1. Introduction
Networks are a natural and powerful representation for relational data, providing access to a large repertoire of analytic tools that may be leveraged to better understand the underlying data in numerous applications. Among the many popular methods developed throughout social network analysis and network science, community detection provides a valuable vehicle for exploring, visualizing, and modeling network data. The identification and characterization of community structures also highlights subgraphs that may be of special interest, depending on the application. Many methods for community detection are available and have been employed meaningfully in applications (see, e.g., reviews [
1,
2,
3,
4,
5,
6]).
Some of the most heavily used computational heuristics for finding communities involve optimizing a quantity known as modularity, which was introduced by Newman and Girvan [
7] and measures the total weight of within-community edges relative to the expected weight in a corresponding “null-model” random graph. For (possibly weighted) undirected networks that are compared to the configuration null model, modularity is given by [
7]
where
are the elements of the adjacency matrix describing the presence (and possibly weights) of edges between nodes
i and
j,
is the strength (weighted degree) of node
i,
is the total edge weight,
indexes the community to which node
i has been assigned,
if
and 0 otherwise, and
is a resolution parameter introduced by Reichardt and Bornholdt [
8] to influence the number and sizes of communities obtained, whereas
in the original formulation [
7]. Tuning the resolution parameter
can reveal community structures at multiple scales, which offers one strategy to overcome the “resolution limit” of modularity [
9] wherein small communities in sufficiently large networks cannot be detected via Equation (
1) given fixed
. See also [
10,
11] for an alternative approach for resolving multiple scales.
Formulae analogous to Equation (
1) exist to define modularity for a variety of other network types, including directed [
12], bipartite [
13], signed [
14,
15] and multilayer networks [
16], with corresponding replacements for the
term to account for the expected weights under different null models. Motivating our present contribution, some of these models introduce additional parameters beyond the resolution parameter
. We emphasize that throughout this work we will use the term “modularity” in its broadest sense to include any of these generalizations as applied appropriately to a given data set. Such generalizations include the use of resolution parameter
, multiple resolution parameters for signed networks, and including one or more interlayer-coupling parameters for multilayer networks. Regardless of the network type, the primary goal in modularity maximization is to determine the community labels
that maximize
Q. Finding the partition with a guarantee of globally optimizing modularity is not computationally feasible except in the smallest networks [
17], and there may be many nearly-optimal partitions [
18].
It is worth noting that in initial exploration of a new data set, there is no a priori notion of what constitutes a “good” value of Q. Furthermore, real community structures may be more complex than those describable by a hard partition of nodes into communities, which fails to account for overlapping communities and insists on assigning every node to a community.
Even with its problems, maximizing modularity remains a highly-used method for community detection, with many software packages available, some of which are computationally very efficient in practice. Moreover, maximizing modularity is one of the few approaches for community detection in networks that has been extended in a principled way [
16] to multilayer networks [
19], though we also call attention to the multilayer extension [
20] of Infomap [
21] and recent developments extending stochastic block models [SBMs] to multilayer networks (see [
22,
23,
24] and, for a general update of developments in SBMs, [
4]). While multilayer modularity provides a means for community detection in multilayer networks using many of the same heuristics and applying some of the same conventional wisdom developed for single-layer networks, the generalization admits at least one more parameter to control the contribution of interlayer connections to modularity relative to that from intralayer connections, e.g., the interlayer coupling
in [
16]. The same multilayer modularity framework can be applied generally to include multiple interlayer coupling parameters controlling the relative contributions of different parts of the multilayer structure, e.g., for data that is both temporal and multiplex. As such, multilayer modularity requires exploring a two-dimensional parameter space in its simplest setting, and higher dimensions in more general cases. For present purposes, we will here only explicitly consider the case of a single interlayer coupling parameter
in
Section 2.1 and
Section 3.4; but this does not put constraints on the coupling topology or relative values, only that there is some selected interlayer coupling tensor that is multiplied by
. Meanwhile, the approach we develop here can be naturally generalized to higher dimensions.
Identifying appropriate values for parameter
(and in the multilayer setting,
) involves running one or more heuristics at various parameter values and comparing the results. Because identifying globally optimal community structure is computationally intractable (both for modularity and most other approaches), these algorithms are usually run stochastically or with random initial conditions to account for entrapment in local extrema. The possibly different community structures found by computational heuristics at a particular
parameter point [
for multilayer networks] are then typically assessed only at that point before moving on to generate results at other parameter values. For instance, one might select the partition with greatest modularity found at that specific value of
or measure some statistic over the partitions that were generated at that
(see, e.g., [
25]). In order to determine whether the obtained community structures are “robust” to the
selection in any sense, one might look for stable plateaus in the number of communities (see, e.g., [
11,
26,
27,
28]), consider another metric such as significance [
29], directly visualize the different community assignments across parameters (as in [
28,
30]), or compare obtained communities with other generally-acceptable labels by some measure such as pairwise counting scores (see, e.g., the discussion in [
31]) or information-theoretic measures like Variation of Information [
32] and Normalized Mutual Information [
33]. A more computationally-demanding approach that directly attacks the problem that there is no
a priori notion of what constitutes a “good” value of modularity is to compare the obtained best modularity at each
with the distribution of modularities obtained by running community detection across some selected random-graph model, either on realizations from a model or from permutations of the data, repeating this process at different
to identify parameter values where the obtained communities are strongest relative to the random cases [
34]. Additionally, one may use a given set of partitions to generate a new partition by ensemble learning [
35] or consensus clustering [
34,
36].
Importantly, in each of these approaches for exploring the parameter space, the optimal partitions associated with each value are typically computed independently of those at other values [and, again, in the multilayer case, ]. Variation in the structure of these partitions and their corresponding modularity can arise from both adjusting the input parameters and importantly, from the stochasticity of the algorithm itself. Often, for close enough values of , the variation in modularity of identified partitions is driven more by the stochasticity of the algorithm rather than the difference in the value of . Because of this independent treatment of the results from different values, a large amount of information that might be useful for further assessing the quality of the obtained partitions is typically thrown away. We propose a different approach, which we call CHAMP, that uses the union of all computed partitions to identify the Convex Hull of Admissible Modularity Partitions in the parameter space. CHAMP identifies the domains of optimality across a set of partitions by ignoring the that was used to compute each partition, finding instead the full domain in for which each partition is optimal relative to the rest of the input partitions (hereafter, we always use the word “optimal” in this restricted sense relative to the set of partitions at hand). Visualizing the geometry of this identification process, each partition is represented as a line in for single-layer networks, and as a plane in the space in the multilayer case with a single interlayer coupling parameter . We find the intersection of the half-spaces above the linear subspaces by computing the convex hull of the dual problem. By identifying the convex hull of the dual problem, we prune that set of partitions to the subset wherein each partition has at least some non-empty domain in the parameter space over which it is has the highest modularity. This pruned subset contains all of the partitions admitted through the dual convex hull calculation. Visually, plotting Q as a function of the parameters, the pruned subset is that which remains in the upper envelope of Q, so that each partition appears along the boundary of the convex space above the envelope in the domain where it provides the optimal Q relative to the input set. The partitions removed by CHAMP do not provide optimal Q for any range of parameters. Meanwhile, the partitions that remain in the pruned set are ‘admissible’ candidates for the true but unknown upper envelope of Q; that is, each of these ‘admissible’ partitions corresponds to some non-empty parameter domain of optimality relative to the input set of partitions. We propose an algorithm to find this convex intersection of half-spaces for single-layer networks and demonstrate its ability to greatly reduce the number of partitions under consideration. We also propose an algorithm for mapping out the two-dimensional domains of optimal modularity for multilayer networks in terms of the dual convex hull problem.
The rest of this paper is organized as follows. We first define the CHAMP algorithm in
Section 2. We then apply CHAMP to example networks in
Section 3, including several, single-layer examples with resolution parameter
and a multilayer network with a
parameter space (
Section 3.4, with additional figures in the
Appendix). We conclude with a brief Discussion (
Section 4).
2. The CHAMP Algorithm (Convex Hull of Admissible Modularity Partitions)
Consider a set of
unique network partitions encoded by the node community assignments
with
. By construction,
if nodes
i and
j are in the same community in partition
(i.e.,
), and 0 otherwise. Let
denote the value of Equation (
1) for given
under partition
. Ignoring the constant multiplicative factor in front of the summation (alternatively, absorbing that factor into the normalization of
and
), Equation (
1) can be written as
where the quantities
and
are the respective within-community sums over
and
for partition
. Importantly,
and
are scalars that depend only on the network data (i.e.,
A), null model (i.e.,
, and partition
. Thus, for a given partition
, Equation (
2) is a linear function of
, which can be visualized as a line in the
plane. (See
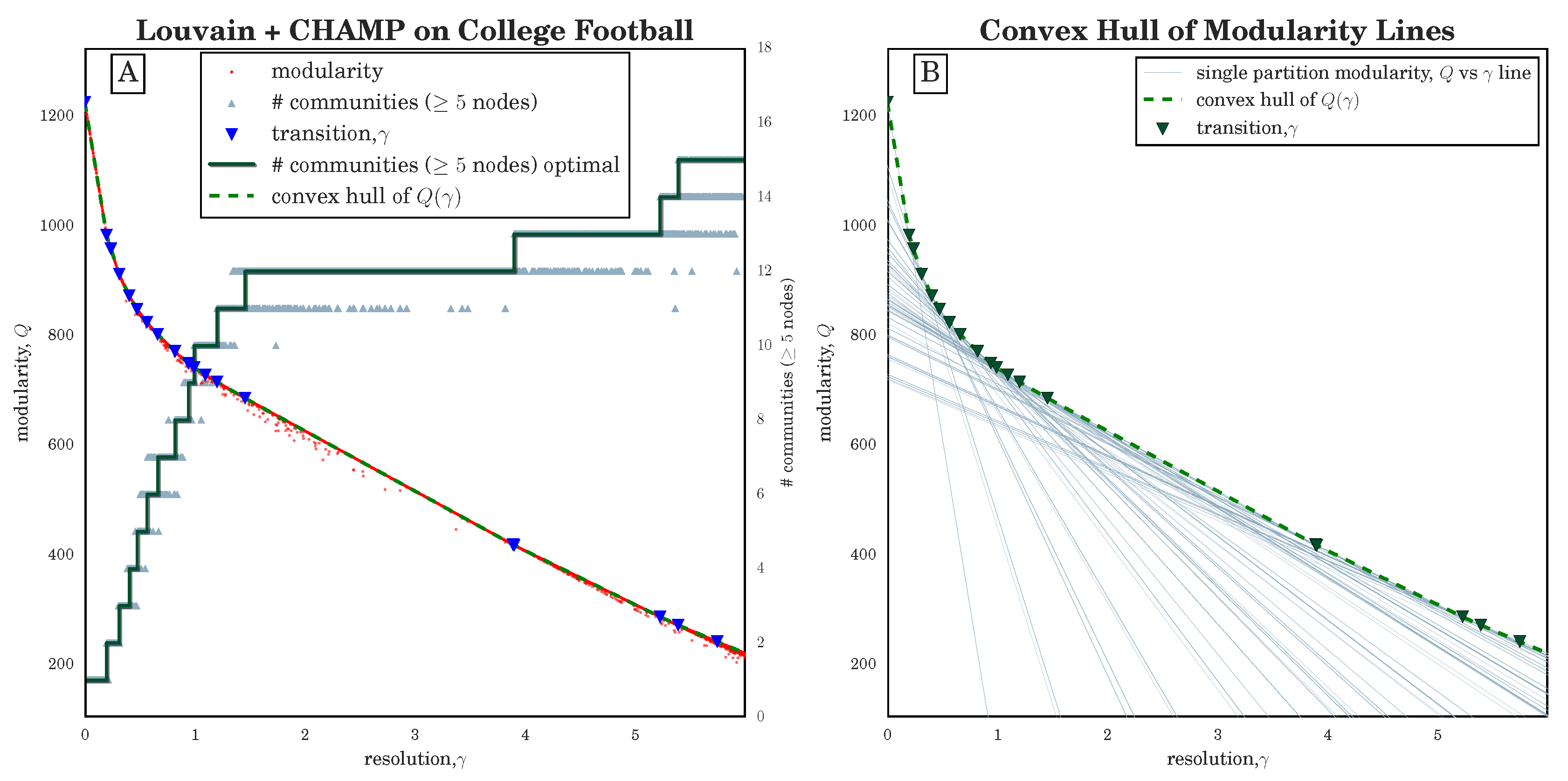
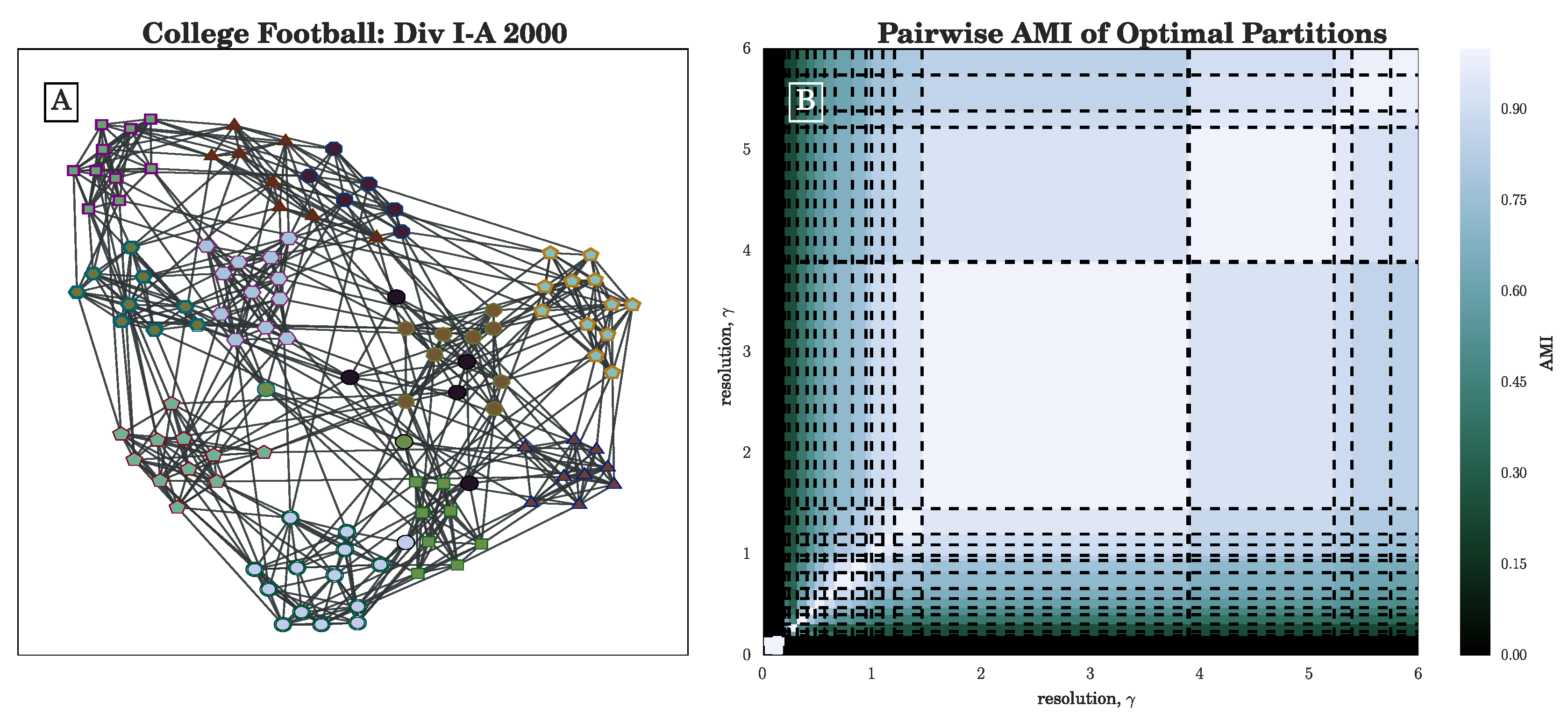
Figure 1B in
Section 3.1 for an illustration of lines
for several partitions of the 2000 NCAA Division I-A college football network [
37,
38].)
We now compare the partitions’ modularity lines
, seeking to identify the optimal partitions that yield the largest modularity values across the
values—that is, the upper-envelope boundary
for the set. We will additionally obtain
-domains over which a given partition is optimal (discarding partitions that are never optimal). Given a finite set of partitions
, the coefficients
and
can be computed individually, independent of how those partitions were obtained. Therefore, a given value of
admits an optimal partition
corresponding to the maximum
from the given set of partitions
. At most values of
, only a single partition provides the maximum (i.e., “dominant”) modularity. When two partitions
and
correspond to identical modularity values [i.e.,
], it is typically because this is the unique intersection of the two corresponding lines. (It is possible to have the case where two different partitions have identical
and
coefficients, and thus have equal
for all
; but in practice we have not observed this situation in our examples. We hereafter ignore this possibility; but if it were to occur in practice, it merely indicates two partitions of equal merit, in the sense of modularity, across all scales.) For a pair of partitions
and
, the intersection point
indicates the resolution
at which one partition becomes more (less) optimal over the other with increasing (decreasing)
. That is, one partition dominates when
, while the other dominates when
. It immediately follows that the
-domain of optimality for a partition must be simply connected. (We note that in higher dimensions, such as for signed or multilayer networks, the same linearity requires that domains of optimality must be convex [
16].)
We leverage these intersections to efficiently identify the upper envelope of modularity for a given set of partitions, and the corresponding dominant partitions (relative to the set) for all
as follows. Starting at
, the partition with maximum
is optimal. For networks with a single connected component, this partition is a single community containing all nodes; for multiple disconnected components, any union of connected components gives the same
, but we select the partition wherein each separate component defines a community. Denoting the optimal partition at
by
, we calculate the intersection points
with the other partitions
where
. Substituting Equation (
2) into this constraint yields
where
for generality. Starting with partition
for
, we identify the smallest intersection point
, which we define as
. We denote the associated partition by
. That is, partition
is optimal for the
-domain
, above which partition
becomes optimal. In the unlikely event that multiple partitions are associated with the
intersection point, the one with smallest
becomes
. Setting
p to
, we iteratively repeat this process until there are no intersections points satisfying
. We thus obtain an ordered sequences of optimal partitions,
, and intersection points
for
. The optimal modularity curve for
, given by the upper envelope of the set
, is then given by the piecewise linear function
Of course, this procedure can be started at any selected of interest, and the analogous procedure for identifying intersections for decreasing could be used to obtain the upper envelope for ; but in practice here we restrict our attention to .
2.1. MultiLayer Networks and Qhull
As noted previously, modularity has also been extended to multilayer networks [
16], for detecting communities across layers in a way that respects the disparate nature of intralayer v. interlayer edges. In order to keep our notation as simple as possible, here we let each node in a layer be indexed by a single subscript,
i or
j. (See [
19,
39] for broader discussion about different notations and their advantages.) The formulation developed in [
16] is then written as follows for the case of a single intralayer coupling parameter with general intralayer null models and interlayer connectivity (again, ignoring multiplicative prefactors in the definition of modularity):
where
,
, and
represent the (possibly weighted) edges, null model, and interlayer connections, respectively, between the node-in-a-layer indexed by
i and that indexed by
j; and
indicates the community assignment. For example, in the ‘supra-adjacency’ representation of a simple multilayer network of multislice type where the same
N nodes appear in each of
L layers, one might order the indices so that
corresponds to the first layer,
corresponds to the second layer, and so on. To emphasize that the formulation of CHAMP is independent of the details of the multilayer network under study, we note here that the only distinction used presently is that
encodes all of the edges,
specifies the within-layer null model contributions, and
describes the known interlayer connections. The key fact here is that
and
make distinct contributions to multilayer modularity, as controlled by two different parameters,
and
. As such, we need to extend CHAMP to simultaneously address both parameters. We will not assume anything here about the values or the topology of the elements of
, only that the role of these interlayer connections in determining multilayer modularity is controlled by a single interlayer coupling parameter,
. Larger values of
promote partitions with larger total within-community interlayer weight, encouraging the identification of partitions with greater spanning across layers (for a detailed analysis of behavior across
, see [
40]). We use the GenLouvain [
41] generalized implementation of the Louvain [
42] heuristic to identify partitions at selected
parameter values in the multilayer network example in
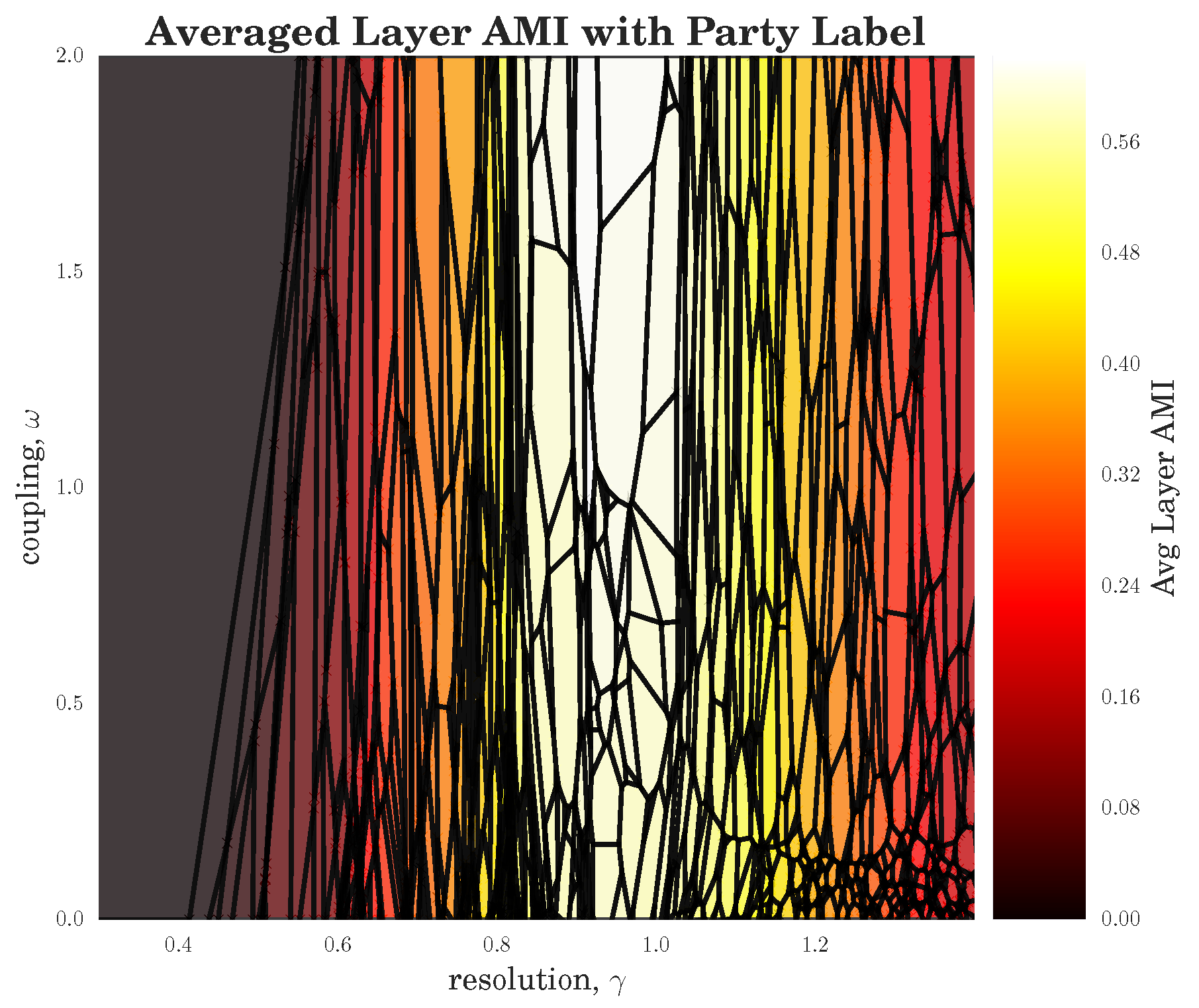
Section 3.4.
Coupling the communities across layers is conceptually intuitive. Unfortunately, introduction of the additional parameter,
makes the previous methods for parameter selection via visual inspection difficult to employ in practice and would seem to greatly complicate the challenge of selecting good values of the parameters. (See [
34] for one approach to addressing this challenge.)
However, because the multilayer modularity function is linear in the parameters
and
, we can again apply the general approach of CHAMP, albeit now in a larger dimensional parameter space. For each partition
, we again define the scalar quantities
and
to be the within-community sums over the adjacency matrix and null model, respectively, and now include a similar sum over the interlayer connections,
:
In this notation, the multilayer modularity of partition
becomes simply
Thus, the partition is represented by the plane in . Analogous to the single-layer case, each point in the two-dimensional parameter space admits an optimal .
Given a set of partitions
, CHAMP calculates the coefficients of the
planes in Equation (
7) and solves a convex hull problem to find the convex intersection of the half-spaces above these partition-representing planes. That is, each partition is represented by a plane dividing
in two, thereby defining a half-space. The intersection of the half-spaces above all of these planes is the convex space of
values greater or equal to all observed quality values, with the boundary specifying the maximum modularity surface of the set. In single-layer networks, we considered ordered
and iteratively identified the next intersection and associated partition for increasing
. In the presence of multiple parameter dimensions here, we instead apply the Qhull implementation [
43,
44] of Quickhull [
45] to solve the dual convex hull problem. In practice, multiple partitions of the network can be identified in parallel, calculating and saving each set of
,
, and
coefficients. These coefficients defining the planes are then input into Qhull. CHAMP thereby prunes
to the subset admitted to the convex hull and identifies the convex polygonal domain in
where each partition is optimal (relative to
).
We note that in practice the runtime for finding the pruned subset of admissible partitions and associated domains of optimality is typically insignificant compared to that of identifying the input set of partitions in the first place. In particular, computing the scalar coefficients of the linear subspace of each partition is a direct calculation for M edges in the network. Meanwhile, the subsequent convex hull problem has no explicit dependence on the network size, depending instead on the number of partitions in the input set.
While we assume here that there is a single interlayer coupling parameter , we emphasize again that we do not restrict ourselves here to a particular form of the interlayer coupling, which might connect nearest-neighbor layers, all-to-all layers, connect only some nodes in one layer to those in another, and might have multiple different weights along different interlayer edges. Rather, we only require here that there is some selected interlayer coupling tensor C that is multiplied by .
Even more complicated interlayer couplings with multiple parameters (e.g., data that is both multiplex and temporal with the freedom to vary the relative weights between these couplings) can in principle be treated analogous to the above in the appropriate higher-dimensional space. With the notation
and
, we can write Equation (
7) as
, specifying linear subspaces of codimension one in higher-dimensional parameter spaces, given appropriate definitions of
and
. However, we do not go beyond two parameters
in our example results here.
For convenience we have implemented and distributed a python package for running and visualizing both the single layer and multilayer CHAMP found at [
46].
4. Discussion
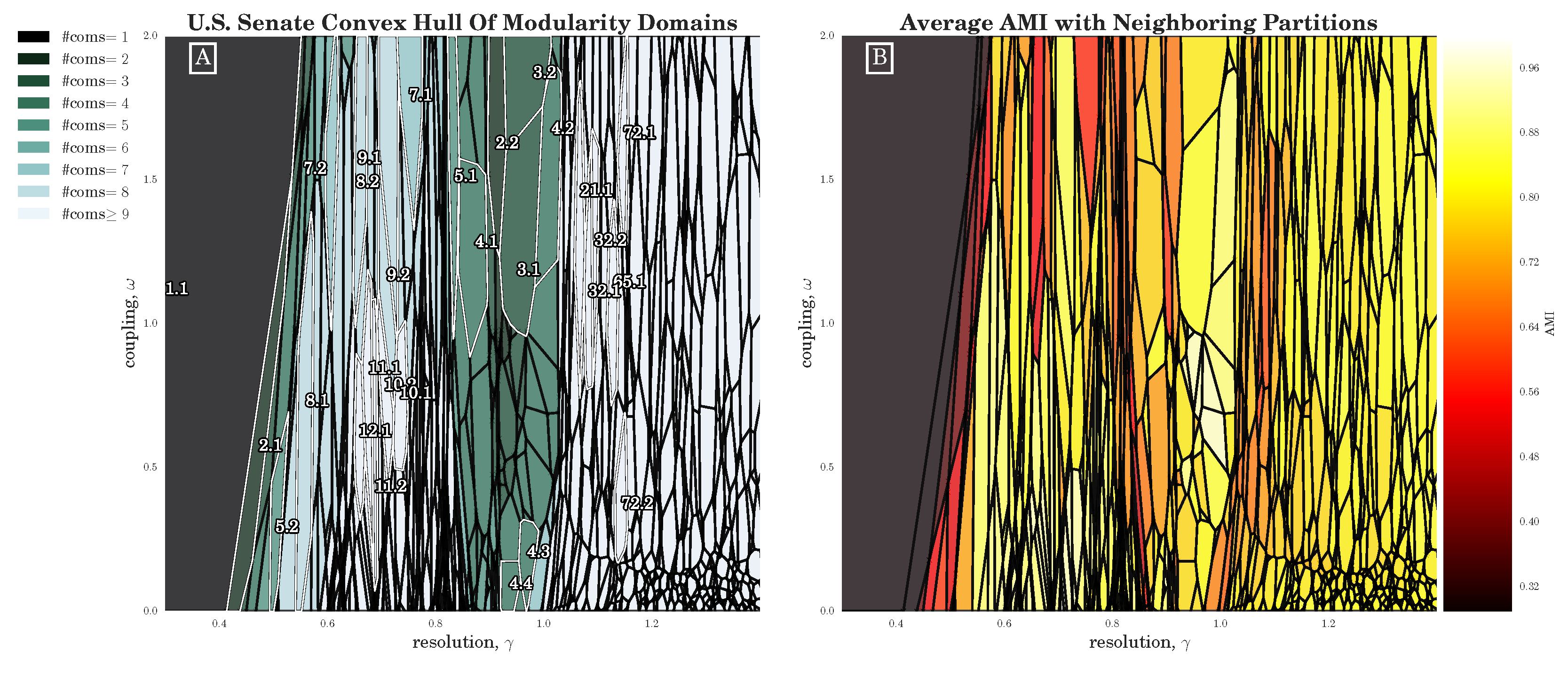
There are a number of features of CHAMP that make it a useful tool for community detection, as we have demonstrated by way of a variety of examples. By eliminating partitions that are non-admissible to the convex hull, CHAMP can greatly reduce the number of partitions remaining for consideration. By assessing the sizes of the domains of optimality of the partitions in the pruned admissible subset, and through direct pairwise comparisons of partitions in the admissible subset, CHAMP provides a framework for identifying stable parameter domains that signal robust community structures in the network.
The set of input partitions can be obtained as a result of a community-detection method across a range of parameter choices (as we explored here) or from the comparison of different community-detection methods. Ideally the input set contains near-optimal partitions with relevance for the application at hand. Because each partition is allowed to compete across the whole space of resolution and coupling parameters, CHAMP can surmount some of the pathologies associated with modularity-based community detection heuristics. For example, CHAMP has uncovered several cases where there is a parameter range over which Louvain consistently identifies suboptimal partitions compared to partitions that Louvain itself identifies at other parameter values. In our study of the Human Protein Reactome network (see
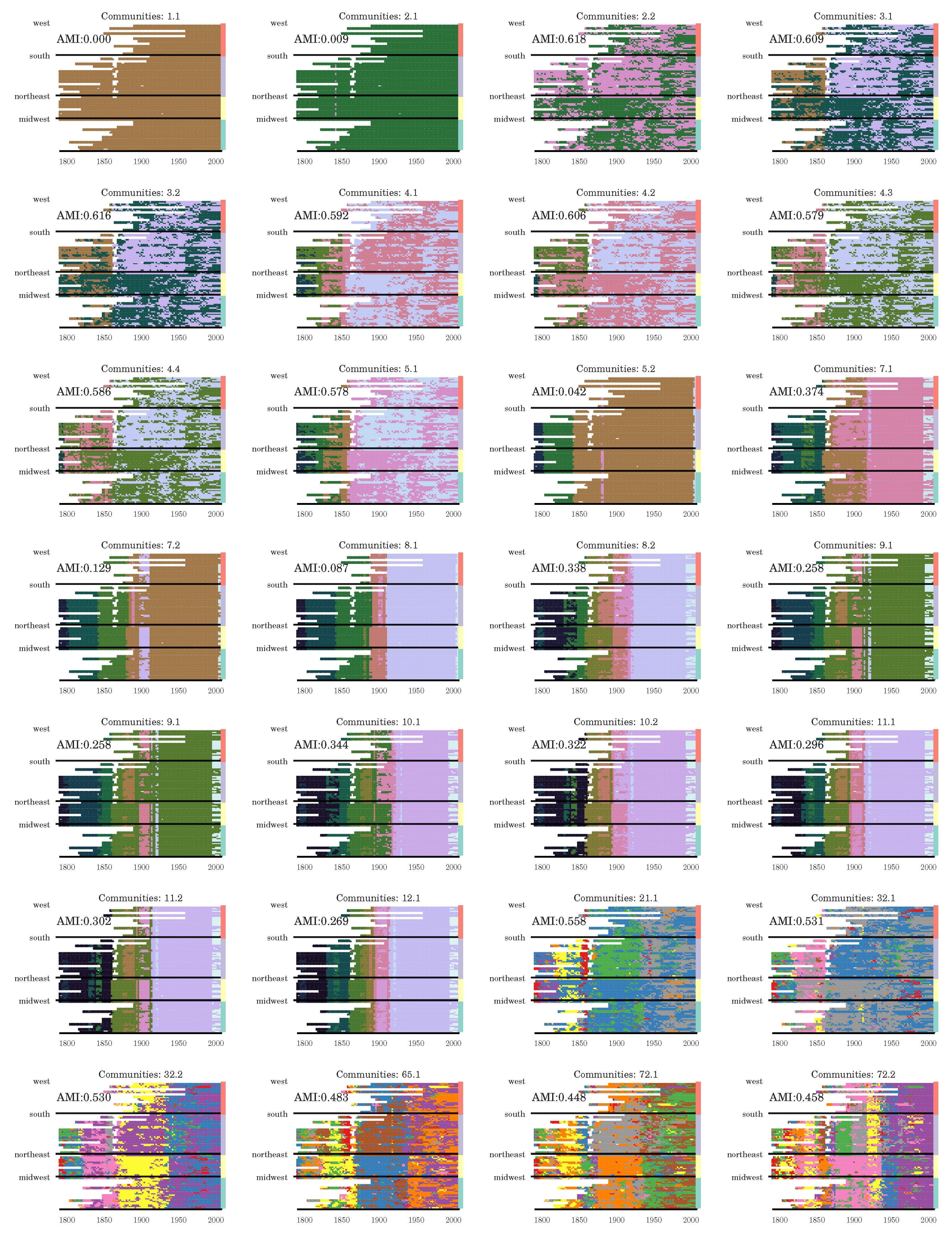
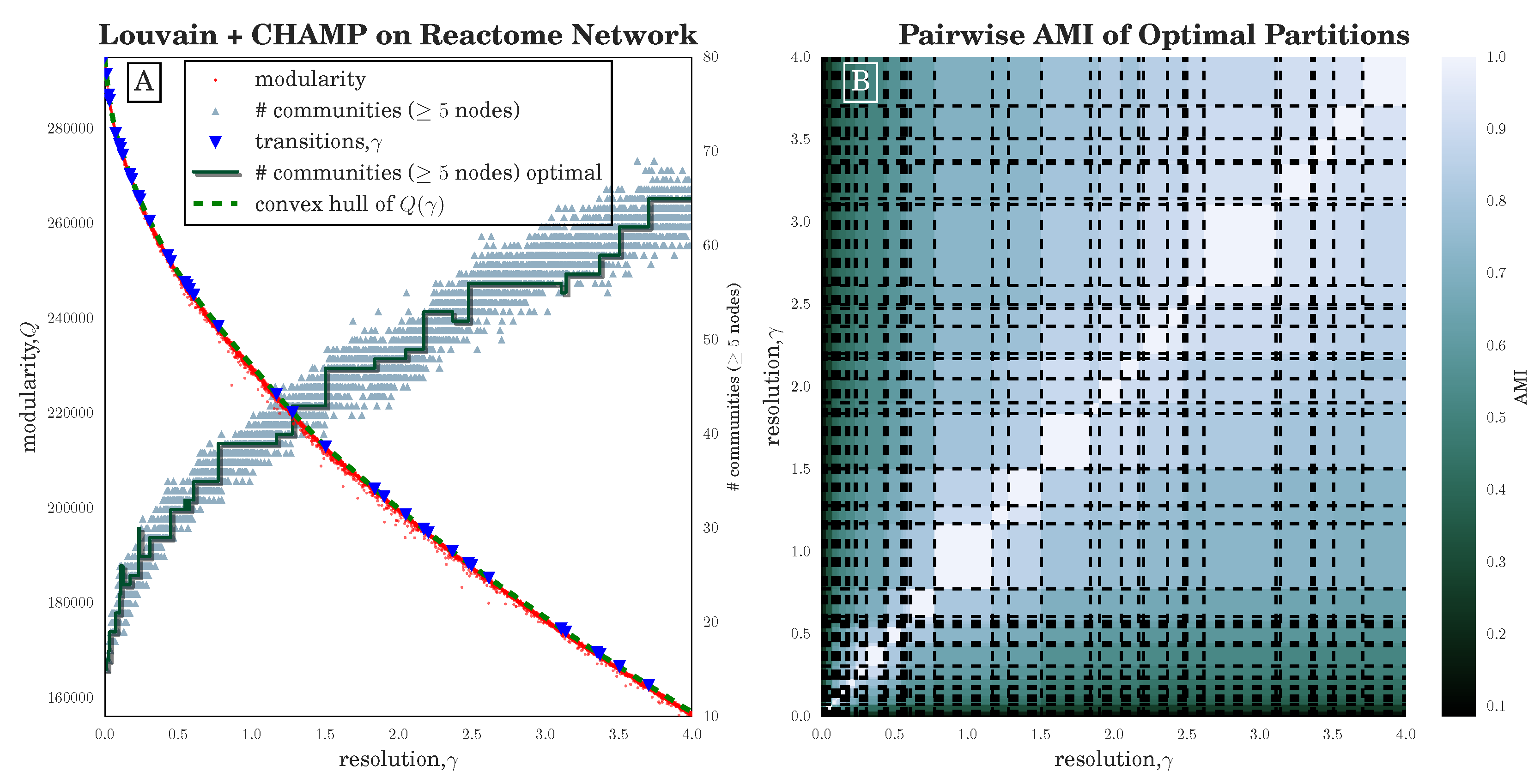
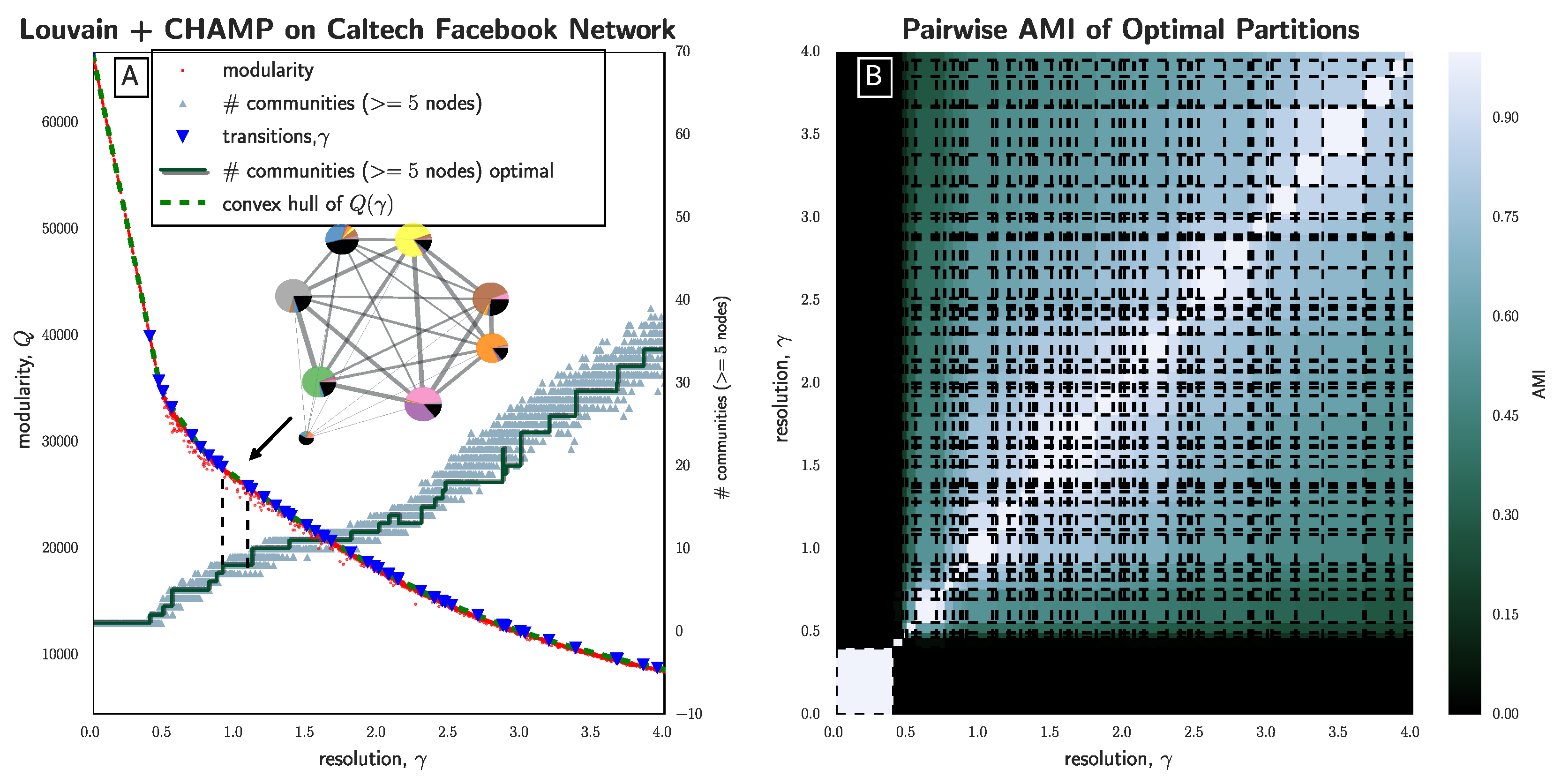
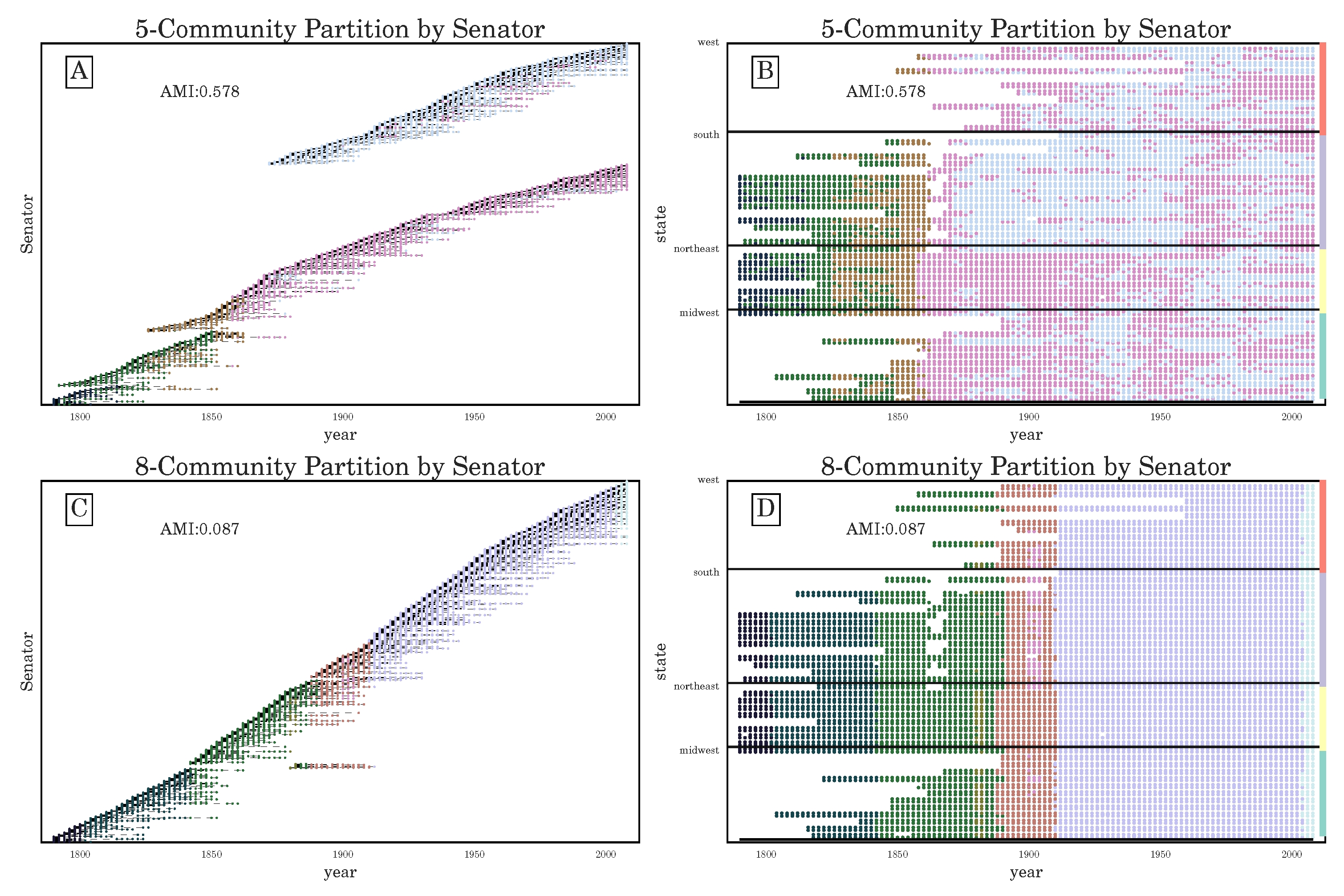
Section 3.2), we have seen that the stochasticity over multiple runs of the heuristic makes finding a plateau in the number of communities challenging; nevertheless, CHAMP is able to identify regions where a single partition is intrinsically stable, regardless of how frequently a particular detection algorithm uncovers such a partition. By identifying a manageable-sized and organized subset of admissible partitions with CHAMP, one can then apply a pairwise measure of similarity such as AMI to adjacent partitions to identify shifts in the landscape of optimal community structure.
We in no way claim that CHAMP resolves all of the problems with modularity-based methods (see, e.g., the discussion in [
3]). And CHAMP is certainly not the only way to try to process different results across various resolution parameters (see again the Introduction). However, by taking advantage of the underlying properties of modularity, including the fact that each partition defines a linear function for
Q in terms of the resolution and interlayer coupling parameters, CHAMP provides a principled method built directly on the definition of modularity to make better sense of the parameter space when modularity methods are employed. In particular, many of the various other proposed approaches assess each partition at the particular parameter value input into the community detection heuristic that found the partition, that is treating each partition as a single point in
. In contrast, CHAMP returns to the underlying definition of modularity with a resolution parameter to recognize that each partition here is more completely represented as a line in
[in the multilayer case, as a plane in
]. The single point is on that line but does not completely explore the potential of that partition to compete against the other identified partitions. By using the full linear subspace associated with each partition, CHAMP prunes away the vast majority of partitions in practice.
Importantly, CHAMP itself is not a method for partitioning a network, and as such its ability to highlight partitions is limited by the set of partitions given as input to the algorithm. Given the many available heuristics, the computational complexity of maximizing modularity [
17], and the potentially large number of near-optimal partitions [
18], it is possible that interesting and important community features may be missing from the provided input set. CHAMP as developed here is restricted to processing hard partitions of nodes into community labels, whereas overlapping communities and background nodes (those not belonging to any community) can be important for some applications. One may also reasonably worry about the potential value of partitions in the input set that are near-optimal over a wide domain of the parameters but yet never achieve admission to the convex hull itself and are thus discarded by the algorithm.
With the introduction of CHAMP presented here, we have left open many other possible uses of this general approach that may be worth exploring. Although we apply Louvain to discover partitions, CHAMP is agnostic to the detection method used to generate the set of partitions. The partitions input into CHAMP do not even need to be generated by modularity-maximizing heuristics; for example, one may also include new partitions as generated by ensemble learning [
35] or consensus clustering [
34,
36]. By comparing the results between sets of partitions generated by different methods, CHAMP might be useful as an additional method for making comparisons between these methods.
Of course, even with a resolution parameter, modularity may not be a good measure for what constitutes a good “community” in some networks, and one could investigate whether other quality functions with parameters might be explored with an analogous approach. Even within the consideration of modularity, it would be interesting to generalize the approach of CHAMP to exploring different scales as resolved with different self-loop weights as proposed in [
10] (see also [
11] for an application of this approach). Unlike the resolution and coupling parameters used here, changing the self-loop weight makes a nonlinear change to modularity. Nevertheless, we believe it may be possible to extend CHAMP to the self-loop method for resolving different scales. It would also be useful to extend CHAMP to methods for community structures with overlap and with background nodes.
In further developing CHAMP, it is important to recognize the inability of many community-detection algorithms to assess the reliability of identified communities versus apparent structures arising in random network models. The particular value of modularity, for example, does not immediately indicate whether an identified partition is significant; in fact, the modularities of many classes of random networks such as trees of fixed degree can be quite high in the asymptotic limit [
58,
59]. Thus, it may be interesting to use CHAMP to further explore and characterize the domains of optimization for partitions of such random networks, to determine the extent to which leveraging such partition stability information can address questions about detected structures and random noise.
Additionally, it would be interesting to study the consistency of optimality domains output from the application of CHAMP to different input sets of partitions in order to possibly provide insight about how quickly the convex intersection of half-spaces shrinks to the underlying true but unknown upper envelope as the set of input partitions grows. For the networks tested here, the numbers of admissible partitions remaining in the pruned subset were only a very small fraction of the numbers in the input sets. In our experience, the numbers of partitions in the final pruned admissible subset appeared to increase slowly as the size of the input set was increased, but the position of the larger domains appeared to remain relatively consistent in practice. The number of initial partitions needed to get a good mapping of the parameter space undoubtedly depends on the structure of the network and the computational heuristics used. It may also be possible to use a variant of CHAMP to iteratively steer the parameters at which additional partitions might be sought. For instance, input parameters that consistently give rise to dominant partitions with broad domains could be targeted for more runs in an iterative fashion.
In summary, we have presented the CHAMP algorithm as a post-processing tool for pruning a set of network partitions down to the admissible subset in the convex hull that optimizes modularity at different parameters. We have demonstrated the utility of CHAMP on various single-layer networks and on a multilayer network, identifying partitions and their associated domains of optimality in the parameter space. Further research may focus on how the sizes of these domains and the comparisons between domains can be best used to ascertain confidence in identified community structures, to explore subgraphs of a network, and to further process the admissible subset for consensus clustering, as well as other uses of the pruned subset identified by CHAMP.