Validation of Automated Chromosome Recovery in the Reconstruction of Ancestral Gene Order
Abstract
1. Introduction
2. RACCROCHE
- In Lines 6–7, the compilation of oriented candidate adjacencies at each of the ancestral nodes of a given binary branching tree phylogeny using the “safe” criterion that such an adjacency must be evidenced in genomes in two or three of the subtrees connected by this node, not just one or none.
- In Lines 8–9, the large set of these candidates is then resolved, at each node, by maximum weight matching (MWM) to give an optimally compatible subset, which ipso facto defines linearly (or circularly) compatible “contigs” of the ancestral genomes to be constructed, thus avoiding the branching segments that plague other methods [10]. Use of MWM for ancestral gene order reconstruction was introduced some time ago, but with modest results [11].
- In Line 10, local sequence matching, satisfying proximity and contiguity conditions, of each ancestral contig on all of the chromosomes of the extant genomes, followed in Line 11 by the construction of a total chromosomal co-occurrence matrix of contigs belonging to each ancestral node.
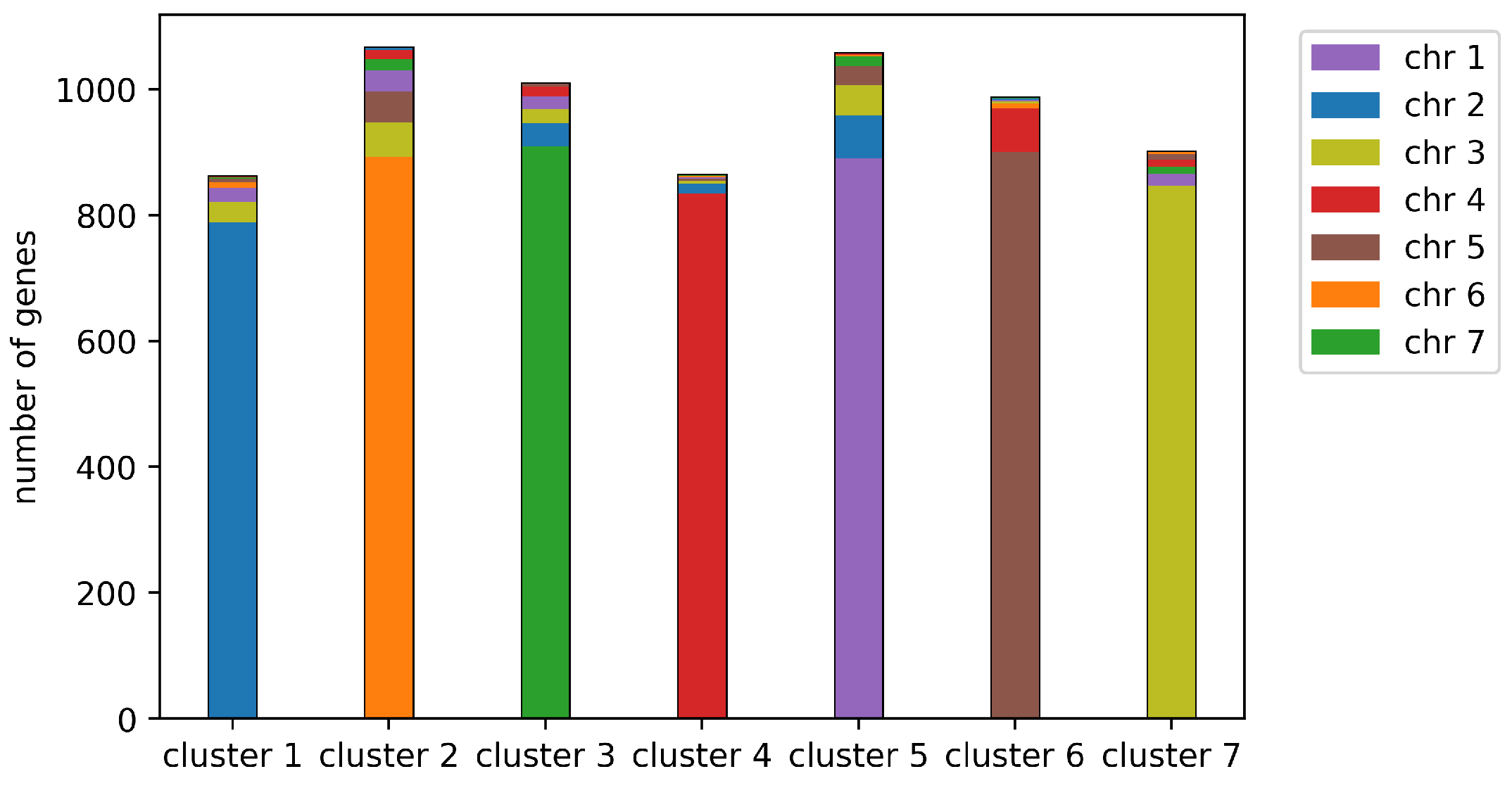
- In Line 12, a clustering applied to the co-occurrence matrix. This is then decomposed into chromosomal sets of closely clustered contigs. Within each contig, the order of the genes is already predetermined by the MWM step. Ordering the contigs along the chromosomes is carried out by a linear ordering algorithm.
| Algorithm 1:RACCROCHE—reconstruction of ancestral contigs and chromosomes |
 |
3. Clustering
- Loss of evolutionary signal due to a lengthy time period between the ancestor and its descendants. This leads to a sparsity of co-occurrence values of non-negligible size, meaning that some contigs do not fit into any cluster at a meaningful level.
- Scale bias. Large contigs will have more co-occurrences than smaller contigs that will be included late, often erroneously (especially with complete-linkage), in the clustering procedure.
- Variable scores. Due to vagaries in deletion and other evolutionary processes, not all high scores reflect true ancestral co-occurrence. Coversely, some co-occurrences cannot be captured due to low scores.
- Inflexible visualization settings. The heat maps color pixels by dividing the range of scores into equal intervals by default. However, this is not useful in comparing heat maps produced by different settings in the construction of contigs or in the use of different similarity or distance measures of contig co-occurrence. One heat map may be simply darker or lighter than the other overall, thus obscuring the real object of comparison, which is how clear-cut and distinct the clusters are and how they are qualitatively different from map areas not corresponding to clusters.
4. Updates to the Clustering
4.1. Update to the Co-Occurrence Measure
4.2. Update to Heat Map Visualization
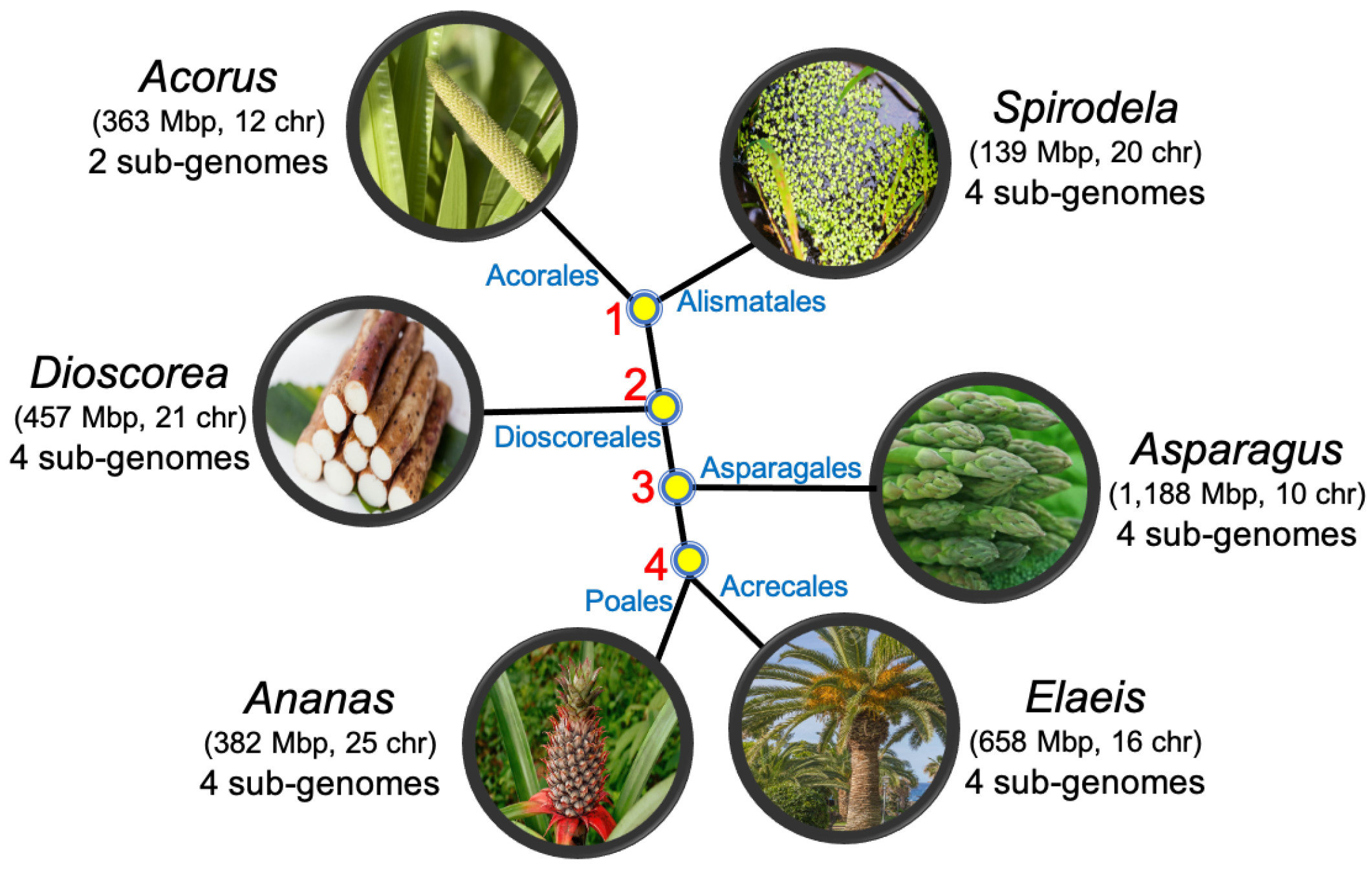
5. The Monocots
- Acorus calamus (sweet flag) from the order Acorales;
- Spirodela polyrhiza (duckweed) from the order Alismatales;
- Dioscorea rotundata (yam) from the order Dioscorales;
- Asparagus officinalis (asparagus) from the order Aspargales;
- Elaeis guineensis (African oil palm) from the order Arecales;
- Ananas comosus (pineapple) from the order Poales.
6. Simulations
6.1. Parameters for Simulations
| Algorithm 2: Estimate optimal gene family sizes in simulated ancestral genomes. |
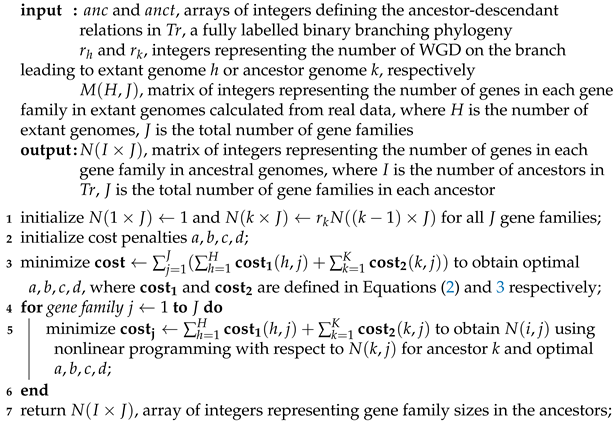 |
6.2. The Simulation Process
| Algorithm 3: The simulation of gene repertoire in extant genomes |
 |
7. Results
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MWM | Maximum weight matching |
| Mya | Million years ago |
| WGD | Whole genome duplication |
| WGT | Whole genome triplication |
References
- Perrin, A.; Varré, J.S.; Blanquart, S.; Ouangraoua, A. ProCARs: Progressive reconstruction of ancestral gene orders. BMC Genom. 2015, 16, S6. [Google Scholar] [CrossRef] [PubMed]
- Rubert, D.P.; Martinez, F.V.; Stoye, J.; Doerr, D. Analysis of local genome rearrangement improves resolution of ancestral genomic maps in plants. BMC Genom. 2020, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, C.; Muffato, M.; Abecassis, J.; Crollius, H. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep. 2015, 10, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jin, L.; Zheng, C.; Leebens-Mack, J.H.; Sankoff, D. RACCROCHE: Ancestral flowering plant chromosomes and gene orders based on generalized adjacencies and chromosomal gene co-occurrences. In Proceedings of the 10th International Conference on Computational Advances in Bio and Medical Sciences, Virtual, 10 December–12 December 2020; Volume 12686. [Google Scholar]
- Lyons, E.; Freeling, M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008, 53, 661–673. [Google Scholar] [CrossRef]
- Lyons, E.; Pedersen, B.; Kane, J.; Freeling, M. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates rosids. Trop. Plant Biol. 2008, 1, 181–190. [Google Scholar] [CrossRef]
- Yang, Z.; Sankoff, D. Natural parameter values for generalized gene adjacency. J. Comput. Biol. 2010, 17, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Sankoff, D. Tests for gene clusters satisfying the generalized adjacency criterion. In Proceedings of the Brazilian Symposium on Bioinformatics, Santo André, Brazil, 28–30 August 2008; Springer: Berlin/Heidelberg, Germany, 2008; pp. 152–160. [Google Scholar]
- Tannier, E.; Bazin, A.; Davín, A.; Guéguen, L.; Bérard, S.; Chauve, C. Ancestral genome organization as a diagnosis tool for phylogenomics. In Phylogenetics in the Genomic Era; Scornavacca, C., Delsuc, F., Galtier, N., Eds.; No Commercial Publisher | Authors Open Access Book; 2020; pp. 2.5:1–2.5:19. Available online: https://hal.archives-ouvertes.fr/hal-02535466/ (accessed on 25 March 2021).
- Zheng, C.; Chen, E.; Albert, V.A.; Lyons, E.; Sankoff, D. Ancient eudicot hexaploidy meets ancestral eurosid gene order. BMC Genom. 2013, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Chanderbali, A. Buxus and Tetracentron genomes help resolve eudicot phylogeny, gamma hexaploidy, and paleogenomics. 2021; In preparation. [Google Scholar]
- Chase, M.W.; Christenhusz, M.; Fay, M.; Byng, J.; Judd, W.S.; Soltis, D.; Mabberley, D.; Sennikov, A.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Jiao, Y.; Li, J.; Tang, H.; Paterson, A.H. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 2015, 26, 2792–2802. [Google Scholar] [CrossRef] [PubMed]
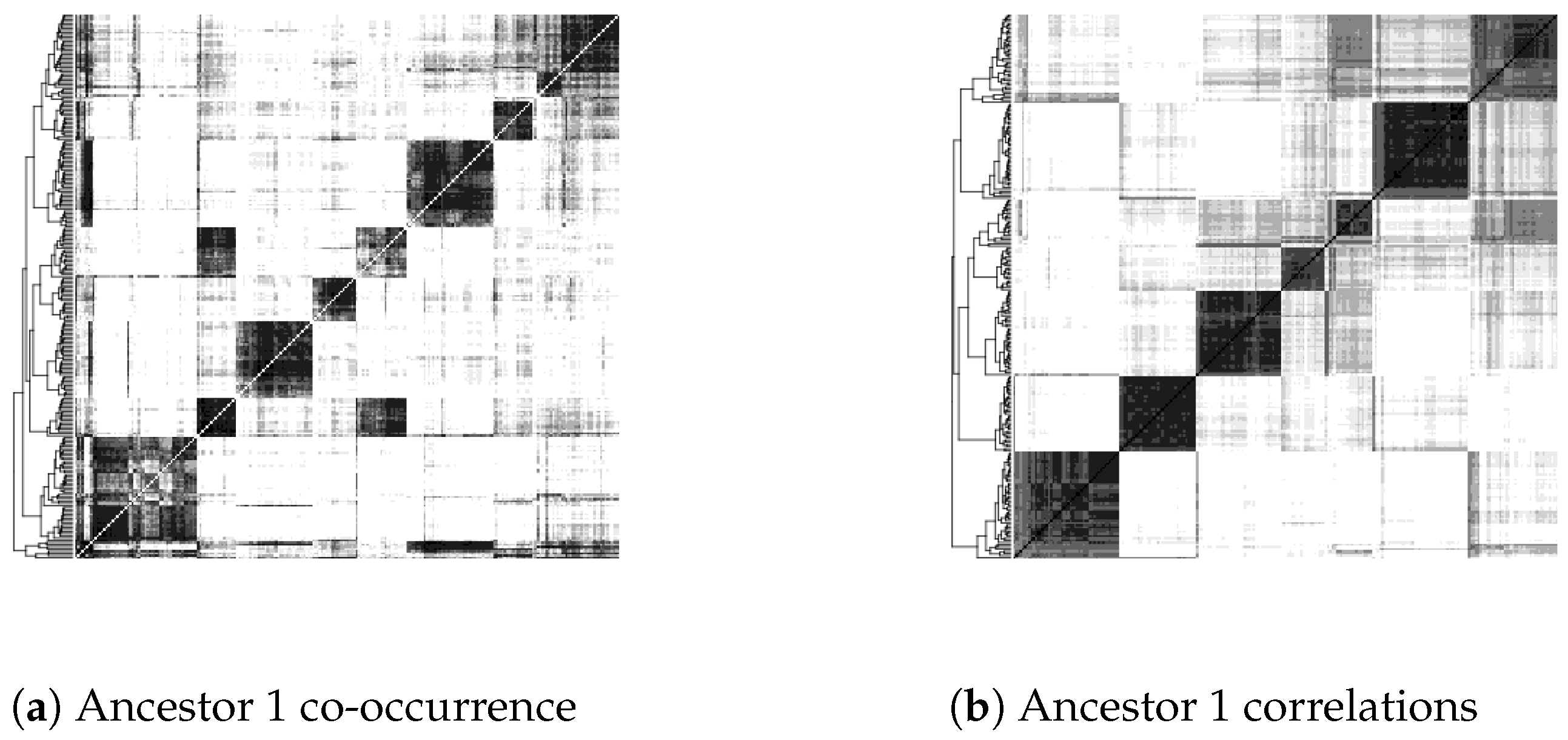
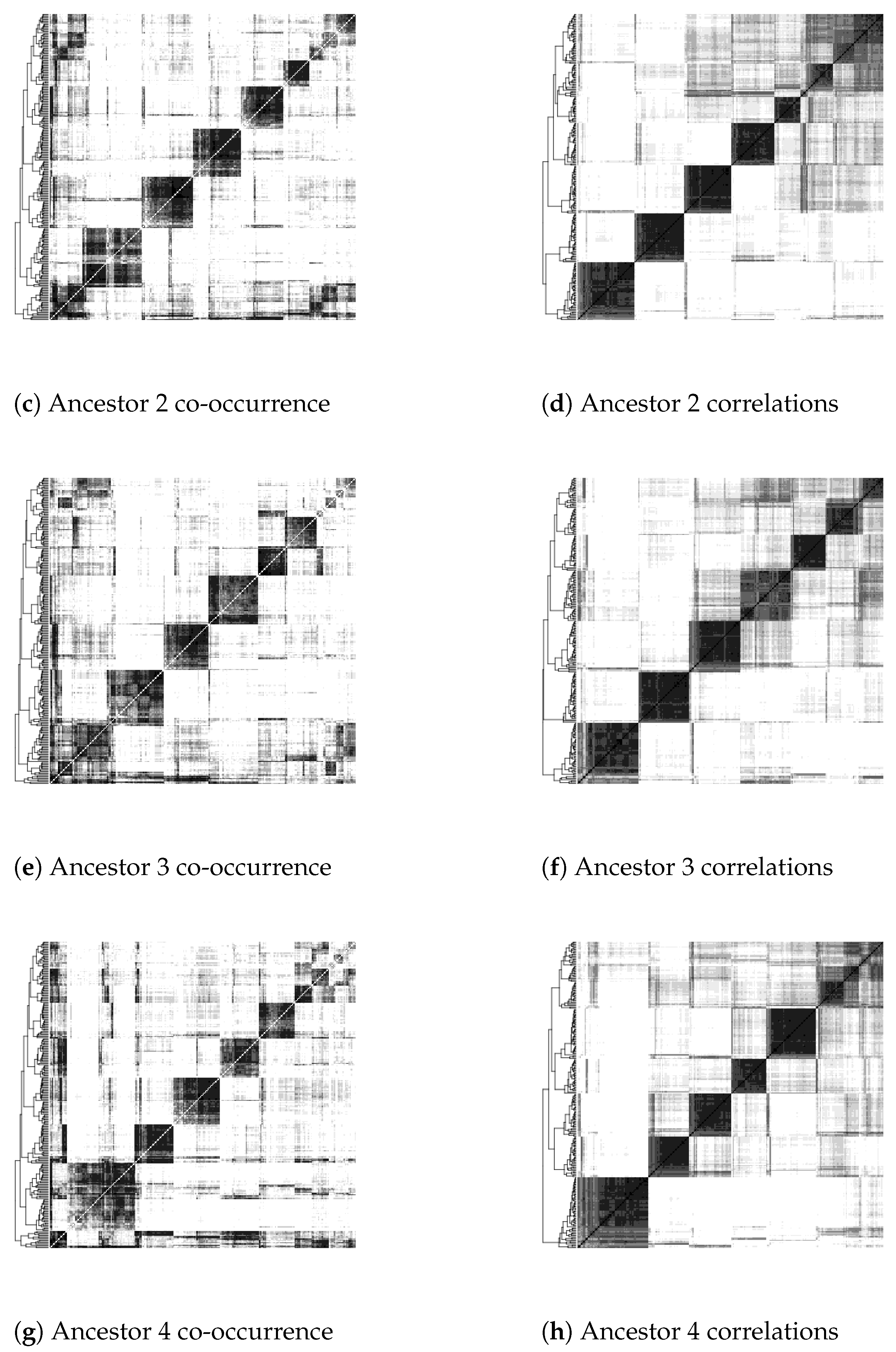
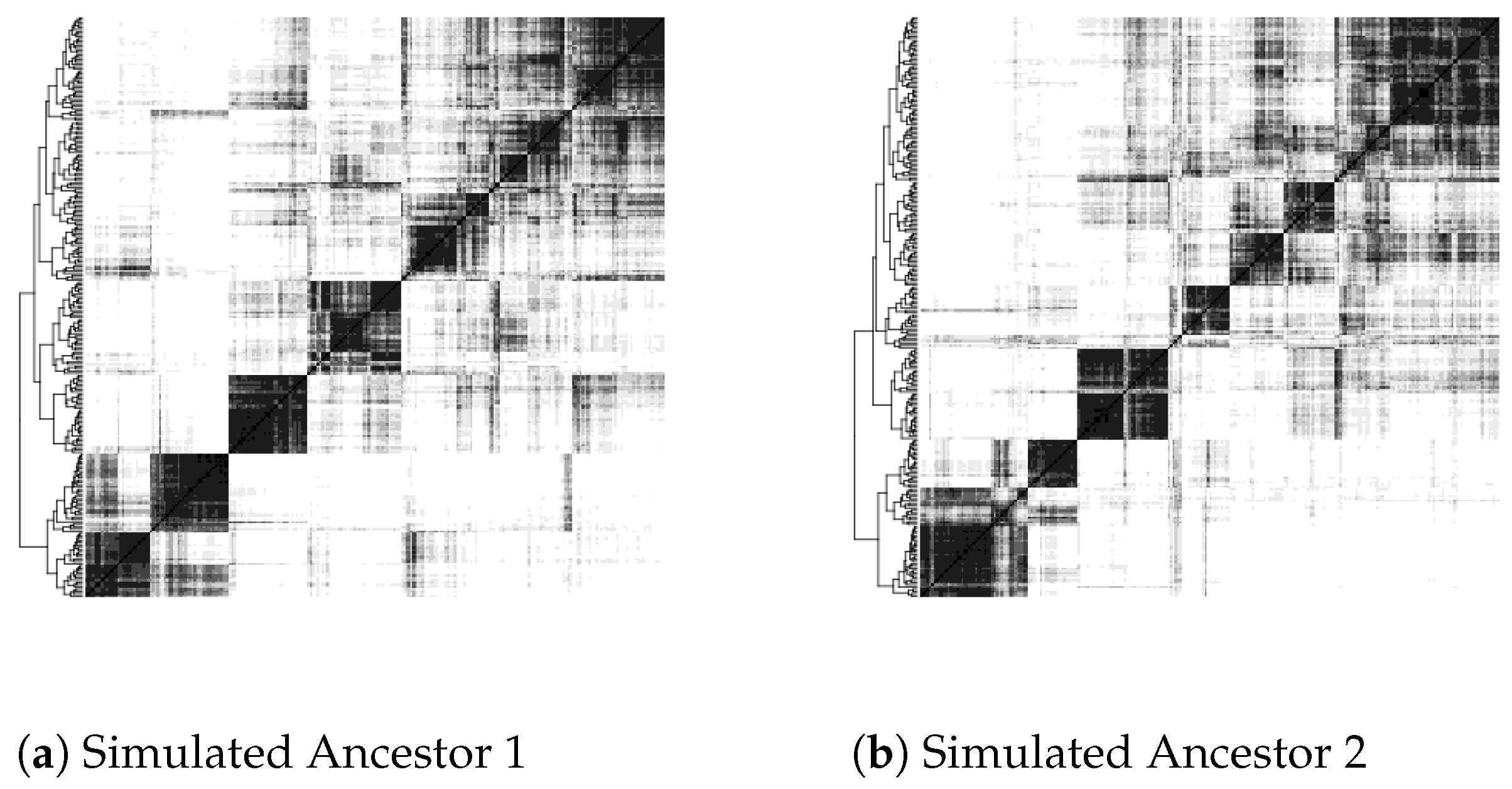



| Greyscale intensity | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Proportion of pixels | 50% | 15% | 10% | 6% | 4% | 4% | 4% | 6.5% | 0.5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Jin, L.; Leebens-Mack, J.H.; Sankoff, D. Validation of Automated Chromosome Recovery in the Reconstruction of Ancestral Gene Order. Algorithms 2021, 14, 160. https://doi.org/10.3390/a14060160
Xu Q, Jin L, Leebens-Mack JH, Sankoff D. Validation of Automated Chromosome Recovery in the Reconstruction of Ancestral Gene Order. Algorithms. 2021; 14(6):160. https://doi.org/10.3390/a14060160
Chicago/Turabian StyleXu, Qiaoji, Lingling Jin, James H. Leebens-Mack, and David Sankoff. 2021. "Validation of Automated Chromosome Recovery in the Reconstruction of Ancestral Gene Order" Algorithms 14, no. 6: 160. https://doi.org/10.3390/a14060160
APA StyleXu, Q., Jin, L., Leebens-Mack, J. H., & Sankoff, D. (2021). Validation of Automated Chromosome Recovery in the Reconstruction of Ancestral Gene Order. Algorithms, 14(6), 160. https://doi.org/10.3390/a14060160







