1. Introduction
Cyber manufacturing is one of the most influential branches of the physical cyber system. Typically, a cyber-manufacturing system focuses on the architecture of the devices used in cyber systems, their data acquisition, retrieval and processing methods, the functional efficiency and security features of data and devices [
1]. At the same time, it deals with related engineering and technological aspects of the devices to make them good-looking, easy to use and strong. Very commonly used cyber devices are smart watches, healthcare and assistance devices of disabled and elderly people [
2]. With the advancement in industry and technology, those devices have started to stand out in terms of their functionality. In today’s world, no one doubts that, in near future, a small device will provide us with multiple services, e.g., we could add our own security, day-to-day work, accounting, asset management, etc. Then, body-worn devices will obtain a different dimension. These devices will be widely used to simplify our day-to-day operations and increase security [
2].
Certainly, these devices will include healthcare services, as well. Current healthcare devices are capable of storing several pieces of casual information about the people who wear them. Some devices may update a user’s information to a limited extend through a set of device-implanted sensors. However, future healthcare devices will show many surprises that are unimaginable compared to the present time. They will be able to perform many physical and pathological experiments on the human body automatically. These devices will automatically start communicating with the designated doctors or any healthcare provider to receive suggestions and consultations. It will not be surprising if a robotic device takes its owner to the doctor or hospital if it senses the deteriorated health condition of the owner. Although we have started to see similar applications in a short range, there are still questions about their capability and the quality of their skills. That limitation is mainly due to the small size and low-cost demand of such devices. However, we expect better services from them by making them capable of processing and storing huge amounts of information.
The healthcare devices will store demographic, pathological, historical and other data of the owner in their memory. This information will be updated regularly. In that case, if we preserve the historical data, at some point in time, that data will become larger, especially for old patients and chronic diseases. It could even be larger than the memory capacity of the device. Again, the information stored on the device will be threatened because the device will be connected to the cyber world and its information can easily be stolen if we store plane data in them. Hence, processing data to a secured format is essential. Moreover, failure to provide data protection will also hamper the cyber-manufacturing process. Therefore, in these days, the security breach on the information will be a big obstacle in the way of obtaining good service. If we do not work now on the security architecture of the cyber-manufacturing world, the cyber-manufacturing method, especially on wearable healthcare devices, will not be implemented [
3]. Consequently, we have to look at three things—(i) a simple architecture for the device, (ii) its information management and (iii) information security. Hence, in this article, we place an emphasis on developing an architecture for future healthcare devices and an efficient algorithm for their data management and security.
Our primary target is to design a simple but effective architecture. We first assume that the intended wearable cyber device is equipped with the necessary sensors to collect and process human health information as well as to store them. The sensors are connected to a memory unit and a signalling unit through the processing unit of the device. The memory unit consists of two parts—a restricted part and a reusable part. The initial information of the person, e.g., the demographic information of a person, will be kept in a restricted part of the memory at the time of manufacturing or in the one-time erasable memory of the device. Generally, this are very ordinary personal information, e.g., name, parent’s name, address, gender, etc. All of this information will be encrypted and stored in that stated memory. The details of the architecture are given at
Section 3.
Our secondary target is to meet the demand of storing a large volume of data, even if it is an ever-growing dataset. As we are working on smart devices where memory size is a concerning issue, we have presented a method of repeatedly implanting secrets in a small-sized cover. As a medium, we have chosen a small DNA/RNA sequence. That technique has helped us in both managing large data and securing them.
The proposed scheme has several distinct features: (i) It presents a simple architecture for cyber devices that will be a boon for production in the industry; (ii) The architecture would be useful for managing its memory, a signaling unit and sensors effectively and building a perfect synchronization among these units; (iii) Uses of restricted memory will allow the devices to be used for other purposes, e.g., in banking, owner identifying, etc.; (iv) As far as we know, no other study has used a small chunk of nucleotides for embedment of a large volume of data, e.g., a chunk of the ten nucleotides is enough to conceive any volume of information; (v) A multi-time implantation method is proposed for the first time in the field of its kind; (vi) There is no other scheme that generalizes its functionality to work with any size of message, e.g., message size of some multiple of the length of DNA; (vii) Shuffling elements in the stego key by the proposed scheme is an innovative idea that is effective for improving the security of data. For this, we think that the proposed method will be a striking one in the field of cyber physical manufacturing, as well as the DNA-based data hiding arena.
The study involved experiments on real medical data, as well as on four separate RNA and three DNA sequences to justify its robustness. Four RNA sequences of COVID-19 and one DNA sequence were taken from the NCBI repository. The RNA sequences of nCoV-19 and DNA sequences are open to all [
4,
5,
6,
7] and, therefore, freely available. Two additional sequences were prepared by us from real data. The proposed method worked successfully in all the RNA/DNA sequences and provided promising results.
The article comprises seven sections.
Section 2 is provided to give a description of the background studies.
Section 3 narrates the proposed cyber physical manufacturing architecture. The proposed multi-time data embedment method is given in
Section 4.
Section 5 analyses the proposed scheme. We discuss the results in
Section 6.
Section 7 concludes the article.
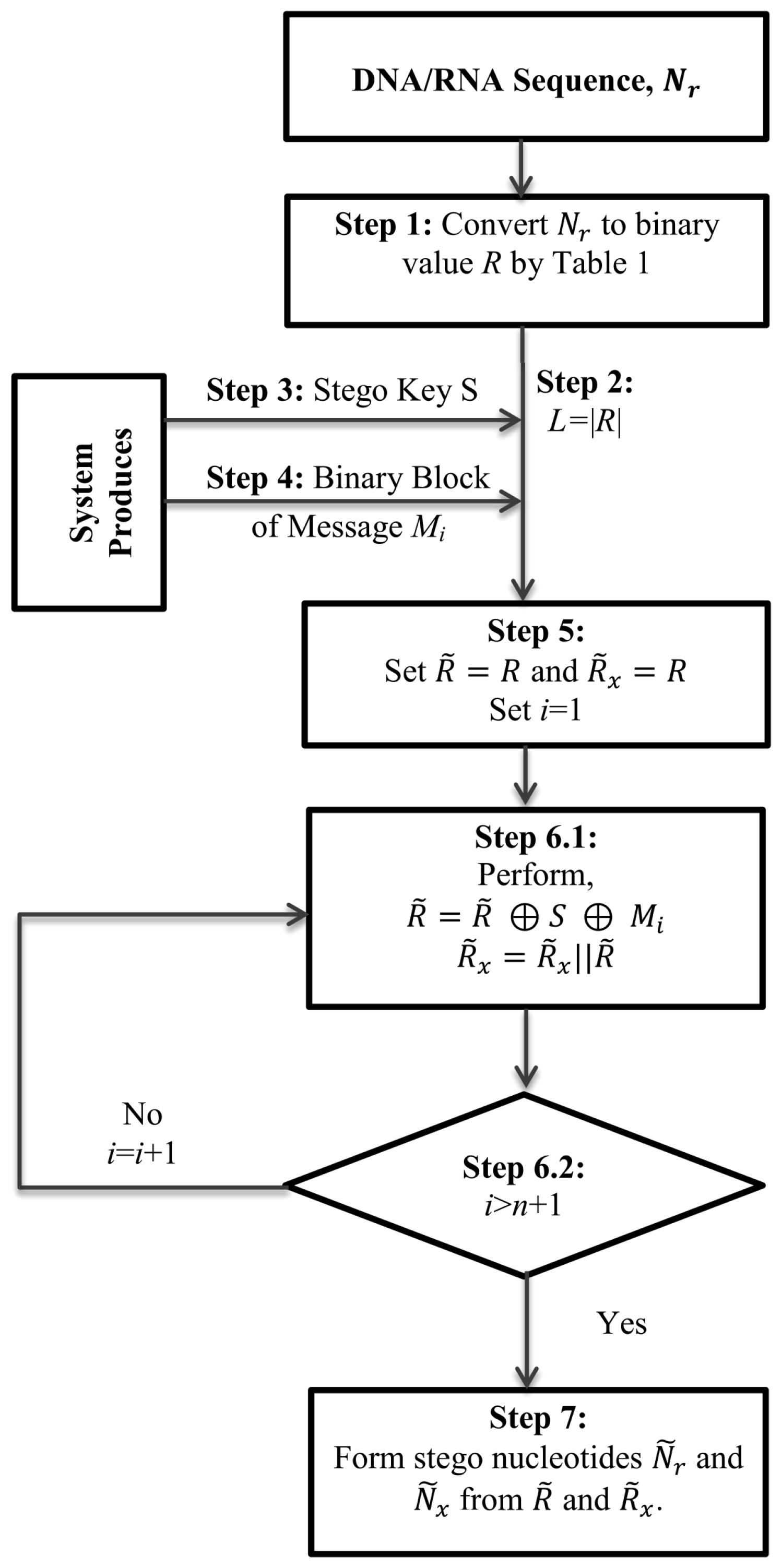
3. Proposed Architecture
The list of main hardware of the target smart devices is a processing unit, a memory unit, a set of sensors for collecting user data, a wireless sensor to connect to the cloud and a set of connections among the units. The memory unit consists of a restricted ROM and a general purpose ROM. These two ROMs could be implanted in the device as either two separate ROMs or in a single ROM. In the second case, a part of the ROM will be declared as a restricted part of the memory. The restricted ROM is writable for a single time only. The demographic information of the device owner will be written there.
The proposed method will convert this demographic information into a DNA sequence. The converted DNA sequence will be stored in an erasable ROM (E-ROM) (or part of one). In all subsequent tasks, it will achieve five aims—(i) it will gather the latest health information of a person through its sensors; (ii) it will collect the previously stored information from E-ROM and will extract all healthcare data; (iii) it will understand the situation of the patient by comparing that information with the most recently collected information; (iv) if that measurement signifies some serious issues that are to be treated soon, it will take the necessary steps to save the patient, otherwise, it will notify the person through a gentle signalling process; (v) it will update the device memory with the latest information. The updating task will be performed by the proposed data hiding technique. The architecture of the proposed cyber-manufacturing system is graphically shown in
Figure 1. The functionality of the hardware equipment of the proposed architecture is described in detail in the section on the analysis of the proposed method with a different figure.
A concern issue of this research work is to secure the information in the second and fifth steps of the above five tasks. For this, during its update task, the scheme will implant the latest information in the previously stored DNA sequence. That is why it is advised to generate a DNA sequence from the owner’s demographic information at the first stage of the device registration.
The proposed scheme will convert the demographic information to a DNA sequence, as explained. That conversion technique will, indeed, make the information useless to outer applications. However, this demographic information might be used for other purposes, e.g., in banking. To make the device usable for applications other than healthcare, we have preserved this demographic information in a restricted ROM as raw data.
5. Analysis of Proposed Method
5.1. Manufacturing Architecture
The hardware architecture of the system is depicted in
Figure 4.
5.1.1. Manufacturing Architecture: Processing Unit
The processing unit of the cyber device stores demographic information of the device owner in the restricted a memory as raw data. At the same time, it converts the demographic information into a DNA sequence and hides the other information in it using our proposed data hiding method. After that, it saves the stego DNA sequence in the main memory. Next, when the processing unit senses a new piece of data from its sensors, the unit collects the most recently saved DNA sequence from the main memory. The processing unit then extracts the secret data from that stego DNA sequence. The unit also generates the original DNA sequence by the proposed reversible data extraction method. The processing unit compares the newly sensed data with the extracted data. It sends the results to the signaling unit.
5.1.2. Manufacturing Architecture: Signaling Unit
The signaling unit is developed to provide one of five notifications. The first four notifications are called in-control signals. The last one is referred to as an out-control signal. According to the demand of the users, the manufacturer can increase the number of in- and out-control signals. The signaling unit applies a mathematical function
f to generate a number from 1, 2, 3, 4, i.e.,
f = 1 or
f = 2 or
f = 3 or
f = 4. Here, we have not given any fixed equation as a part of that function. Rather, it is left to the manufacturer so that they can set their own function and fitted parameters according to this proposal or their modified one. The signaling unit may generate one of the four notifications automatically based on a function’s generated value
f. When the signaling unit generates
f = 1, the cyber device creates a gentle notification. That means that there is no problem in the sensed data.
f = 2 indicates some minor changes in the health statistics. The device signals accordingly. Again,
f = 3 indicates major changes in the health conditions. The person demands extra care. If the signaling unit measures
f = 4, the person is in danger. They should consult a doctor immediately. The signaling unit then automatically calls a designated doctor. Notification 5 is not generated by the auto system of the signaling unit. This is done after receiving a request from a doctor. That request automatically triggers the signaling unit to set
f = 5. When the device realizes a value of
, it sends the report of the last-sensed and the extracted historical data to the doctor. A list of those notifications is shown in
Figure 5.
5.2. Analysis on Data Hiding Algorithm
To analyze, the data hiding method first considers a small DNA sequence. Let us consider the sequence TTTGTTGAGT.
Hence, DNA : TTTGTTGAGT;
DNA to Binary, R: 11111110111110001011;
Secret Key, S: 00001111110111000001;
Message, M: 101101111000111001100010011101110111010111111110001011100001.
Divide M into chunks of length of L (here, it is 20)
Then, M = 10110111100011100110-00100111011101110101-11111110001011100001
The embedding program returns the following results:
Stego DNA, i.e., : GCTTTTATGA;
Concatenated Stego DNA, i.e., : CACGGGGGTACGTGAAACGAGCTTTTATGA.
In the following, the processing steps and their results are demonstrated.
5.3. Execution of Data Implantation Module
In the following, we explain the execution steps of the data implantation module.
Execution cycle 1: 11-11-11-10-11-11-10-00-10-11
Using Equation (
2),
11-11-11-10-11-11-10-00-10-11 ‖ 01-00-01-10-01-10-10-10-11-00
Execution cycle 2: 11-11-11-10-11-11-10-00-10-11 ‖ 01-00-01-10-01-10-10-10-11-00 ‖ 01-10-11-10-00-00-00-01-10-00
Execution cycle 3: 11-11-11-10-11-11-10-00-10-11 ‖ 01-00-01-10-01-10-10-10-11-00 ‖ 01-10-11-10-00-00-00-01-10-00 ‖ 10-01-11-11-11-11-00-11-10-00
= GCTTTTATGA
= TTTGTTGAGT-CACGCGGGTA-CGTGAAACGA-GCTTTTATGA
The result is tabulated in
Table 5.
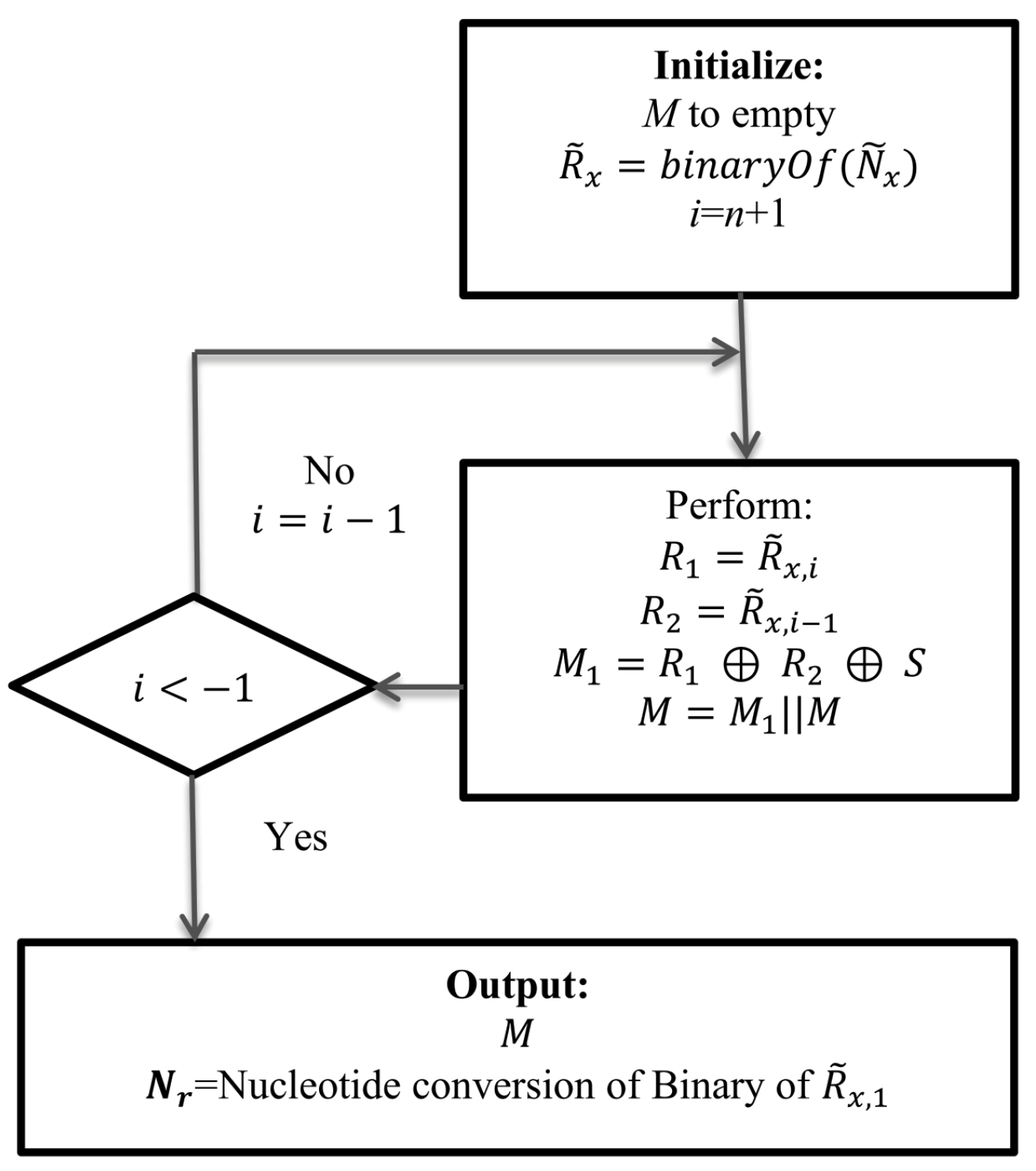
5.4. Execution of Data Extraction Module
Data extraction rules work at receiver end. The receiver end has all the necessary information to extract the implanted data and to reconstruct the cover DNA/RNA. It has
= TTTGTTGAGT-CACGCGGGTA-CGTGAAACGA-GCTTTTATGA;
Secret key, S = 00001111110111000001.
Then, the receiver generates , where 11-11-11-10-11-11-10-00-10-11 ‖ 01-00-01-10-01-10-10-10-11-00 ‖ 01-10-11-10-00-00-00-01-10-00 ‖ 10-01-11-11-11-11-00-11-10-00
The execution steps are shown below:
Execution cycle 1: Using Equation (
3):
10-01-11-11-11-11-00-11-10-00 (last chunk of
). Using Equation (
4):
01-10-11-10-00-00-00-01-10-00 (second chunk of
from the last). Using Equation (
5), we can compute new
and
. Here,
is the value of
in the data implantation module. The result is shown in
Table 6.
Using Equation (
6),
M is computed.
11-11-11-10-00-10-11-10-00-01
Execution cycle 2: Now, again, using Equations (
3)–(
5), we compute
,
and
. In this cycle, the value of
is the same as the value of
in the data implantation module. The result is depicted in
Table 7 01-10-11-10-00-00-00-01-10-00
01-00-01-10-01-10-10-10-11-00
= 00-10-01-11-01-11-01-11-01-01 ‖ 11-11-11-10-00-10-11-10-00-01
Execution cycle 3: Using Equations (
3)–(
5), we can compute
,
and
. This
is the same as the
of the data implantation module. The result is depicted in
Table 8 01-00-01-10-01-10-10-10-11-00
11-11-11-10-11-11-10-00-10-11 Equation (
6) yields:
10-11-01-11-10-00-11-10-01-10 ‖ 00-10-01-11-01-11-01-11-01-01 ‖ 11-11-11-10-00-10-11-10-00-01
After the execution of the i loop in the data extraction part of the Method, we will find . Hence, 11-11-11-10-11-11-10-00-10-11, DNA conversion of R is = TTTGTTGAGT. Thus, the scheme retrieves the hidden message and rebuilds the cover DNA/RNA.
5.5. Boosting Up Vulnerability
One noticeable point of the scheme is that the cover DNA is found at the first block of . The same is true in the case of RNA. According to the above example, cover DNA is TTTGTTGAGT. As the cover DNA is always found as a first block of , in case of stolen key, it cannot preserve the security of cover DNA. Therefore, we introduce a shuffling policy of blocks of . We have not explained that policy in the previous section due to better understanding. Moreover, it is not a necessary part of the algorithm for its functionality.
We know that
consists of
stego DNA blocks.
Table 9 shows the method of shuffling
stego blocks among themselves. The first column of the table shows the current block numbers of
. These block numbers are randomly rearranged. After shuffling, block positions have changed. The second column shows the position of blocks after shuffling, e.g., the blocks, 0, 1, 2, 3, and so on, are moved to the 1, 0, 2, 0, … index positions, respectively. The indices of all blocks after their movements are tabulated at the third column. The fourth and fifth column show the equivalent binary of moved position and converted nucleotides of the binaries. These nucleotides are interpreted in a top-down manner, and thus, the read values are ACATAGAACGCATCTATTCTTGGTGGGACCGC. Say, it is
. We make that
as a part of stego key. In that case,
will increase in size by more than 64 bits. Finally,
will be modified by
by
, where ‖ stands for concatenation. Then, the new sequence will be as
GCTTTTATGA-TTTGTTGAGT-CGTGAAACGA-CACGCGGGTA.
At the receiver end, the decoder first separates from the last 32 nucleotides of . For this example, it is ACATAGAACGCATCTATTCTTGGTGGGACCGC. At that stage, it is necessary to know the number of blocks that the stego nucleotides contain. That could be part of a negotiated stego key or might be managed by adding two additional nucleotides. In our case, we communicate that information between encoder and decoder through stego key. Indeed, it is the cycle number, i.e., n. In our above example, we used 4 blocks. Hence, the decoder will separate first eight nucleotides of the ACATAGAACGCATCTATTCTTGGTGGGACCGC, i.e., ACATAGAA. The decimal conversion of that ACATAGAA will produce 1-3-2-0. That result states that the 2nd block, 4th block, 3rd block and 1st block are the reading sequences. The readers will generate by . Thus, = TTTGTTGAGT-CACGCGGGTA-CGTGAAACGA-GCTTTTATGA. From that stage, the process of extracting secrets and recovering cover DNA is explained in earlier discussions.
7. Conclusions
The proposed cyber-manufacturing system is very simple but effective. It would be easy for the industry to manufacture such devices. The research also provides strong data security, as well as enhances the embedding capacity. The proposed multi-time data embedment scheme allows an application to implant any number of bits of secret message in a small DNA sequence and even in a small portion of a DNA sequence, known as a DNA strand. Hence, a small memory is enough to make the scheme functional. Thus, this opens up a unique opportunity for data hiding-based healthcare applications as well as for the manufacturing industries.
Moreover, the scheme could be used in message communication, identifying a device and recognizing the source of data. It will work in both offline and in the cloud. This will be a very useful scheme for an organization whereby the organization works with DNA. The medical and criminology departments of a country work on DNA and/or RNA. They just have to manage a huge volume of information for their patients and criminals. It would be a very notable application to hide the large volume of their secrets in their official DNA/RNA. From a managerial point of view, this contribution will be a smart one in handling their data. In our future work, we hope to work more on manufacturing architecture. We will work to improve the hardware and software architectures of such cyber devices to make them more user-friendly, efficient and functional. Additionally, we intend to embed personal information and one’s DNA strand in a smart wearable device in our next research. Then, criminals and unknown dead bodies could be identified easily.