficult to
a priori define the number of clusters to be found.
A novel method to find initial cluster prototypes (initialization) while concurrently determining the optimal number of clusters to be found, is presented in this paper. Contributions of this paper are as follows.
- (1)
A subset method is introduced to create the best subset of the dataset which still preserves the underlying structure of the original dataset. This is performed using a measure based on sparse sampling of the data, a concept widely used in clusterability studies of datasets. The reduced subset is then used instead of the original dataset to initialize cluster prototypes and find the optimal number of clusters.
- (2)
A coevolutionary scheme is presented which evolves candidate solutions that encode for both cluster prototypes as d-dimensioned real-valued numbers as well as the number of clusters. A multi-population differential evolution algorithm is presented with each population using a randomly assigned variant of the differential evolution algorithm. An elite population that does not evolve candidate solutions directly but participates in the evolution of the other populations is fundamental to the proposed scheme.
- (3)
A cluster-validity index is used as the fitness function to guide the evolution. The cluster validity index is calculated by first computing memberships using the information encoded in the evolutionary candidate vector and then using the memberships to find the index as a single-step operation. Using the reduced subset method, the implementation is less resource intensive compared to using the entire dataset.
The rest of the paper is organized as follows. We start with providing a background of FCM with a review of recent literature in the determination of the optimal number of clusters and the methods for cluster prototype initialization in
Section 2. In
Section 3 we present the subset selection method, which is central to the proposed method, followed by the multi-population coevolutionary differential evolution framework in
Section 4. In
Section 5, we present simulation results using synthetic data and datasets from the University of California Irvine’s Machine Learning (UCI ML) repository. In
Section 6, we present a case study with a real-world dataset from the domain of machine health monitoring. In
Section 7, we conclude the paper and present some directions for future research.
2. Background
Fuzzy c-means or FCM is a partitional clustering algorithm based on the notion of fuzzy membership of objects in a dataset to a cluster. FCM is useful when the natural partitions in a dataset are not evident or very well defined. A data object
xk has a certain membership
uik (which takes values between zero and one, not including 0 or 1) in a cluster
Ci, which is seen as the partial (fuzzy) belongingness of the data point to that cluster, subject to the constraint that the sum of memberships across all clusters is unity and the contribution of memberships of all data points to any particular cluster is always less than the cardinality of the dataset
n (
k = 1,2,…,
n).
The fuzzy sum-of-squared-error objective function is the least squares estimator function,
The exponent
m is called the fuzzifier which determines the fuzziness of the partition,
and
is the distance between cluster prototype
and data object
where
A is the norm matrix. The identity norm matrix
A =
I yields Euclidean distance and results in spherical clusters while other norms are used for elliptical clusters, etc. In this paper, the Euclidean distance has been used. The FCM-AO minimizes the functional in Equation (2) by iteratively calculating cluster prototypes and updating memberships until there is no change in the termination criterion, usually as measured by a change in memberships between two successive iterations.
The equations presented for FCM are from [
4]. The algorithm often starts by initializing cluster prototypes randomly, then calculating memberships using Equation (4), followed by recalculating prototypes using Equation (3) until convergence. The final membership matrix
is sensitive to the initialization of cluster prototypes. In addition, the iterative optimization scheme assumes that the number of clusters
c is known. The only parameter to tune is the fuzzifier
m which depending on the fuzzification required is often set to
m = 2 for most clustering problems (especially one involving spherical clusters).
2.1. Determination of Optimal Number of Clusters
For two-dimensional datasets, the number of clusters can be determined by simple visualization. For higher dimensional data, the dataset can be mapped to a two-dimensional plane using dimensionality reduction techniques such as principal component analysis (PCA), kernel, locally linear embedding (LLE), diffusion maps, and sparse dictionary representations. These techniques do not, however, provide a mapping that conserves the internal partitional structure of the dataset. More popular are cluster validity methods that quantify the goodness of a partition after the clustering method produces the partition. Different partitions for different values of c are created and then the goodness of partition is compared using a measurable index. The value of the index reaching a maximum or minimum or an inflection point is often a good indicator of the goodness of partition and the corresponding number of clusters is the optimal number of clusters. We use several of these cluster validity measures in this paper. However, these are used not to determine an optimal value for c, but instead to compare the performance of different algorithms.
The first cluster validity index was proposed by Zadeh called the degree of separation which was later refined as the concept of partition coefficient and the closely related partition entropy by Bezdek [
6]. Lee proposed a fuzzy clustering validity index using the distinguishableness of clusters measured by the object proximities [
7]. Based on Shannon entropy and fuzzy variation theory, Zhang and Jiang proposed a fuzzy clustering validity index taking into account of the geometry structure of the dataset [
8]. Saha et al. presented an algorithm based on differential evolution for automatic cluster detection, which evaluated the validity of the clustering result [
9]. Yue et al. partitioned the original data space into a grid-based structure and proposed a cluster separation measure based on grid distances [
10]. Based on the idea that a natural partition is one that creates well-separated compact clusters, measures of compactness and separation were proposed as part of several cluster validity measures. These include Fukuyama–Sugeno index
FS [
11], Xie–Beni index
XB [
12], Bensaid index
SC [
13], Tang index
VT [
14], Kwon index
VK [
15],
PBMF index [
16],
PCAES index [
17],
WL index [
18], Wang index
VW [
19],
CWB index [
20], and Zhu index
Vz [
21].
Since these measures compare and quantify the goodness of the partition after the partition is generated, they are not practical for problems involving large datasets (high cardinality
n) or high number of dimensions
d. Lately, some new clustering algorithms have been proposed that adjust the number of clusters while the partition is being generated. Competitive agglomeration [
22] and split hierarchical clustering [
23] have been used to guide the optimization of the number of clusters in a partitional clustering process. A similarity-based clustering method that combines single-point iteration with hierarchical clustering to determine the number of clusters is proposed in [
24]. A Mercer kernel-based clustering [
25] estimated the number of clusters by the eigenvectors of a kernel matrix. A clustering algorithm based on maximal
θ-distant subtrees [
26] detected any number of well-separated clusters of any shape.
2.2. Initialization of Cluster Prototypes
In most implementations of FCM-AO, the cluster centers are randomly initialized. The random initialization affects the accuracy and running time of the algorithm, especially in datasets where natural clusters overlap. Based on the initial choice of cluster prototype, the algorithm can either quickly converge or get trapped in local minima. A subtractive clustering algorithm has been used to find the initial cluster centers in [
27,
28]. However, there are several parameters that need to be set to get the desired outcome, which are not trivial. A cluster prototype initialization based on a density cluster algorithm is presented in [
29]. A fuzzy entropy algorithm is proposed with a hybrid FCM algorithm to initialize cluster centers in [
30]. An improved FCM algorithm based on morphological reconstruction and affiliation filtering (FRFCM) which can determine the number of clusters while optimizing initial cluster prototypes has been presented in [
31].
Metaheuristic methods such as evolutionary algorithms including genetic algorithms [
32,
33], particle swarm optimization, and differential evolution [
34] have been used for cluster prototype initialization. A detailed recent review of metaheuristics used to solve the problem of cluster prototype initialization for FCM in the area of image segmentation can be found in [
35]. A single population differential evolution algorithm called automatic differential evolution-based fuzzy clustering (ADEFC) was used to optimize initial cluster prototypes and number of clusters in [
9]. The multiple population genetic algorithm (MPGA) to optimize cluster prototypes using multiple evolving populations was proposed in [
36]. Each subpopulation is allowed to evolve independently; however, a migration operator is used to relate the individually evolving populations. Recently, the derivative multi-population genetic algorithm (DMGA) was proposed in [
37] which first initializes the population by using a derivative operator before each subpopulation is evolved using canonical genetic operators. The probabilities of the genetic operators are dynamically selected by an adaptive probability fuzzy control operator. The quality of the initial cluster prototypes is shown to be superior to those found using MPGA. However, these methods are very resource intensive for big datasets.
3. Subset Selection
A co-evolutionary algorithm recognizes the diversity of the possible candidate solutions in the population. It emphasizes the correlation between the domain of the problem being solved and the candidate solutions. Evolutionary algorithms are prone to get trapped in local optima, and performance depends on fine tuning a few or many parameters. Co-evolutionary systems, on the other hand, have been shown to have the ability to avoid local optima by dividing the solution space and also use different evolutionary strategies to support either a competitive system or a cooperative system. Their strength lies in the divide-and-conquer decomposition strategy and in their implicit parallelism. These algorithms can be divided into competitive co-evolutionary systems and cooperative co-evolutionary systems and include systems such as co-evolution of predator and prey systems, competing species, among others [
38,
39,
40]. These systems can balance the exploration and exploitation capability by utilizing cooperative or competitive mechanisms among different subpopulations using different strategies for different subpopulations. They can also improve convergence properties (not necessarily speed) by facilitating information exchange among the subpopulations.
There are two important reasons for using coevolutionary algorithms for clustering problems: (1) evolution of candidate solutions on a subset of the larger dataset improves the convergence properties of the individual subpopulation. This is because a judiciously selected subset of a larger dataset can capture the structure of the larger dataset and since it is smaller than the original dataset, an evolutionary algorithm can converge faster. (2) Coevolution, both competitive and cooperative, ensures that excellent candidates which in a single-strategy one-population evolution may not be able to evolve for several generations which is an implicit strain on resources, now have a better chance of getting selected for evolution.
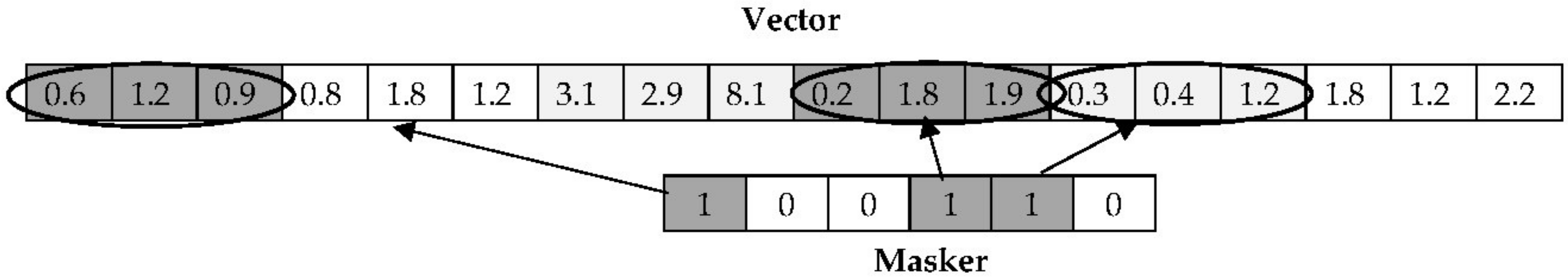
The proposed co-evolutionary framework is described here. The dataset of size
n to be clustered is first divided into SP distinct subsets such that in each subset there are
n/SP objects. If
n is not evenly divisible by SP, then the largest number <
n evenly divisible by SP is used. For example, if
n = 528 and SP = 10, we select 520 objects at random from the dataset and divide them into 10 subsets with 52 objects in each. Every subset is mutually exclusive so that a data object is available to only one subset. This ensures an even coverage of the entire dataset. The algorithm to create mutually exclusive subsets of the data is shown as Algorithm 1.
| Algorithm 1: Dividing dataset into subsets |
Input: Dataset data of size n and dimensions d: X = {x1, x2, … xn}, where xj = {xj1, xj2, … xjd}, SP
Calculate rem = remainder(n/SP). If rem ! = 0, discard rem datapoints randomly from data and retain (n − rem) datapoints as data For k = 1 to SP Select (n − rem)/SP datapoints at random from data to populate subset Pk Remove the same (n − rem)/SP points from data: data = data − Pk End
Output: SP Subsets P = {P1, P2, … PSP}, rem |
Of these distinct datasets, only the one with a similar underlying structure as the original dataset is then selected. This is performed by employing a sparse sampling statistic based on a test of spatial randomness. Sparse sampling tests are based on sampling origins randomly assigned in a sampling window and the underlying structure of the data can be quantified by nearest neighbor measurement of sampling origins to points and comparing them to the random nearest neighbor measurement of paired points within the sampling window. Several tests involving sampling origins have been proposed in the literature, mostly from the field of biological statistics, based on a multitude of tests such as the Hopkins [
41,
42], Holgate [
43], T-square [
44], Eberhardt [
45] and Cox-Lewis [
46]. These have been used as a test of clusterability, clustering tendency, and cluster validation [
47,
48]. In this paper, we use the Hopkins statistic which is by far the easiest of the statistical measures to implement. For a dataset
X of size
n in
d-dimensions,
n0 sampling origins
Y are placed at random in a sampling window such that
n0 <<
n. The sampling origins
Y are
d dimensional as well. Two types of distances are defined:
uk is the distance of the sampling origin
yk to its nearest datapoint in
X, and
wk is the distance of a randomly chosen datapoint
xk to its nearest neighbor in
X. The Hopkins statistic [
41] is defined as,
The statistic, therefore, compares the nearest-neighbor distribution of randomly selected locations in the dataset, to the nearest-neighbor distribution among objects in the data. If the underlying structure of dataset
X is random, then on an average the sum of
uk would be equal to the sum of
wk over
d dimensions and therefore
H will be close to 0.5. However, if dataset
X has distinct separable clusters (without any notion of how many clusters there are), then on an average the sum of
uk will greatly exceed the sum of
wk and therefore
H will be very close to unity. In most cases, real-world datasets will fall somewhere within this spectrum of complete underlying randomness and distinct separable clusters—as a result, Hopkins statistic
H will range between 0.5 and 1.0 for most datasets. If it is close to 0.5, the dataset is more random than clustered, and if close to 1.0 then there are distinct separable clusters. It is easy to see why the Hopkins statistic has been so popular for use as a measure of clustering tendency (the question of whether a dataset has an underlying cluster structure). In this paper, we use the statistic simply as a quantification of the underlying structure and compare it to the underlying structure of the subsets. The subset that under repeated spare sampling has a Hopkins statistic value close to that of the entire dataset most likely captures the underlying structure of the dataset. This is shown as Algorithm 2 below.
| Algorithm 2: Identifying subset with similar underlying structure to the original dataset |
Input: Dataset data of size (n − rem), SP subsets P = {P1, P2, … PSP} each of size (n − rem)/SP, number of sampling origins n0 for data, convergence threshold εPopulate n0 datapoints at random in data Calculate Hopkins statistic H for data diff = ε, best_subset = Ø Repeat For k = 1 to SP Populate n0/SP datapoints at random in Pk Calculate Hopkins statistic Hk for subset Pk If |H − Hk| < diff, diff = |H − Hk| and best_subset = k End Until (best_subset ! = Ø)
Output: Subset Pbest_subset |
5. Simulation Results
The proposed method is compared to some standard algorithms from the literature and their parameters used in this study are listed below.
We do not present these algorithms in detail here and the reader is referred to the original papers. The proposed algorithm will henceforth be referred to as Cooperative DE-FCM or CDE-FCM. The parameters are SP = 10, number of subpopulations = 9 (one for each instance of DE), generations
G = 20, number of candidate vectors in each subpopulation
p = 15. The individual parameter settings of the DE variants are listed in
Table 1 and
Table 2. We compared the performance of these algorithms using cluster validity measures listed in
Section 4.5 on two synthetic datasets and four datasets from the UCI ML repository [
62]. All algorithms are implemented using MATLAB 2022a on an Intel
® Core™ i7-8650U CPU with 8 cores at 1.90 GHz.
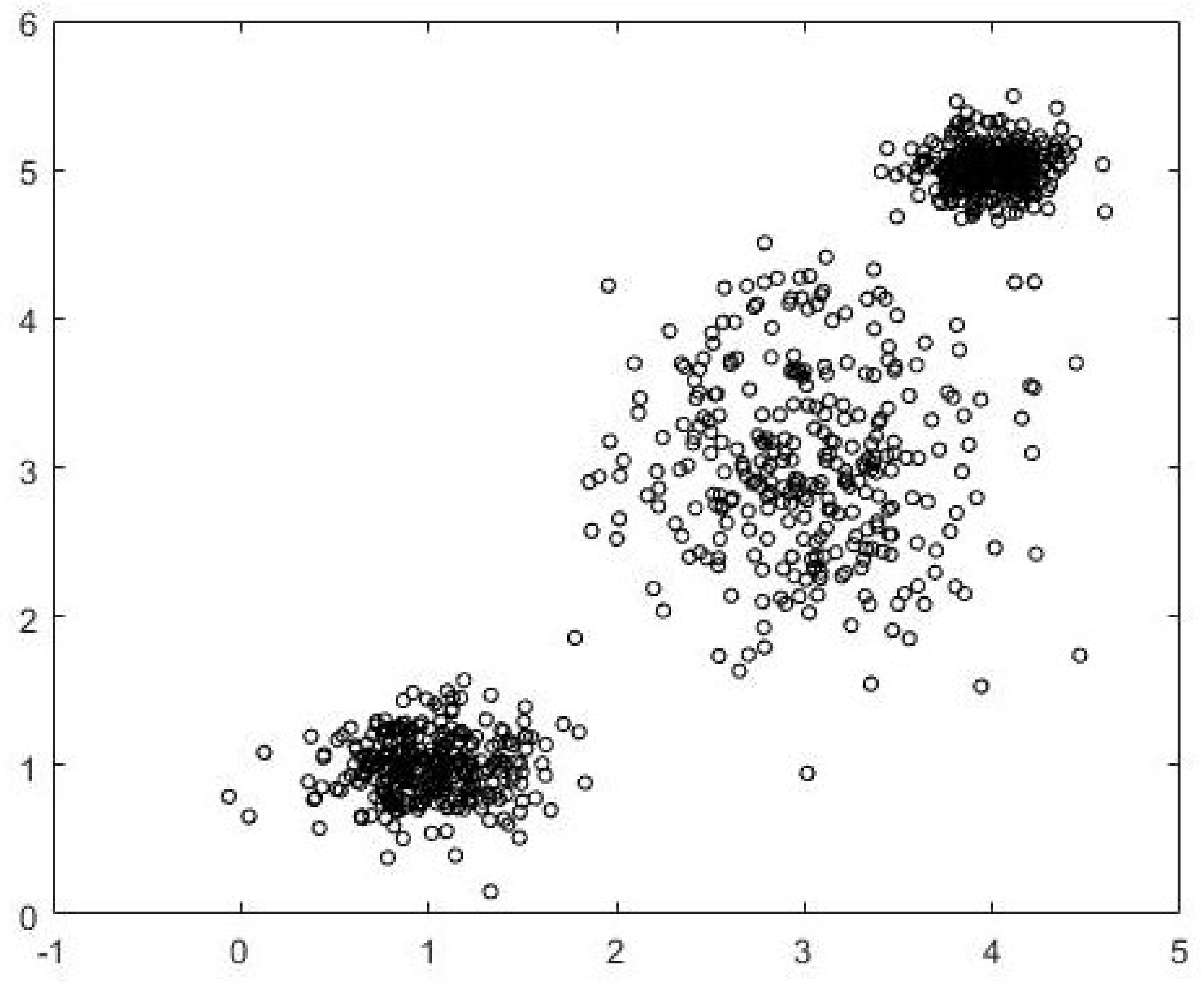
Data1 is a 900-point dataset in two dimensions. There are three well-separated and compact clusters with 300 data points in each cluster. The clusters are generated using normal distributions centered at (1,1), (3,3), and (4,5). The dataset is shown in
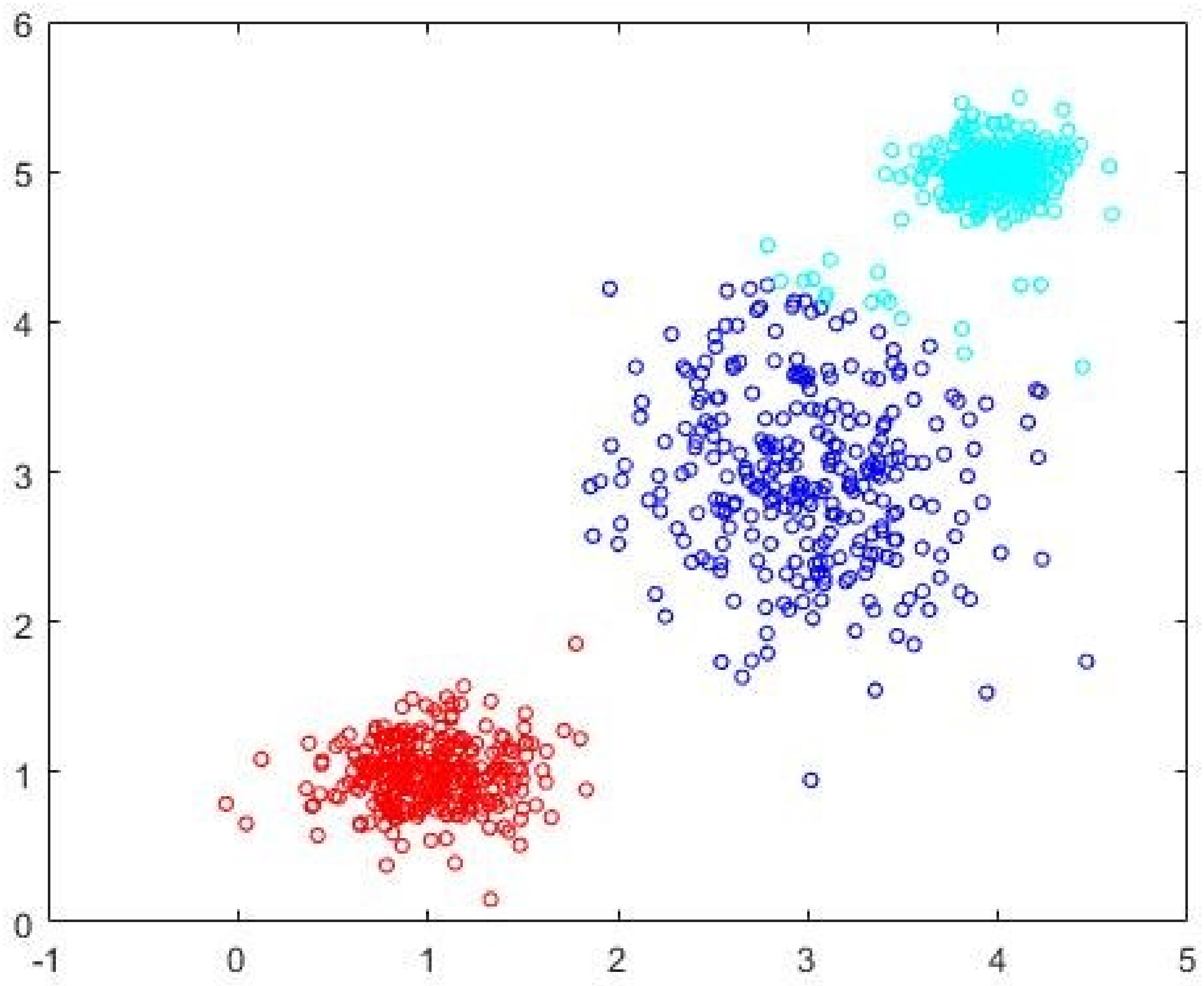
Figure 2. The three-cluster FCM-AO partition is shown in
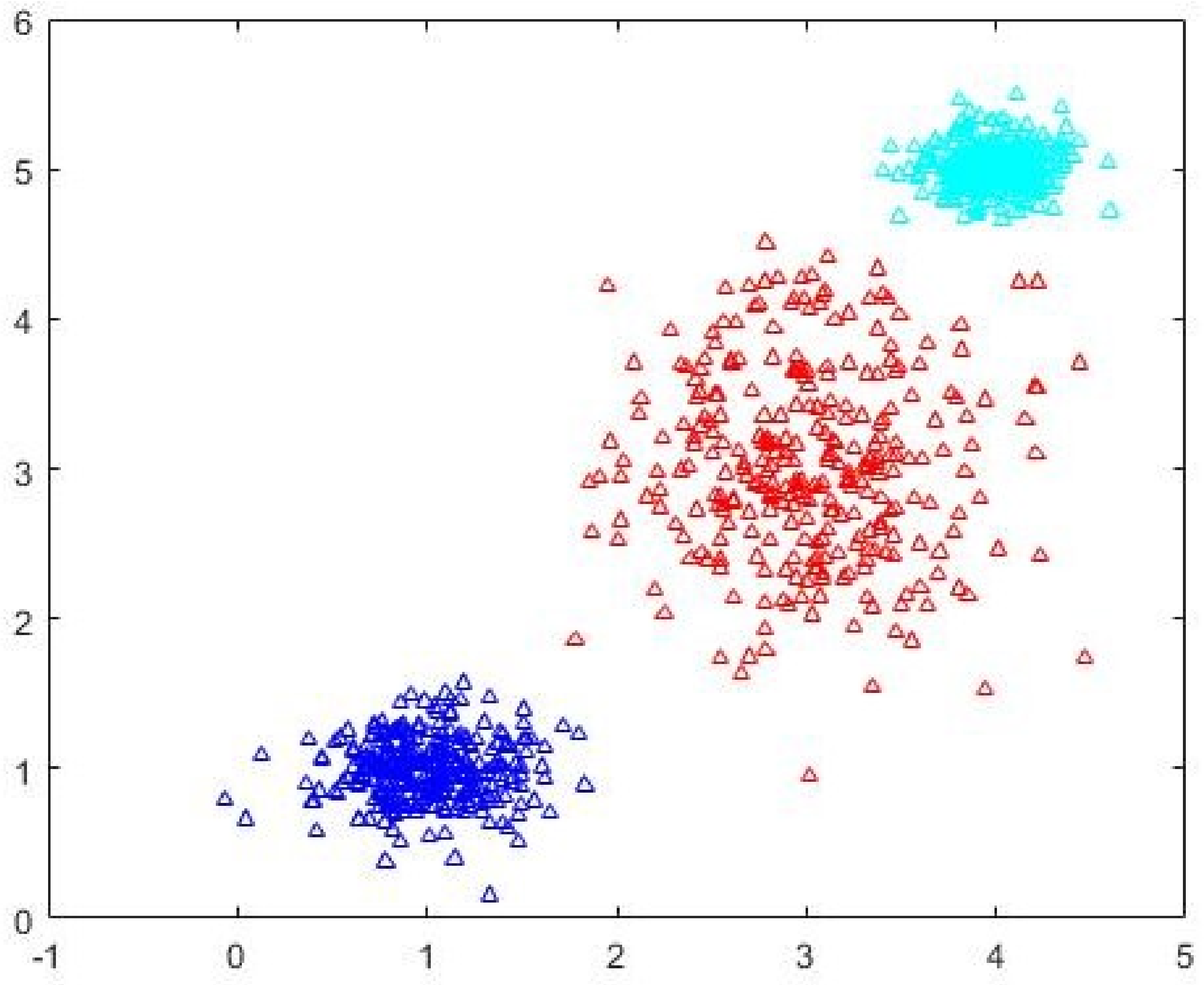
Figure 3 and the three-cluster CDE-FCM after cluster prototype initialization is shown in
Figure 4. The quality of the CDE-FCM can be seen to be marginally better than randomly initialized FCM-AO.
FCM-AO, PSO-V, GAKFCM, and EwFCM are run for
c = 2 to
c = 30. For ADEFC and CDE-FCM the vector representation is of length 60 with
d = 2 and
cmax = 30. The run-time parameters are chosen such as the convergence criteria for all algorithms are roughly the same. For comparison, the run-time of the different algorithms is presented in
Table 3. As can be seen, there is not much to be gained by using sophisticated schemes to address the problems of prototype initialization and unknown number of clusters for a small
well-separated dataset as Data1. The naïve version of FCM-AO with random initializations works as well as any other scheme and in a fraction of the time.
Data2 is a 2000-point dataset in two dimensions. Unlike
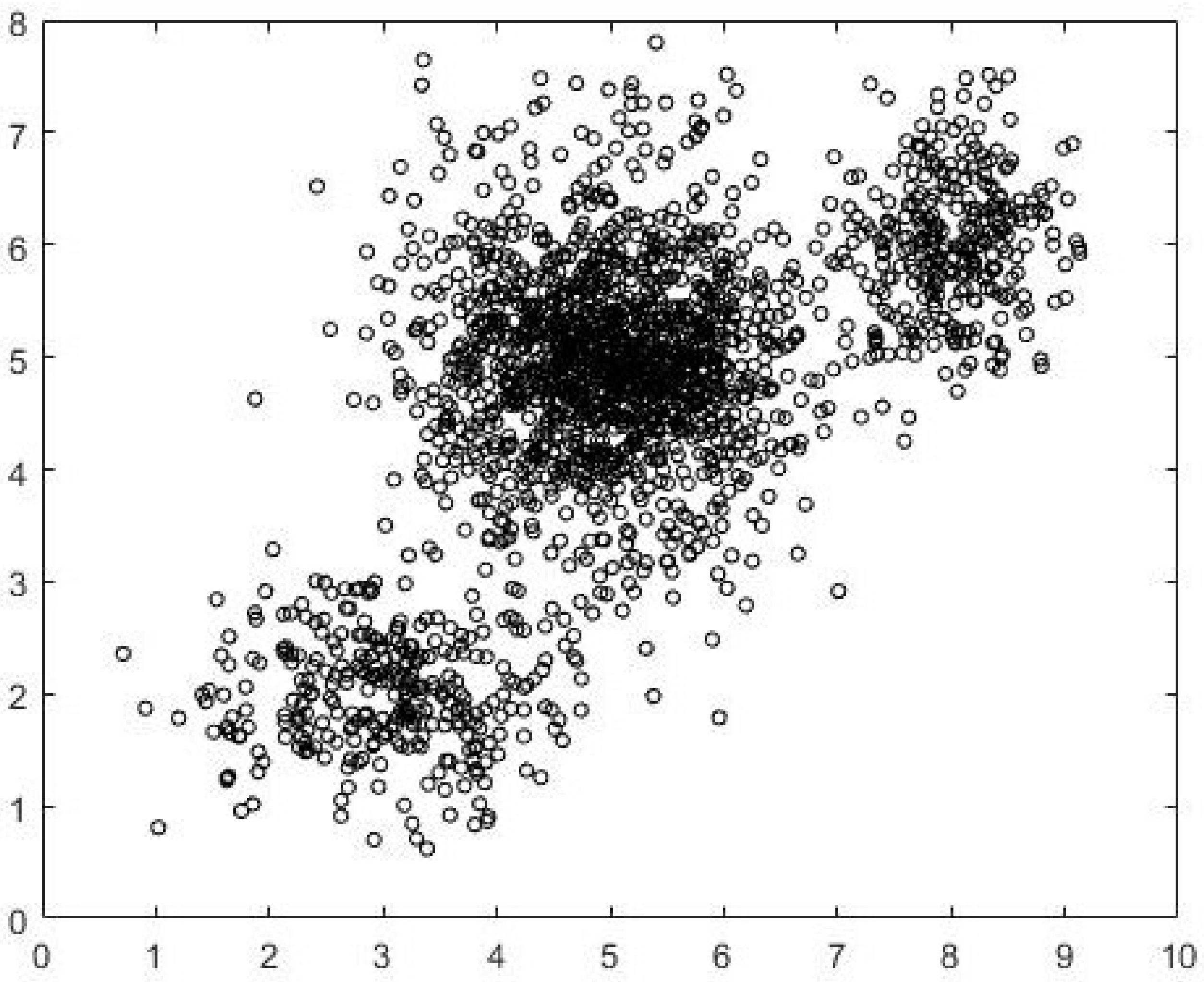
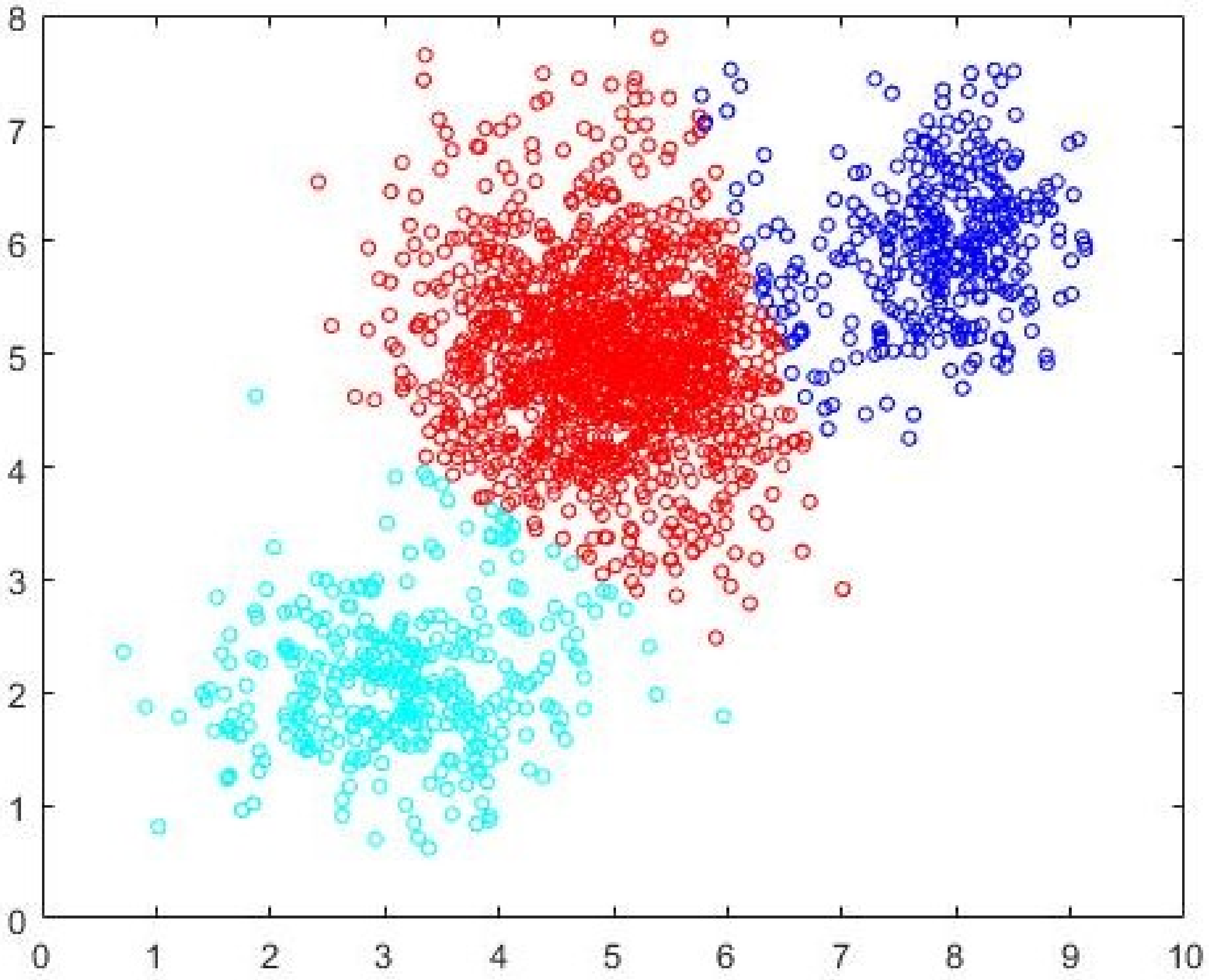
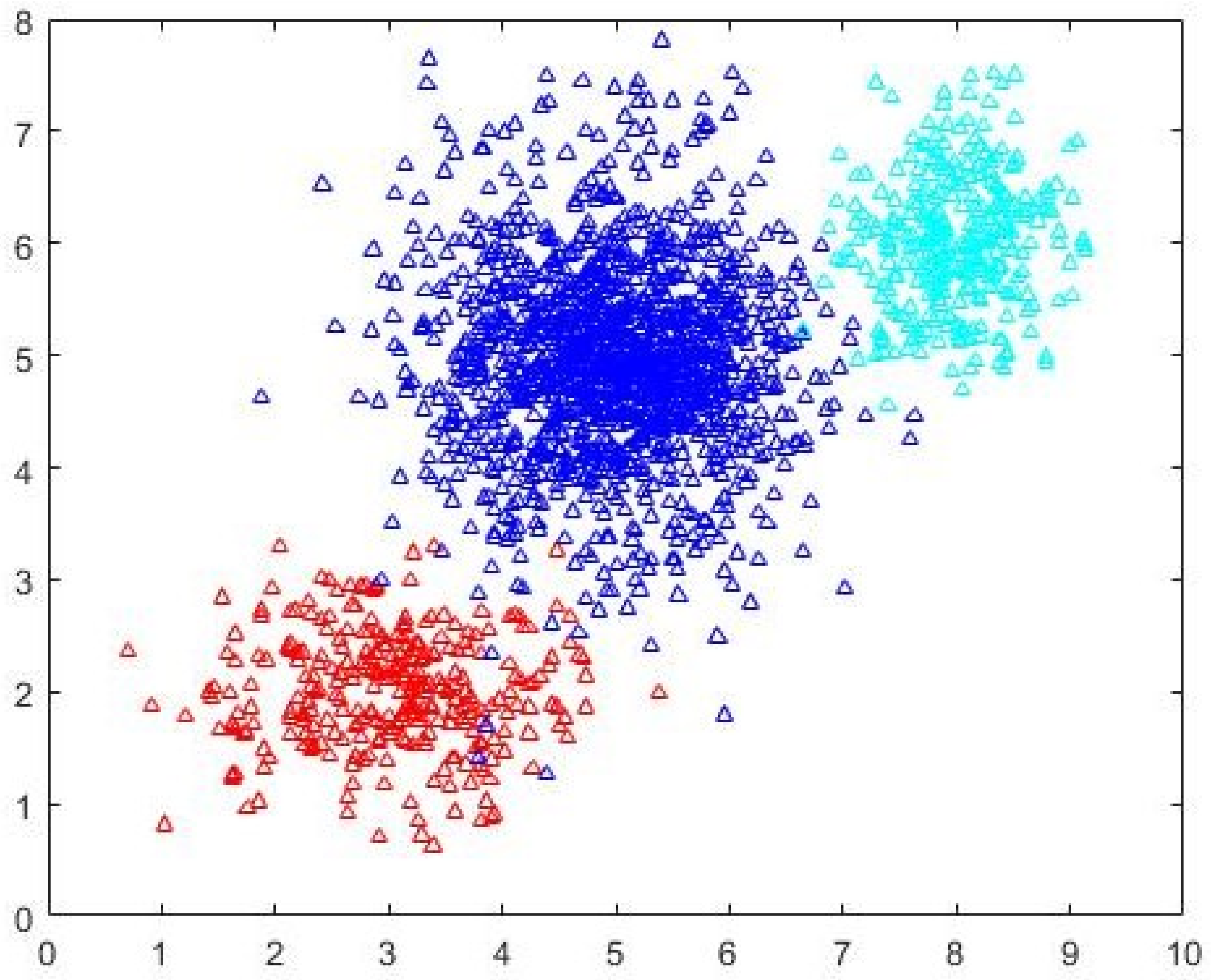
Data1, the separation between the clusters is not well defined. The data is generated using three normal distributions centered at (3,2), (5.5,5.5) and (8.5,6). The intercluster variance is greater than that in
Data1. The dataset is shown in
Figure 5. FCM-AO, PSO-V, GAKFCM and EwFCM are run for
c = 2 to
c = 45. For ADEFC and CDE-FCM, the vector representation is of length 90 with
d = 2 and
cmax = 45. FCM-AO, GAKFCM, and EwFCM identify
c = 5 as the most-optimal cluster. On the other hand, the evolutionary algorithm-based approaches, viz. PSO-V, ADEFC and the proposed CDE-FCM correctly identify the
c = 3 partition. In fact, the terminating elite population in CDE-FCM has 22% individuals that encode for
c = 3 and 10% that encode for
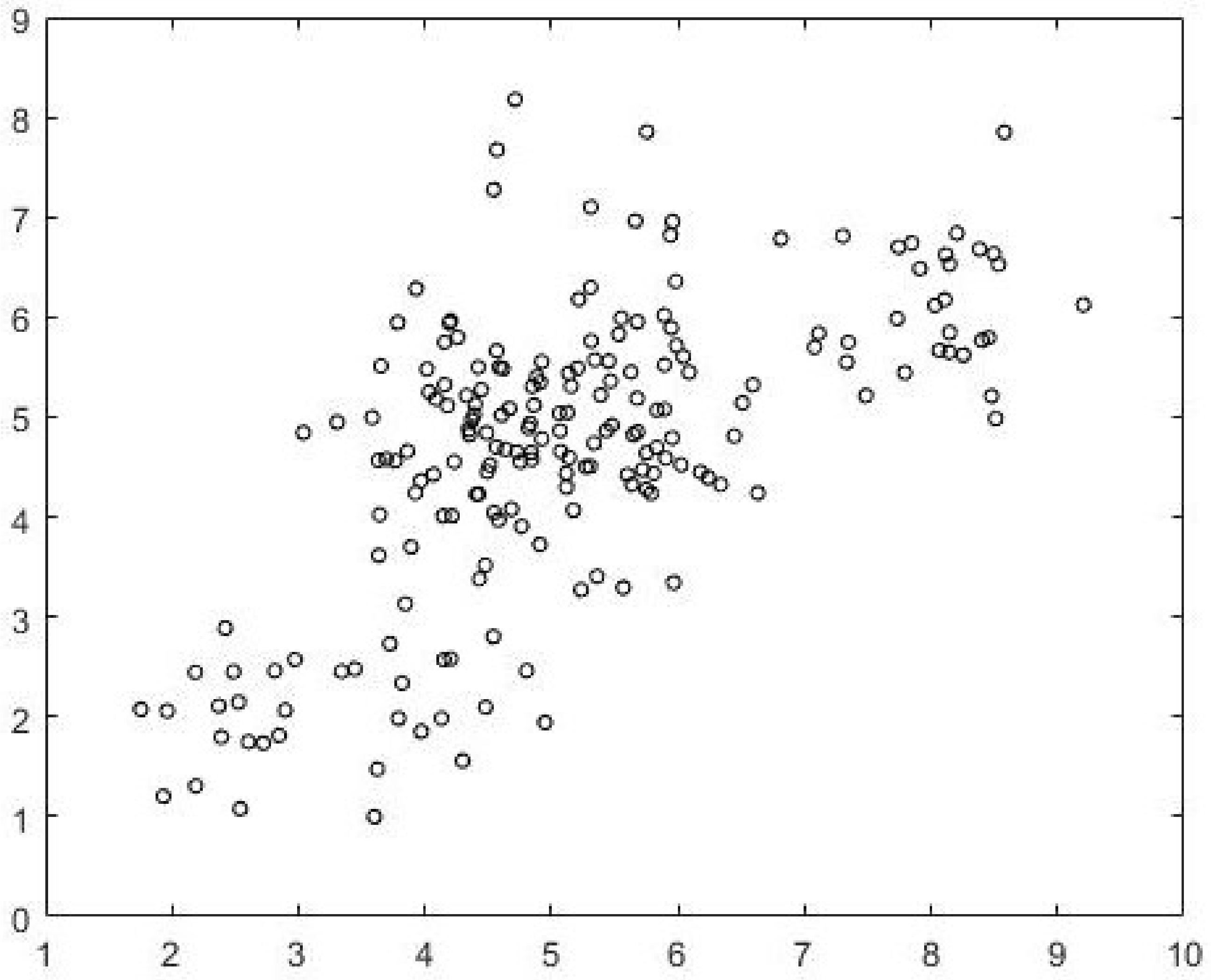
c = 5. The best subset of cardinality 200 as identified by Algorithm 2 is shown in
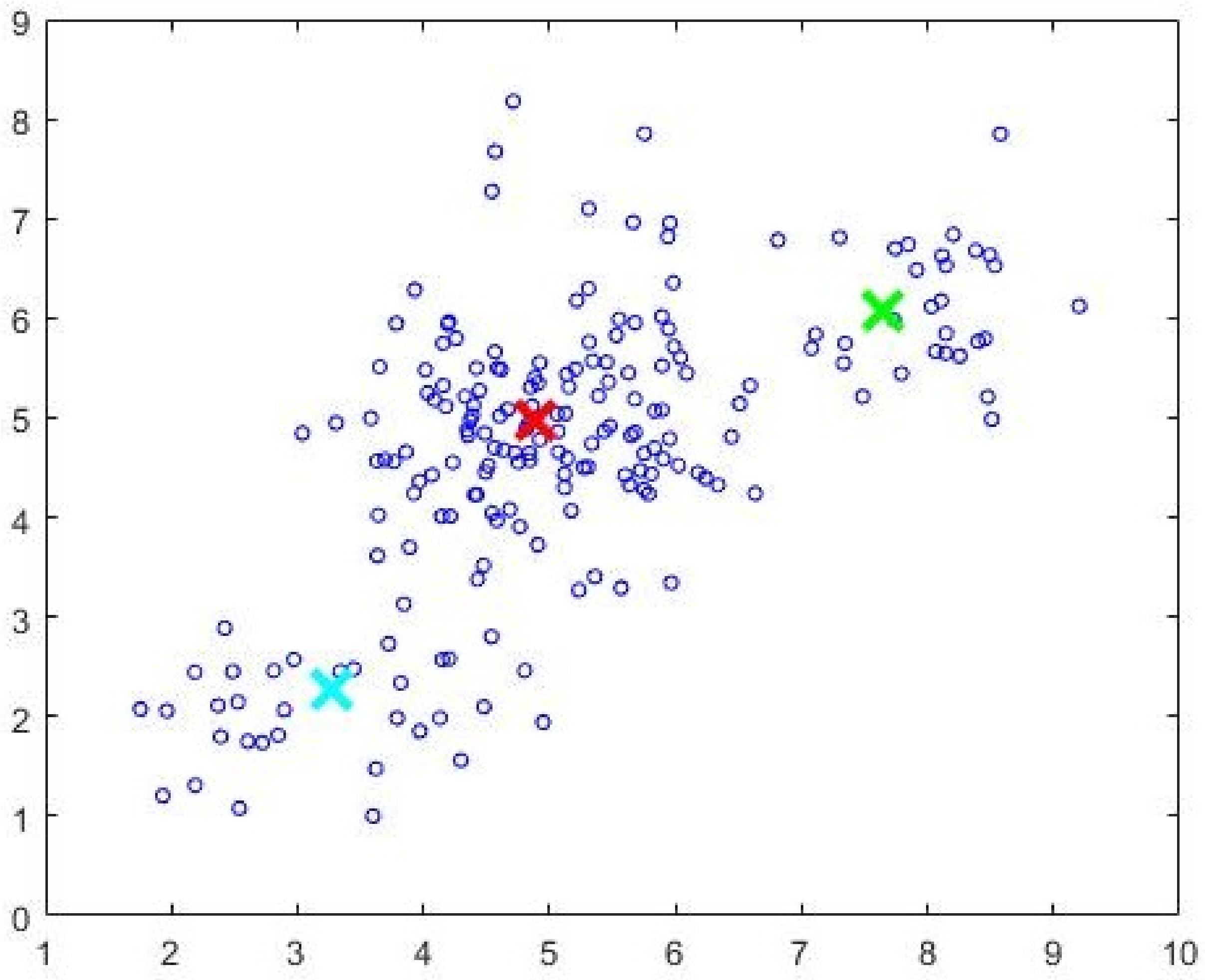
Figure 6 and the initial cluster prototypes evolved by the CDE (Algorithm 3) are shown in
Figure 7. The
c = 5 partition as identified by FCM-AO as the most optimal partition is shown in
Figure 8 and the
c = 3 partition as identified by FCM-AO is shown in
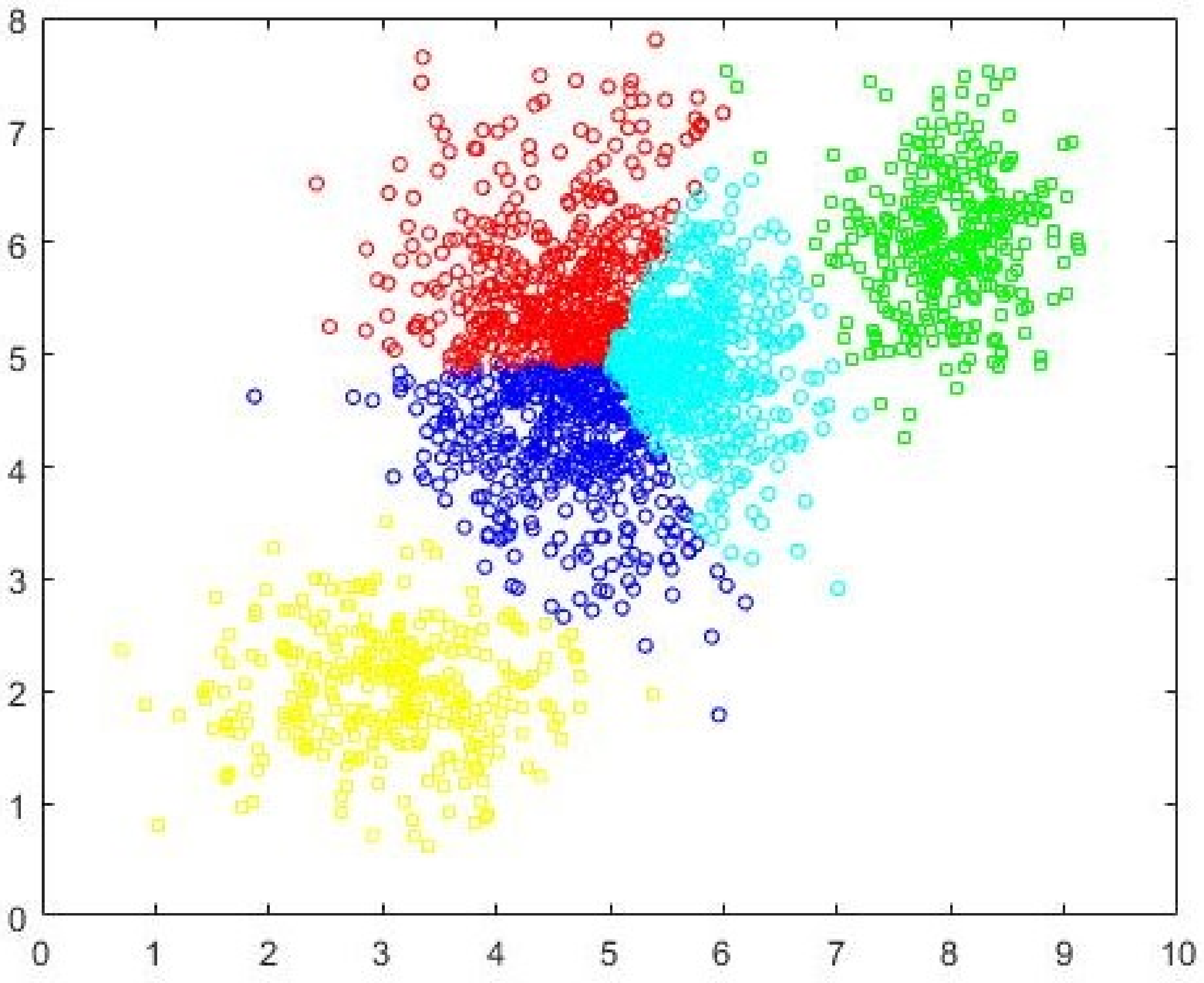
Figure 9. The correct
c = 3 partition as identified by CDE-FCM with evolved initial prototypes is shown in
Figure 10. By simple visual inspection, it can be said that the quality of the partition in
Figure 10 is better than that in
Figure 9 (same
c = 3). The fact that FCM-AO with randomly initialized cluster prototypes identifies
c = 5 as better than
c = 3 on the basis of all cluster validity measures shows the promise of the proposed method. After the initial cluster prototypes are identified by Algorithm 3, the FCM-AO converged on an average in 4.5 iterations over 10 implementations. This is in comparison to the naïve FCM-AO with random initialization which took at least 25 iterations for an average of 28 iterations with the same convergence criterion of
ε = 0.001.
Iris data consists of
n = 150 datapoints divided into three types of Iris flowers-Setosa, Virginica, and Versicolor with 50 samples in each class. Each sample has four associated features: sepal length, petal length, sepal width, and petal width. FCM-AO, PSO-V, GAKFCM and EwFCM are run for
c = 2 to
c = 12. For ADEFC and CDE-FCM the vector representation is of length 48 with
d = 4 and
cmax = 12. The performances of different algorithms as measured by the cluster validity criteria are listed in
Table 4. Two classes (Versicolor and Virginica) are known to be linearly inseparable from each other although Setosa is linearly separable from the other classes [
63] and therefore many algorithms identify the
c = 2 solution as the most optimal one. Almost 20% of the candidate vectors in the terminating elite population of CDE-FCM encoded for
c = 2 while 18% encoded for
c = 3 with an equal number encoding for
c = 9. The cluster prototypes identified by CDE-FCM in almost all candidates (>85%) in the terminating elite population are almost optimal, and a further application of FCM-AO converges on average in 4.1 iterations over 10 independent runs of the algorithm. On the other hand, the best-performing instance of the naïve FCM-AO with random initializations took 22 iterations.
Cancer data has 683 datapoints in two classes (malignant and benign). The data has 9 features (attributes)—clump thickness, cell size uniformity, cell shape uniformity, single epithelial cell size, bare nuclei, bland chromatin, normal nucleoli, and mitoses. There are 440 instances belonging to the benign cluster and 243 instances in the malignant cluster. The cancer dataset is known to be linearly inseparable and it is often difficult for clustering algorithms to achieve high levels of accuracy with this dataset. FCM-AO, PSO-V, GAKFCM and EwFCM are run for
c = 2 to
c = 26. For ADEFC and CDE-FCM the vector representation is of length 234 with
d = 9 and
cmax = 26. The performances of different algorithms as measured by the cluster validity criteria are listed in
Table 5. Little more than 18% of the candidate vectors in the terminating elite population of CDE-FCM encoded for
c = 2 while approximately 15% of candidate vectors encoded for
c = 8. The cluster prototypes identified by CDE-FCM in 72% of the candidates in the terminating elite population are almost optimal, and a further application of FCM-AO converges on average in 8.5 iterations over 10 independent runs of the algorithm. On the other hand, the best-performing instance of the naïve FCM-AO with random initializations took 31 iterations.
Glass data has 214 different glass samples with 9 features (refractive index, weight percent of corresponding oxide of Na, Mg, Al, Si, K, Ca, Ba, and Fe). There are 6 different types of glasses—building window (float), building window (non-float), vehicle window (float), container glass, tableware glass and headlamp glass. FCM-AO, PSO-V, GAKFCM and EwFCM are run for
c = 2 to
c = 15. For ADEFC and CDE-FCM the vector representation is of length 135 with
d = 9 and
cmax = 15. The performances of different algorithms as measured by cluster validity criteria are listed in
Table 6. Almost all cluster validity indices agreed, and all algorithms produced very similar partitions at either
c = 5,
c = 6, and
c = 8.
Wine dataset has 178 samples of wine differentiated by 13 attributes—percent contents of alcohol, malic acid, ash, magnesium, total phenols, flavonoids, nonflavonoid phenols, proanthocyanins, alkalinity of ash, color intensity, OD280/OD315 of diluted wines (protein content), and proline content. The wines are categorized into three classes (red, white, and rosé) with 59 instances in the first cluster, 71 in the second, and the rest in the third cluster. FCM-AO, PSO-V, GAKFCM, and EwFCM are run for
c = 2 to
c = 13. For ADEFC and CDE-FCM the vector representation is of length 169 with
d = 13 and
cmax = 13. The performances of different algorithms as measured by cluster validity criteria are listed in
Table 7. The clusters are well-defined, and all the cluster validity measures reach their minimum at either
c = 3 or
c = 4.
Wine Quality dataset has 1898 samples of wines divided into 11 classes (quality scores ranging from 0 to 10). The data are defined over 11 attributes—fixed acidity, volatile acidity, citric acid, residual sugar, chlorides, free SO
2, total SO
2, density, pH, sulphates, and alcohol content. This is the largest of the UCI datasets tested for this work both in terms of cardinality and dimensions. FCM-AO, PSO-V, GAKFCM and EwFCM are run for
c = 2 to
c = 45. For ADEFC and CDE-FCM the vector representation is of length 495 with
d = 11 and
cmax = 45. The performances of different algorithms as measured by cluster validity criteria are listed in
Table 8. FCM-AO, PSO-V, GAKFCM, and EwFCM identify at least 8 of the classes with moderately high accuracy. Accuracy decreases for
c = 11 as is evident from the cluster validity measures. ADEFC identifies
c = 10 clusters, with an accuracy below 90% while the proposed CDE-FCM identifies 12 clusters with initial cluster centers almost optimal for 8 of the real classes. The FCM-AO implemented with cluster prototypes identified by CDE-FCM converges on an average of 12 iterations over 10 independent runs, while FCM-AO randomly initialized converges after an average of 33 iterations for
c = 8 and 37 iterations for
c = 11.
6. Case Study—Rolling Bearing Fault Analysis
Rolling element bearings are commonly used in supporting rotor components and assemblies in rotating machinery. Bearing defects can lead to undesirable vibrations, noise, or machine failure. Bearing fault diagnosis has been a subject of great importance in machine condition monitoring, predictive maintenance, and machine failure prevention and analysis [
64]. Bearings conditions are presented in [
65]. Techniques used in fault severity evaluation in rolling bearings are reviewed and discussed in [
66]. The review is mainly focused on data-driven approaches such as signal processing for extracting the fault signatures associated with the fault degradation, and the approaches that are used to identify degradation patterns. Modern predictive maintenance techniques are increasingly adopting data analysis techniques such as pattern recognition and machine learning for bearing fault diagnosis. In this case study using the proposed cluster initialization method, wavelet analysis is used to process vibration signals from three bearing cases—no fault, inner race fault, and ball fault, under varying rotating speeds.
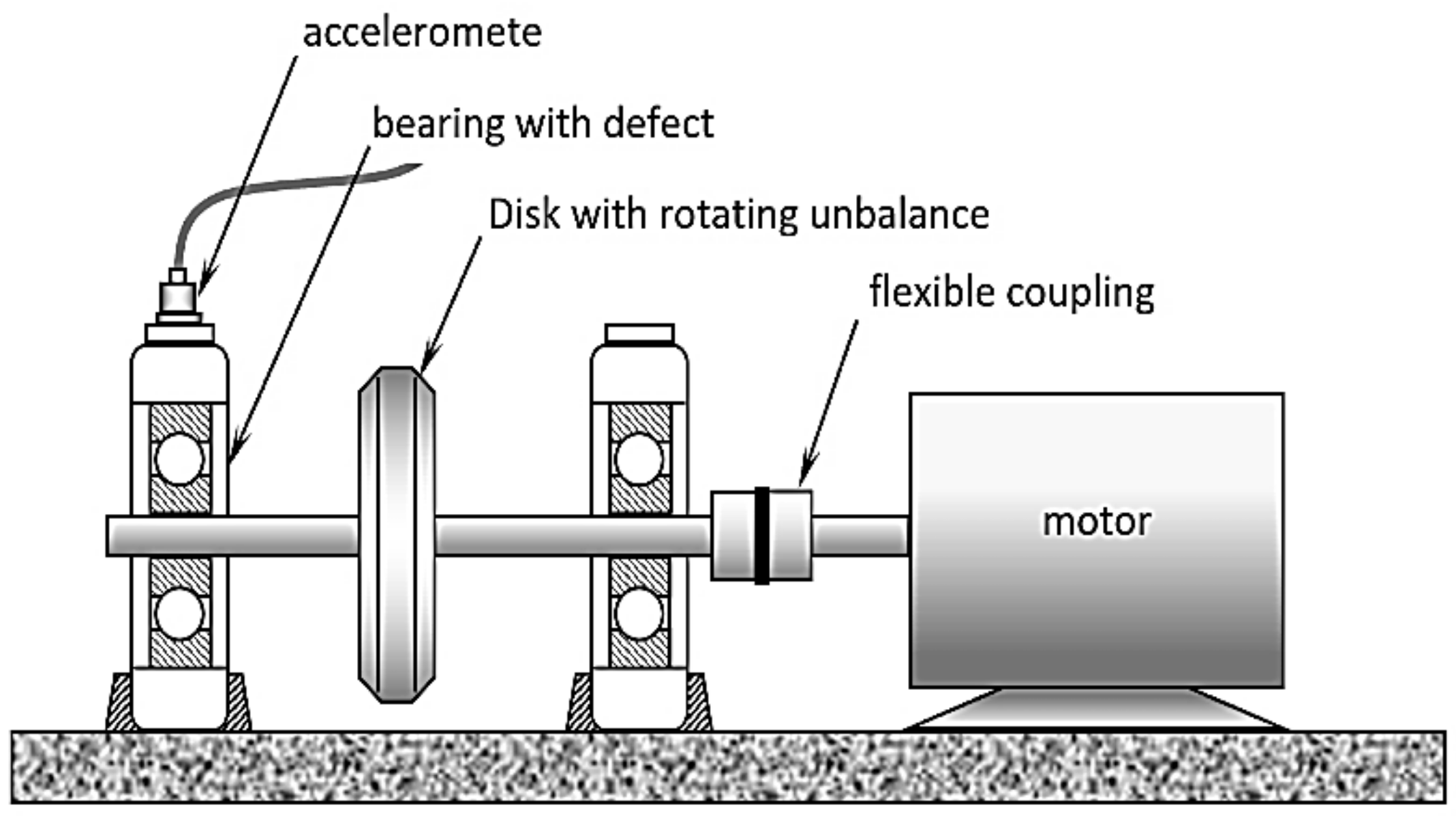
A schematic of the experimental setup is shown in
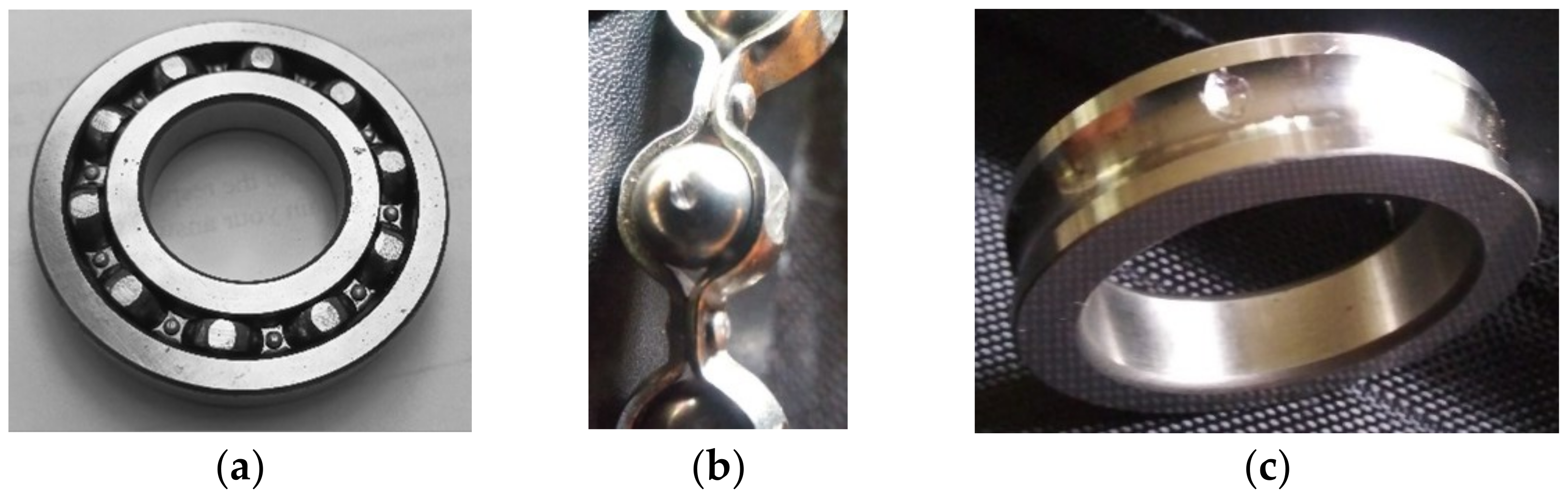
Figure 11. The faults are introduced as a single rough surface spot simulating pitting wear and are created by using a small grinder as shown in
Figure 12. The rotor is run at 10 different speeds (from 500 rpm to 1400 rpm) in increments of 100 rpm. At each speed level, vibration signals are acquired using a PCB accelerometer (model PCB 302A) which is mounted on the outboard test bearing as indicated in
Figure 10. The sampling rate is 10,000 samples/s. A radial load of about 2000 lb was kept constant during all tests. A small unbalance mass is added to the rotor to introduce sustained periodic vibration excitation.
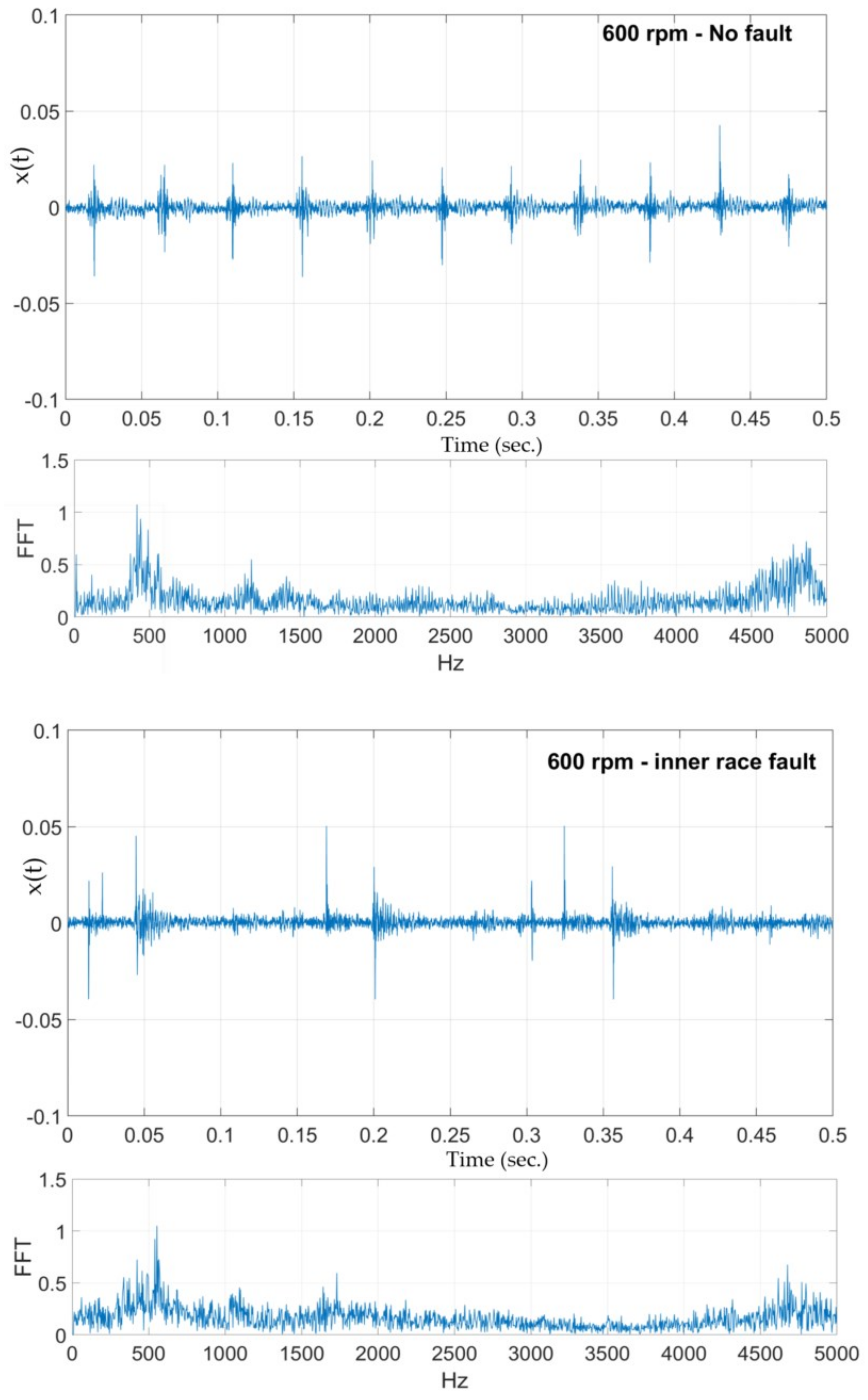
Three sets of samples are collected, each for 0.5 s. The analysis is performed for each of the three sets, followed by sets obtained using a 50% overlap between the non-overlapping sets resulting in five sample sets (3 non-overlapping sets and 2 overlapping sets) for a single operating condition—speed and type of fault (10 different speeds and 3 fault conditions including no fault). The raw time series is analyzed for statistical features such as mean, variance, standard deviation, skewness, and kurtosis. The times series are also subject to fast Fourier transform (FFT) and continuous wavelet transform (CWT) techniques.
Figure 13 shows two sample raw vibration time series and their corresponding frequency spectrum obtained using FFT. After experimenting with several wavelet transform functions, the ‘Mexican hat’ and the Coiflet wavelet functions were used. The CWT implementation was performed using MATLAB’s wavelet toolbox. MATLAB’s CWT can be considered as a filter that scales a mother wavelet function along the time axis. At each scale, the CWT function will superimpose the scaled mother wavelet wave form over a segment of the signal under analysis. The similarities and differences between the form of the wavelet wave and the signal being analyzed are determined by CWT as,
where
represents the CWT mother wavelet function which is shifted in time by
T and dilated or contracted by a factor and then correlated with the vibration signal represented by
x(
t).
All feature attributes are scaled and normalized across the column before clustering. The datasets are unlike others used in the paper—high dimensional data with relatively low cardinality. The idea is to see if the fault conditions can themselves be or if any combination of operating speed and fault condition can be partitioned. The datasets are described below:
BearingData1 is three non-overlapping segments with 16 FFT averages, n = 90, d = 16
BearingData2 is three non-overlapping segments with 64 Mexican Hat averaged wavelet coefficients, n = 90, d = 64
BearingData3 is three non-overlapping segments with 64 Coiflet-averaged wavelet coefficients, n = 90, d = 64
BearingData4 is three non-overlapping segments with kurtosis, skewness, RMS, and crest factors for 10 wavelet approximations and 10 wavelet details, n = 90, d = 80.
We also use a low dimensional data with non-overlapping and overlapping regions of the raw data (50% overlap). BearingData5 is three non-overlapping and two overlapping segments with four statistical features of the raw time signal—mean, skewness, standard deviation, and kurtosis, n = 150, d = 4.
The performance of the proposed method is compared with FCM-AO with random initialization of cluster prototypes using the four cluster validity indices. FCM-AO is implemented for
c = 2 to
c = 10. The vector representation used in CDE-FCM is
d × 10. The results are tabulated in
Table 9.
The proposed algorithm CDE-FCM almost always outperforms FCM-AO. In cases where the three natural groupings are not found, CDE-FCM finds approximately 6 clusters which are decoded as two subclusters based on speed (high and low) in most cases. The accuracy, precision, and recall rate of both FCM-AO and CDE-FCM are similar meaning they uncover very similar clusters although with FCM-AO it is often difficult to ascertain the optimal number of clusters.
A comparative evaluation is performed using sensitivity analysis. In clustering evaluation, a true positive (
TP) is defined as the decision that assigns two similar data objects in the same cluster and a true negative (
TN) is a decision that assigns two dissimilar objects to different clusters. These are both desirable outcomes. The errors can either be a false-positive (
FP) decision for an assignment of two dissimilar objects to the same cluster or a false-negative (
FN) decision when two similar objects are assigned to different clusters. These metrics are evaluated using pairwise measurements—for a dataset of cardinality
n, there are
n(
n − 1)/2 pairs of objects. The precision (
P) and recall (
R) are defined as,
The Rand index
RI measures the percentage of decisions that are correct (also called the accuracy). The
F-score is a measure of the harmonic mean of the method’s precision and recall. The
F-score is implemented as
F1 in this paper (which gives equal weight to precision and recall) [
67].
Results of the sensitivity analysis comparing CDE-FCM with FCM-AO are provided in
Table 10. As can be seen, the proposed method CDE-FCM achieves accuracies of approximately 75% with two of the five datasets. The best accuracy of FCM-AO approaches 75% for only one of the five datasets. The
F1 score equally weighing precision and recall for CDE-FCM is also better than FCM-AO for all five datasets. This is a promising result that shows the superiority of the proposed method over the original FCM with randomized initialization.