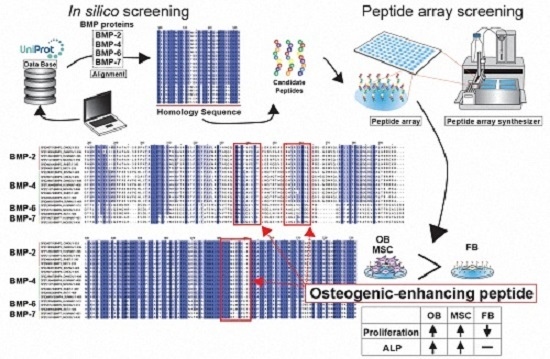
Screening of Osteogenic-Enhancing Short Peptides from BMPs for Biomimetic Material Applications
Abstract
:1. Introduction
2. Results
2.1. Conditions of Peptide Array Screening
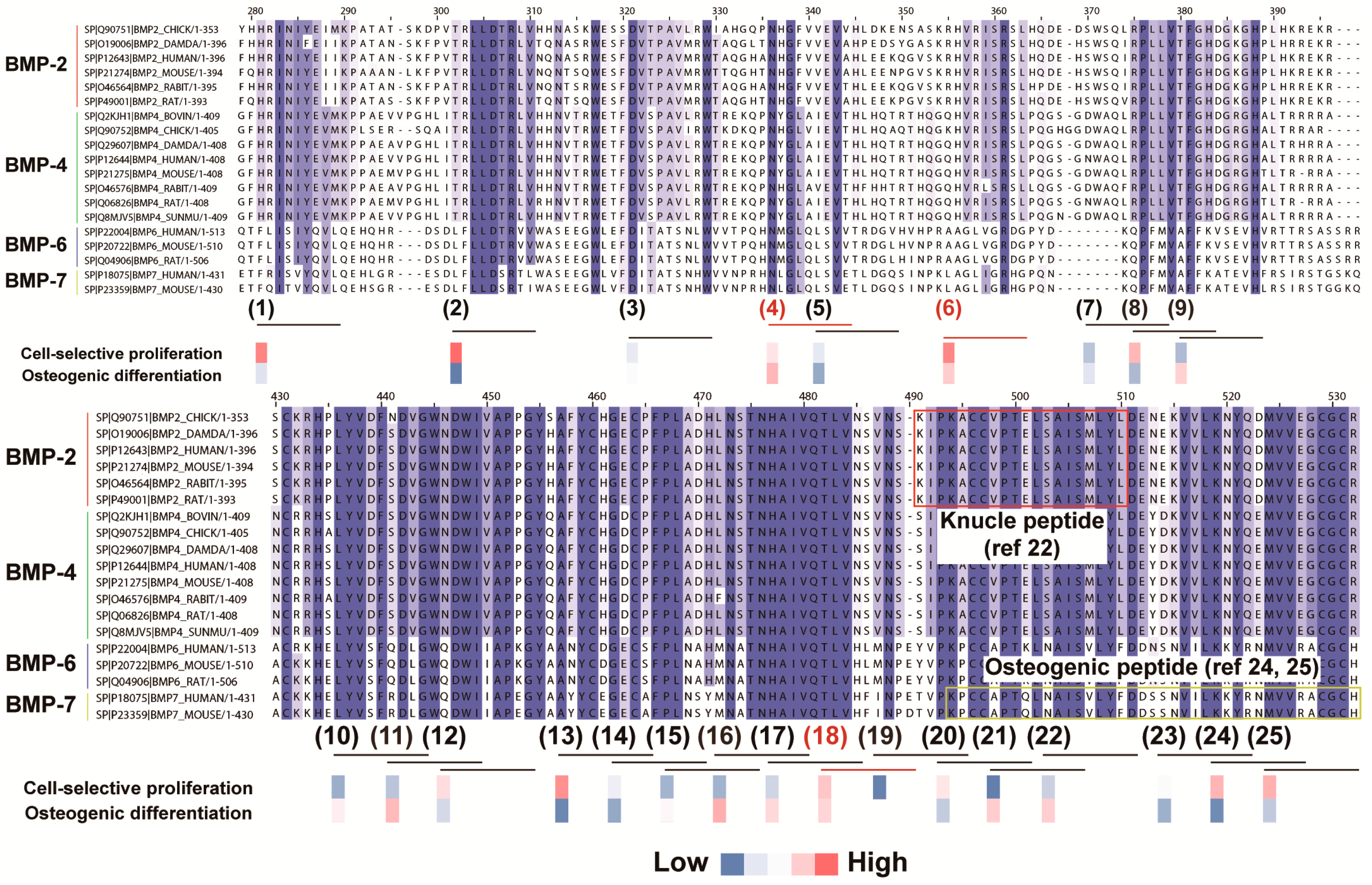
2.2. In Silico Screening to Determine Candidate Peptides
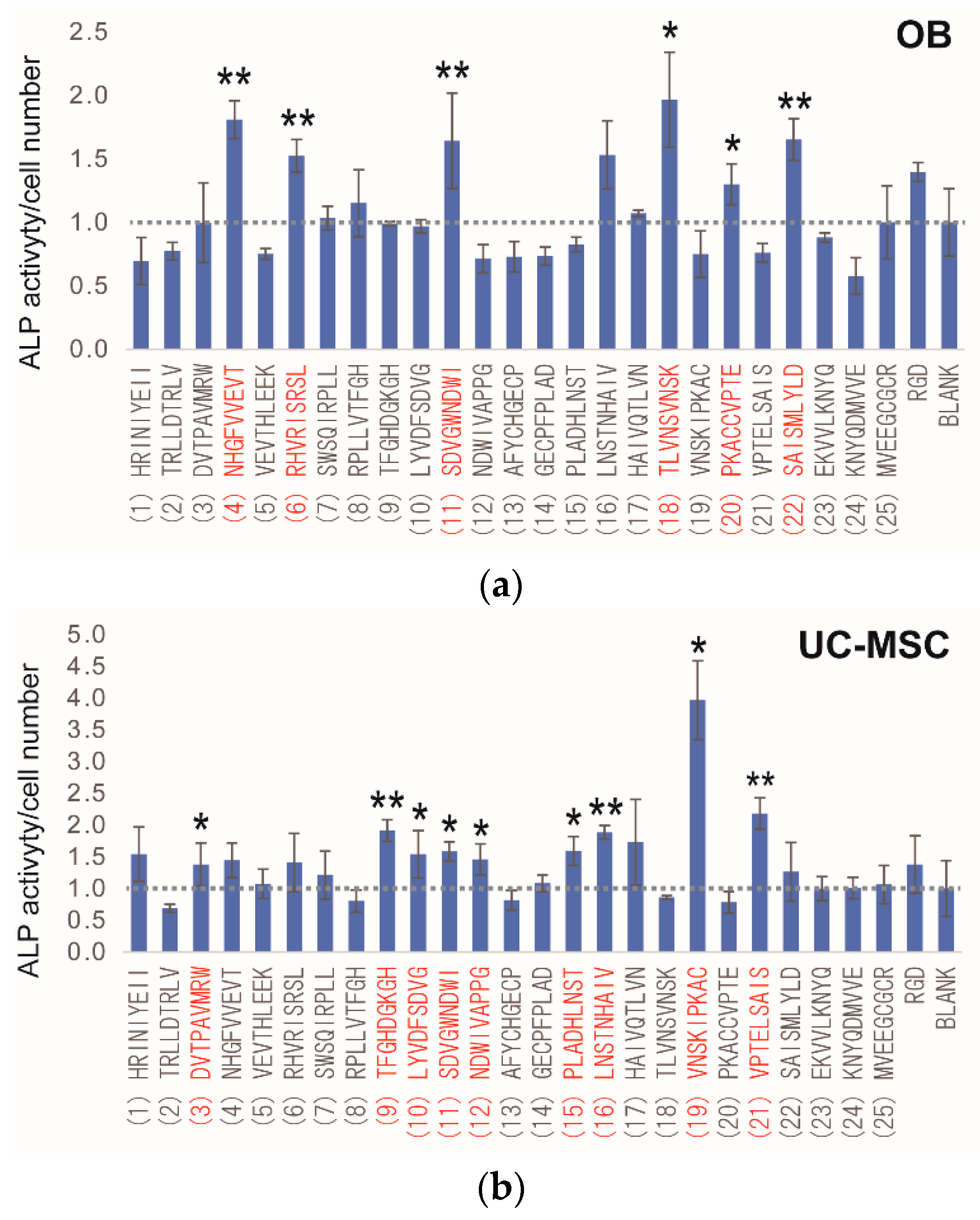
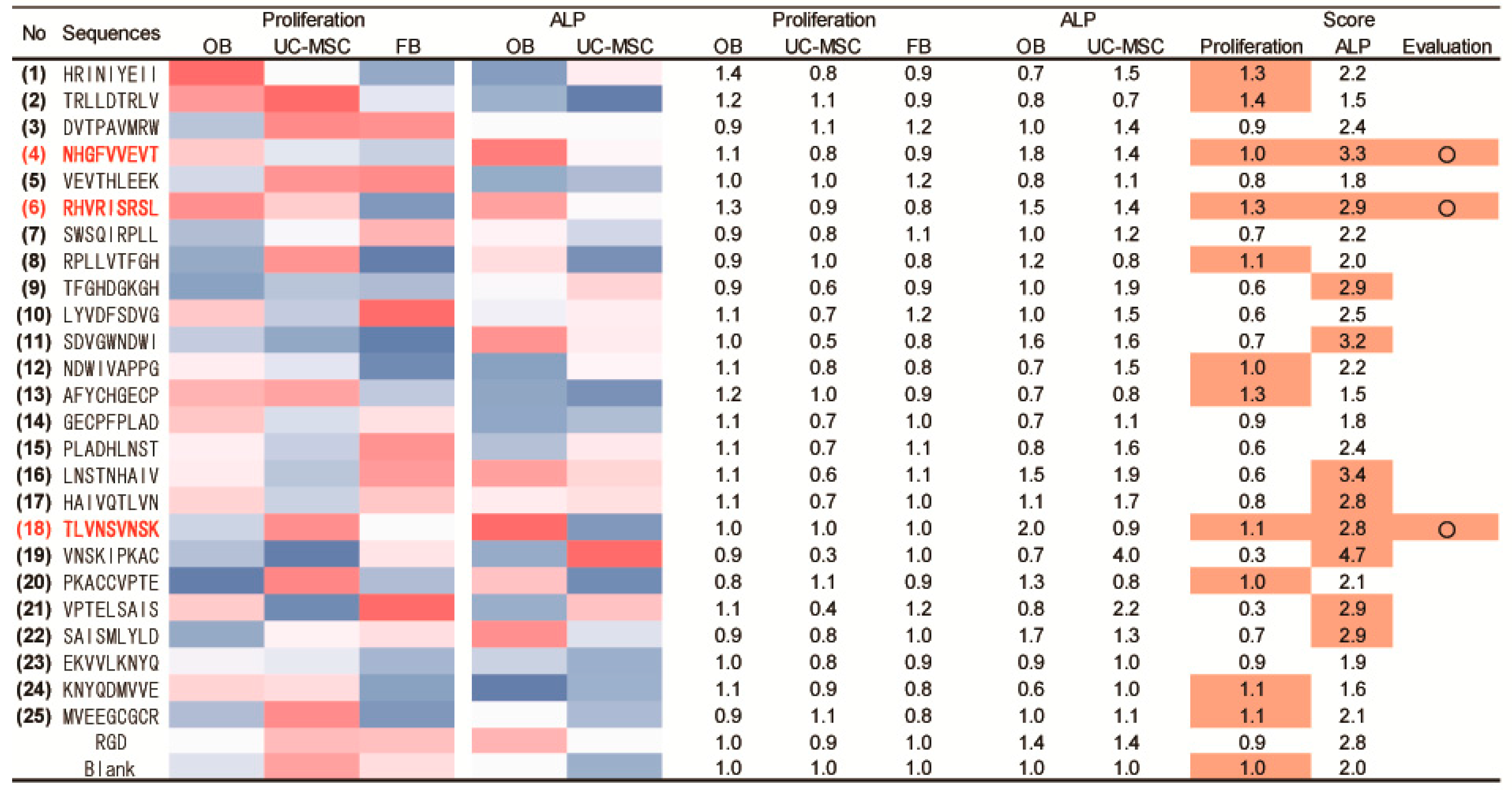
2.3. Peptide Array Screening for Osteogenic Proliferation Peptides
2.4. Peptide Array Screening for Osteogenic Differentiation Peptides
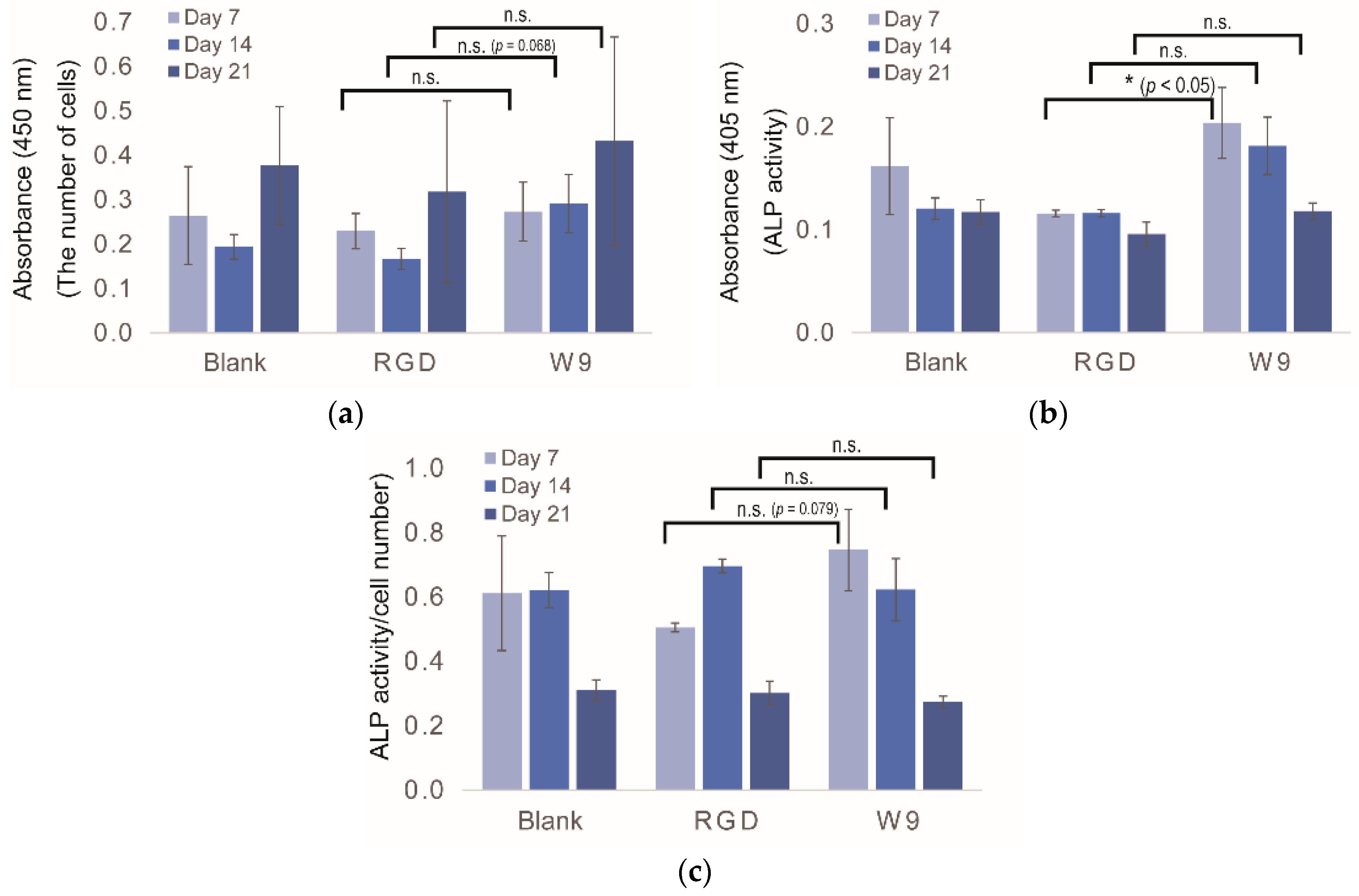
2.5. Determination of Osteogenic-Enhancing Peptides
3. Discussion
4. Materials and Methods
4.1. In Silico Screening for Candidate Peptides
4.2. Cell Culture
4.3. Peptide Array Synthesis
4.4. Cell Proliferation Assay Using the Peptide Array
4.5. ALP Activity Assay Using the Peptide Array
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Iwai, S.; Shimizu, H.; Suzawa, Y.; Akashi, M.; Yura, Y. Hydroxyapatite agarose composite gels as a biochemical material for the repair of alveolar bone defects due to cleft lip and palate. J. Oral Maxillofac. Surg. 2015, 27, 637–644. [Google Scholar] [CrossRef]
- Alhan, C.; Ariturk, C.; Senay, S.; Okten, M.; Gullu, A.U.; Kilic, L.; Karabulut, H.; Toraman, F. Use of bone wax is related to increased postoperative sternal dehiscence. Kardiochir. Torakochirurgia Pol. 2014, 11, 385–390. [Google Scholar] [PubMed]
- Fu, X.M.; Oshima, H.; Araki, Y.; Narita, Y.; Mutsuga, M.; Okada, N.; Tsunekawa, T.; Usui, A. A comparative study of two types of sternal pins used for sternal closure: Poly-l-lactide sternal pins versus uncalcined hydroxyapatite poly-l-lactide sternal pins. J. Artif. Organs 2013, 16, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, R.F.; Nielsen, P.H.; Terp, K.A.; Soballe, K.; Andersen, G.; Hasenkam, J.M. Effect of hemostatic material on sternal healing after cardiac surgery. Ann. Thorac. Surg. 2014, 97, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Shikinami, Y.; Matsusue, Y.; Nakamura, T. The complete process of bioresorption and bone replacement using devices made of forged composites of raw hydroxyapatite particles/poly l-lactide (F-u-HA/PLLA). Biomaterials 2005, 26, 5542–5551. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, A.; Seyedjafari, E.; Soleimani, M.; Ahmadbeigi, N.; Dinarvand, P.; Ghaemi, N. A comparison between osteogenic differentiation of human unrestricted somatic stem cells and mesenchymal stem cells from bone marrow and adipose tissue. Biotechnol. Lett. 2011, 33, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Kern, S.; Eichler, H.; Stoeve, J.; Kluter, H.; Bieback, K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells 2006, 24, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Ciavarella, S.; Dammacco, F.; De Matteo, M.; Loverro, G.; Silvestris, F. Umbilical cord mesenchymal stem cells: Role of regulatory genes in their differentiation to osteoblasts. Stem Cells Dev. 2009, 18, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.Z.; Ma, Q.J.; Cui, F.Z.; Zhong, Y.F. Human umbilical cord mesenchymal stem cells: Osteogenesis in vivo as seed cells for bone tissue engineering. J. Biomed. Mater. Res. A 2009, 91, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tang, M.; Weir, M.D.; Detamore, M.S.; Xu, H.H.K. Osteogenic media and rhBMP-2-induced differentiation of umbilical cord mesenchymal stem cells encapsulated in alginate microbeads and integrated in an injectable calcium phosphate-chitosan fibrous scaffold. Tissue Eng. Part A 2011, 17, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Urist, M.R. Bone—Formation by autoinduction. Science 1965, 150, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Kanakaris, N.K.; Petsatodis, G.; Tagil, M.; Giannoudis, P.V. Is there a role for bone morphogenetic proteins in osteoporotic fractures? Injury 2009, 40, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Carragee, E.J.; Hurwitz, E.L.; Weiner, B.K. A critical review of recombinant human bone morphogenetic protein-2 trials in spinal surgery: Emerging safety concerns and lessons learned. Spine J. 2011, 11, 471–491. [Google Scholar] [CrossRef] [PubMed]
- Moussa, M.; Carrel, J.P.; Scherrer, S.; Cattani-Lorente, M.; Wiskott, A.; Durual, S. Medium-term function of a 3D printed TCP/HA structure as a new osteoconductive scaffold for vertical bone augmentation: A simulation by BMP-2 activation. Materials 2015, 8, 2174–2190. [Google Scholar] [CrossRef]
- Pountos, I.; Georgouli, T.; Henshaw, K.; Bird, H.; Jones, E.; Giannoudis, P.V. The effect of bone morphogenetic protein-2, bone morphogenetic protein-7, parathyroid hormone, and platelet-derived growth factor on the proliferation and osteogenic differentiation of mesenchymal stem cells derived from osteoporotic bone. J. Orthop. Trauma 2010, 24, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Karfeld-Sulzer, L.S.; Siegenthaler, B.; Ghayor, C.; Weber, F.E. Fibrin hydrogel based bone substitute tethered with BMP-2 and BMP-2/7 heterodimers. Materials 2015, 8, 977–991. [Google Scholar] [CrossRef]
- Zhang, X.S.; Linkhart, T.A.; Chen, S.T.; Peng, H.R.; Wergedal, J.E.; Guttierez, G.G.; Sheng, M.H.C.; Lau, K.H.W.; Baylink, D.J. Local ex vivo gene therapy with bone marrow stromal cells expressing human BMP4 promotes endosteal bone formation in mice. J. Gene Med. 2004, 6, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.S.; Long, M.W.; Hankenson, K.D. Osteogenic differentiation of human mesenchymal stem cells is regulated by bone morphogenetic protein-6. J. Cell. Biochem. 2006, 98, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Senta, H.; Park, H.; Bergeron, E.; Drevelle, O.; Fong, D.; Leblanc, E.; Cabana, F.; Roux, S.; Grenier, G.; Faucheux, N. Cell responses to bone morphogenetic proteins and peptides derived from them: Biomedical applications and limitations. Cytokine Growth Factor Rev. 2009, 20, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Saito, A.; Suzuki, Y.; Ogata, S.; Ohtsuki, C.; Tanihara, M. Activation of osteo-progenitor cells by a novel synthetic peptide derived from the bone morphogenetic protein-2 knuckle epitope. Biochim. Biophys. Acta 2003, 1651, 60–67. [Google Scholar] [CrossRef]
- He, X.Z.; Ma, J.Y.; Jabbari, E. Effect of grafting rgd and BMP-2 protein-derived peptides to a hydrogel substrate on osteogenic differentiation of marrow stromal cells. Langmuir 2008, 24, 12508–12516. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, K.; Rheude, B.; Kim, Y.J.; White, K.; Dee, K.C. In vitro mineralization studies with substrate-immobilized bone morphogenetic protein peptides. J. Oral Implantol. 2003, 29, 57–65. [Google Scholar] [CrossRef]
- Chen, Y.P.; Webster, T.J. Increased osteoblast functions in the presence of BMP-7 short peptides for nanostructured biomaterial applications. J. Biomed. Mater. Res. A 2009, 91, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Lock, J.; Liu, H. Nanomaterials enhance osteogenic differentiation of human mesenchymal stem cells similar to a short peptide of BMP-7. Int. J. Nanomed. 2011, 6, 2769–2777. [Google Scholar]
- Kim, H.K.; Kim, J.H.; Park, D.S.; Park, K.S.; Kang, S.S.; Lee, J.S.; Jeong, M.H.; Yoon, T.R. Osteogenesis induced by a bone forming peptide from the prodomain region of BMP-7. Biomaterials 2012, 33, 7057–7063. [Google Scholar] [CrossRef] [PubMed]
- Furuya, Y.; Inagaki, A.; Khan, M.; Mori, K.; Penninger, J.M.; Nakamura, M.; Udagawa, N.; Aoki, K.; Ohya, K.; Uchida, K.; et al. Stimulation of bone formation in cortical bone of mice treated with a receptor activator of nuclear factor-kappab ligand (RANKL)-binding peptide that possesses osteoclastogenesis inhibitory activity. J. Boil. Chem. 2013, 288, 5562–5571. [Google Scholar] [CrossRef] [PubMed]
- Bab, I.; Gazit, D.; Chorev, M.; Muhlrad, A.; Shteyer, A.; Greenberg, Z.; Namdar, M.; Kahn, A. Histone H4-related osteogenic growth peptide (OGP): A novel circulating stimulator of osteoblastic activity. EMBO J. 1992, 11, 1867–1873. [Google Scholar] [PubMed]
- Spreafico, A.; Frediani, B.; Capperucci, C.; Leonini, A.; Gambera, D.; Ferrata, P.; Rosini, S.; Di Stefano, A.; Galeazzi, M.; Marcolongo, R. Osteogenic growth peptide effects on primary human osteoblast cultures: Potential relevance for the treatment of glucocorticoid-induced osteoporosis. J. Cell. Biochem. 2006, 98, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.Y.; Shao, L.; Zhao, H.M.; Zhong, Z.H.; Liu, J.Y.; Hao, C.G. Osteogenic growth peptide accelerates bone healing during distraction osteogenesis in rabbit tibia. J. Int. Med. Res. 2011, 39, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Fiocchi, C. Intestinal fibrosis in inflammatory bowel disease—Current knowledge and future perspectives. J. Crohns Colitis 2008, 2, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.R.; Brittan, M.; Alison, M.R. The role of bone marrow-derived cells in fibrosis. Cells Tissues Organs 2008, 188, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J. Inflammation, fibrosis, and modulation of the process by mesenchymal stem/stromal cells. Matrix Boil. 2016, 51, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Frank, R. Spot-synthesis—An easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 1992, 48, 9217–9232. [Google Scholar] [CrossRef]
- Frank, R. The spot synthesis technique—Synthetic peptide arrays on membrane supports—Principles and applications. J. Immunol. Methods 2002, 267, 13–26. [Google Scholar] [CrossRef]
- Kato, R.; Kaga, C.; Kunimatsu, M.; Kobayashi, T.; Honda, H. Peptide array-based interaction assay of solid-bound peptides and anchorage-dependant cells and its effectiveness in cell-adhesive peptide design. J. Biosci. Bioeng. 2006, 101, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Kato, R.; Kaga, C.; Kanie, K.; Kunimatsu, M.; Okochi, M.; Honda, H. Peptide array-based peptide-cell interaction analysis. Mini-Rev. Org. Chem. 2011, 8, 171–177. [Google Scholar] [CrossRef]
- Kanie, K.; Kato, R.; Zhao, Y.Z.; Narita, Y.; Okochi, M.; Honda, H. Amino acid sequence preferences to control cell-specific organization of endothelial cells, smooth muscle cells, and fibroblasts. J. Pept. Sci. 2011, 17, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Kanie, K.; Narita, Y.; Zhao, Y.Z.; Kuwabara, F.; Satake, M.; Honda, S.; Kaneko, H.; Yoshioka, T.; Okochi, M.; Honda, H.; et al. Collagen type IV-specific tripeptides for selective adhesion of endothelial and smooth muscle cells. Biotechnol. Bioeng. 2012, 109, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, F.; Narita, Y.; Yamawaki-Ogata, A.; Kanie, K.; Kato, R.; Satake, M.; Kaneko, H.; Oshima, H.; Usui, A.; Ueda, Y. Novel small-caliber vascular grafts with trimeric peptide for acceleration of endothelialization. Ann. Thorac. Surg. 2012, 93, 156–163. [Google Scholar] [CrossRef] [PubMed]

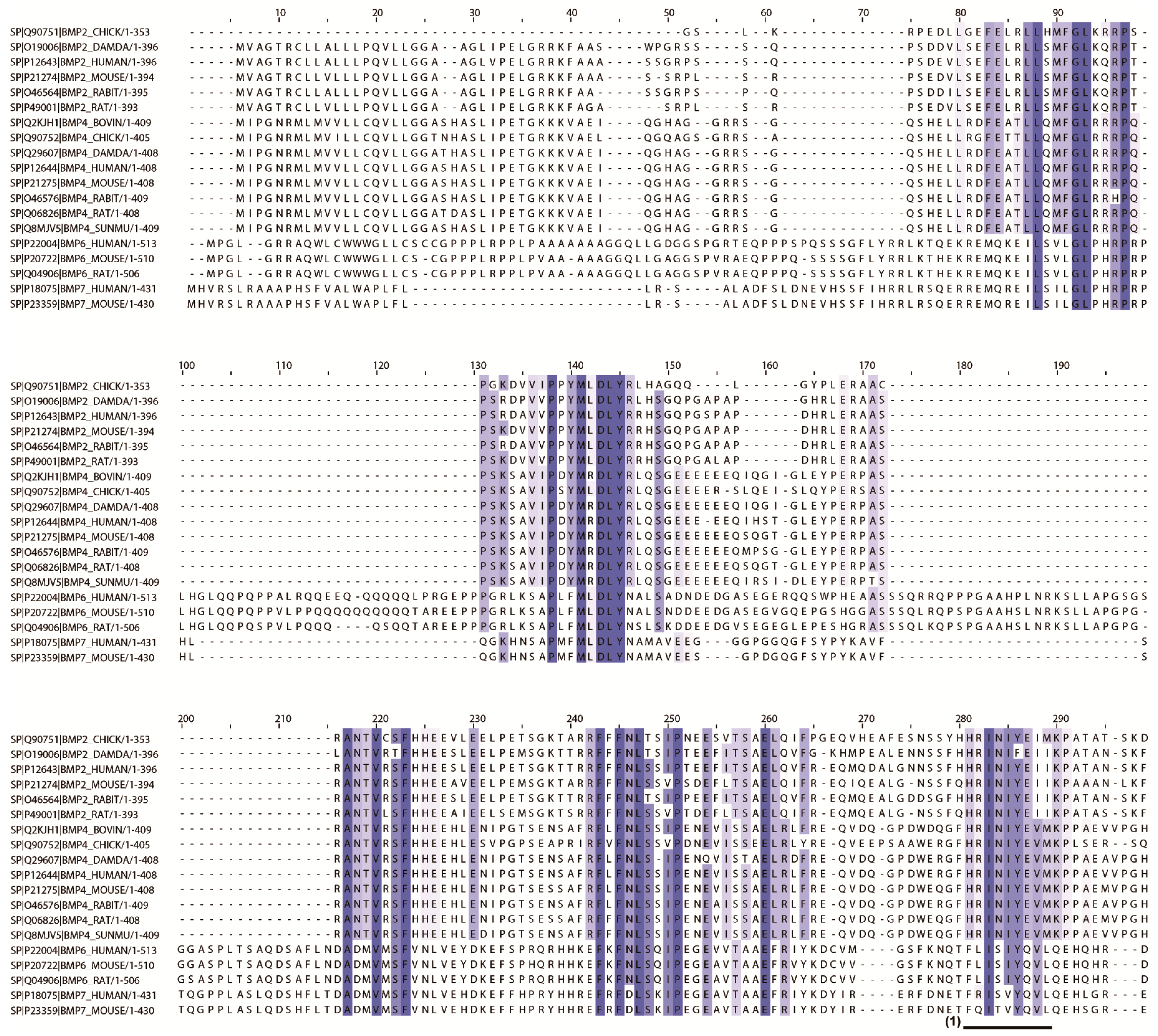


| No. | Entry | Entry Name | Protein Names | Organism | Length |
|---|---|---|---|---|---|
| 1 | Q90751 | BMP2_CHICK | Bone morphogenetic protein 2 (BMP-2) (fragment) | Gallus gallus (chicken) | 353 |
| 2 | O19006 | BMP2_DAMDA | Bone morphogenetic protein 2 (BMP-2) | Dama dama (fallow deer) (Cervus dama) | 396 |
| 3 | P12643 | BMP2_HUMAN | Bone morphogenetic protein 2 (BMP-2) (bone morphogenetic protein 2A) (BMP-2A) | Homo sapiens (human) | 396 |
| 4 | P21274 | BMP2_MOUSE | Bone morphogenetic protein 2 (BMP-2) (bone morphogenetic protein 2A) (BMP-2A) | Mus musculus (mouse) | 394 |
| 5 | O46564 | BMP2_RABIT | Bone morphogenetic protein 2 (BMP-2) | Oryctolagus cuniculus (rabbit) | 395 |
| 6 | P49001 | BMP2_RAT | Bone morphogenetic protein 2 (BMP-2) (bone morphogenetic protein 2A) (BMP-2A) | Rattus norvegicus (rat) | 393 |
| 7 | Q2KJH1 | BMP4_BOVIN | Bone morphogenetic protein 4 (BMP-4) | Bos taurus (bovine) | 409 |
| 8 | Q90752 | BMP4_CHICK | Bone morphogenetic protein 4 (BMP-4) | Gallus gallus (chicken) | 405 |
| 9 | Q29607 | BMP4_DAMDA | Bone morphogenetic protein 4 (BMP-4) | Dama dama (fallow deer) (Cervus dama) | 408 |
| 10 | P12644 | BMP4_HUMAN | Bone morphogenetic protein 4 (BMP-4) (bone morphogenetic protein 2B) (BMP-2B) | Homo sapiens (human) | 408 |
| 11 | P21275 | BMP4_MOUSE | Bone morphogenetic protein 4 (BMP-4) (bone morphogenetic protein 2B) (BMP-2B) | Mus musculus (mouse) | 408 |
| 12 | O46576 | BMP4_RABIT | Bone morphogenetic protein 4 (BMP-4) | Oryctolagus cuniculus (rabbit) | 409 |
| 13 | Q06826 | BMP4_RAT | Bone morphogenetic protein 4 (BMP-4) (bone morphogenetic protein 2B) (BMP-2B) | Rattus norvegicus (rat) | 408 |
| 14 | Q8MJV5 | BMP4_SUNMU | Bone morphogenetic protein 4 (BMP-4) (sBmp4) | Suncus murinus (Asian house shrew) (musk shrew) | 409 |
| 15 | P22004 | BMP6_HUMAN | Bone morphogenetic protein 6 (BMP-6) (VG-1-related protein) (VG-1-R) (VGR-1) | Homo sapiens (human) | 513 |
| 16 | P20722 | BMP6_MOUSE | Bone morphogenetic protein 6 (BMP-6) (VG-1-related protein) (VGR-1) | Mus musculus (mouse) | 510 |
| 17 | Q04906 | BMP6_RAT | Bone morphogenetic protein 6 (BMP-6) (VG-1-related protein) (VGR-1) | Rattus norvegicus (rat) | 506 |
| 18 | P18075 | BMP7_HUMAN | Bone morphogenetic protein 7 (BMP-7) (osteogenic protein 1) (OP-1) (eptotermin alfa) | Homo sapiens (human) | 431 |
| 19 | P23359 | BMP7_MOUSE | Bone morphogenetic protein 7 (BMP-7) (osteogenic protein 1) (OP-1) | Mus musculus (mouse) | 430 |
| No. | Peptide Sequences |
|---|---|
| (1) | HRINIYEII |
| (2) | TRLLDTRLV |
| (3) | DVTPAVMRW |
| (4) | NHGFVVEVT |
| (5) | VEVTHLEEK |
| (6) | RHVRISRSL |
| (7) | SWSQIRPLL |
| (8) | RPLLVTFGH |
| (9) | TFGHDGKGH |
| (10) | LYVDFSDVG |
| (11) | SDVGWNDWI |
| (12) | NDWIVAPPG |
| (13) | AFYCHGECP |
| (14) | GECPFPLAD |
| (15) | PLADHLNST |
| (16) | LNSTNHAIV |
| (17) | HAIVQTLVN |
| (18) | TLVNSVNSK |
| (19) | VNSKIPKAC |
| (20) | PKACCVPTE |
| (21) | VPTELSAIS |
| (22) | SAISMLYLD |
| (23) | EKVVLKNYQ |
| (24) | KNYQDMVVE |
| (25) | MVEEGCGCR |
| RGD | |
| Blank (no peptide) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanie, K.; Kurimoto, R.; Tian, J.; Ebisawa, K.; Narita, Y.; Honda, H.; Kato, R. Screening of Osteogenic-Enhancing Short Peptides from BMPs for Biomimetic Material Applications. Materials 2016, 9, 730. https://doi.org/10.3390/ma9090730
Kanie K, Kurimoto R, Tian J, Ebisawa K, Narita Y, Honda H, Kato R. Screening of Osteogenic-Enhancing Short Peptides from BMPs for Biomimetic Material Applications. Materials. 2016; 9(9):730. https://doi.org/10.3390/ma9090730
Chicago/Turabian StyleKanie, Kei, Rio Kurimoto, Jing Tian, Katsumi Ebisawa, Yuji Narita, Hiroyuki Honda, and Ryuji Kato. 2016. "Screening of Osteogenic-Enhancing Short Peptides from BMPs for Biomimetic Material Applications" Materials 9, no. 9: 730. https://doi.org/10.3390/ma9090730
APA StyleKanie, K., Kurimoto, R., Tian, J., Ebisawa, K., Narita, Y., Honda, H., & Kato, R. (2016). Screening of Osteogenic-Enhancing Short Peptides from BMPs for Biomimetic Material Applications. Materials, 9(9), 730. https://doi.org/10.3390/ma9090730