A Sensitive DNAzyme-Based Chiral Sensor for Lead Detection
Abstract
:1. Introduction
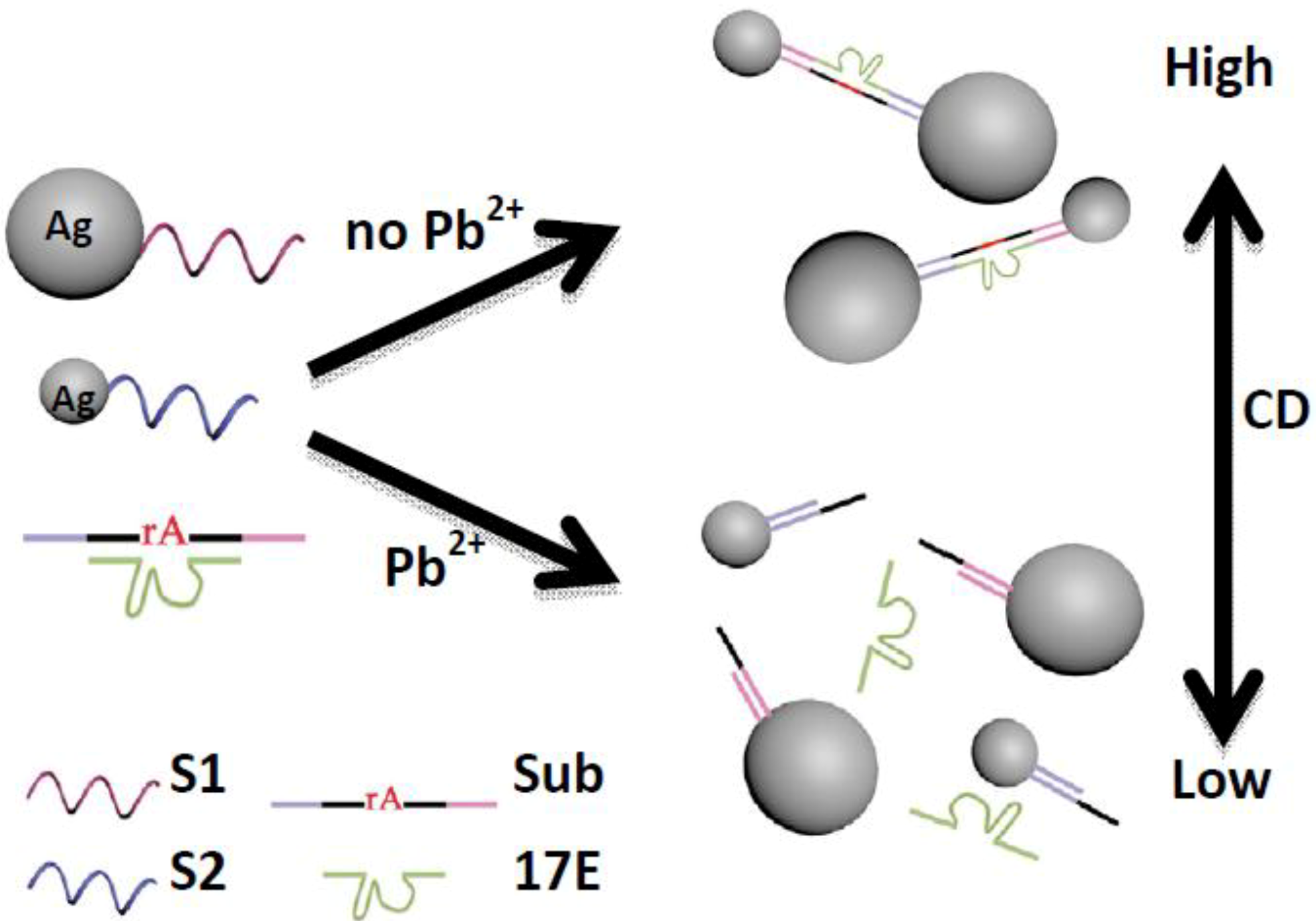
2. Results and Discussion


| Spiked level (ng∙mL−1) | Detected level Mean a ± SD b (ng∙mL−1) | Recovery (%) |
|---|---|---|
| 5 | 4.81 ± 0.44 | 96 |
| 2 | 1.97 ± 0.15 | 98 |
| 1 | 0.94 ± 0.04 | 94 |
| 0.5 | 0.48 ± 0.05 | 95 |
| 0.2 | 0.19 ± 0.04 | 97 |
| 0.1 | 0.10 ± 0.01 | 100 |
| 0.05 | 0.05 ± 0.01 | 100 |
3. Materials and Methods
3.1. Materials and Instruments
- S1: 5'-TCACAGATGAGT-SH-3';
- S2: 5'-SH-CACGAGTTGACA-3';
- 17E: 5'-CATCTCTTCTCCGAGCCGGTCGAAATAGTGAGT-3';
- Sub: 5'-ACTCATCTGTGAACTCACTAT(rA)GGAAGAGATGTGTCAACTCGTG-3'.
3.2. Synthesis of Silver NPs
3.3. AgNPs Functionalized with DNA
3.4. Construction of the DNAzyme-Based Sensor
3.5. Specificity Tests
3.6. Recovery in Tap Water Samples
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Godwin, H.A. The biological chemistry of lead. Curr. Opin. Chem. Biol. 2001, 5, 223–227. [Google Scholar] [CrossRef]
- Marsden, P.A. Increased body lead burden-cause or consequence of chronic renal insufficiency? N. Engl. J. Med. 2003, 348, 345–346. [Google Scholar] [CrossRef] [PubMed]
- Magyar, J.S.; Weng, T.C.; Stern, C.M.; Dye, D.F.; Rous, B.W.; Payne, J.C.; Bridgewater, B.M.; Mijovilovich, A.; Parkin, G.; Zaleski, J.M.; Penner-Hahn, J.E.; Godwin, H.A. Reexamination of lead (II) coordination preferences in sulfur-rich sites, implications for a critical mechanism of lead poisoning. J. Am. Chem. Soc. 2005, 127, 9495–9505. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, S.V.; Aimo, L.; Oteiza, P.I. Aluminium and lead, molecular mechanisms of brain toxicity. Arch. Toxicol. 2008, 82, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Grinshtein, I.L.; Vilpan, Y.A.; Vasilieva, L.A.; Kopeikin, V.A. Direct atomic absorption determination of cadmium and lead in strongly interfering matrices by double vaporization with a two-step electrothermal atomizer. Spectrochim. Acta Part B Atomic Spectrosc. 2001, 56, 261–274. [Google Scholar] [CrossRef]
- Aydin, F.A.; Soylak, M. Separation, preconcentration and inductively coupled plasma-mass spectrometric (ICP-MS) determination of thorium (IV), titanium (IV), iron (III), lead (II) and chromium (III) on 2-nitroso-1-naphthol impregnated MCI GEL CHP20P resin. J. Hazard. Mater. 2010, 173, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [PubMed]
- Breaker, R.R. DNA enzymes. Nat. Biotechnol. 1997, 15, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004, 73, 791–836. [Google Scholar] [CrossRef] [PubMed]
- Keiper, S.; Vyle, J.S. Reversible photocontrol of deoxyribozyme-catalyzed RNA cleavage under multiple-turnover conditions. Angew. Chem. 2006, 118, 3384–3387. [Google Scholar] [CrossRef]
- Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 2008, 37, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, J. Functional DNA nanotechnology, emerging applications of DNAzymes and aptamers. Curr. Opin. Biotechnol. 2006, 17, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, J. Smart nanomaterials inspired by biology, dynamic assembly of error-free nanomaterials in response to multiple chemical and biological stimuli. Acc. Chem. Res. 2007, 40, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, Y. Functional DNA directed assembly of nanomaterials for biosensing. J. Mater. Chem. 2009, 19, 1788–1798. [Google Scholar] [CrossRef]
- Xiang, Y.; Wang, Z.; Xing, H.; Lu, Y. Expanding DNAzyme functionality through enzyme cascades with applications in single nucleotide repair and tunable DNA-directed assembly of nanomaterials. Chem. Sci. 2013, 4, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 6642–6643. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Accelerated color change of gold nanoparticles assembled by DNAzymes for simple and fast colorimetric Pb2+ detection. J. Am. Chem. Soc. 2004, 126, 12298–12305. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Rowe, A.A.; Plaxco, K.W. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J. Am. Chem. Soc. 2007, 129, 262–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Chang, H.-T.; Shiang, Y.C.; Hung, Y.L.; Chiang, C.K.; Huang, C.C. Colorimetric assay for lead ions based on the leaching of gold nanoparticles. Anal. Chem. 2009, 81, 9433–9439. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Lin, Z.; Chen, L.; Qiu, B.; Chen, G. A sensitive and specific electrochemiluminescent sensor for lead based on DNAzyme. Chem. Commun. 2009, 40, 6050–6052. [Google Scholar] [CrossRef]
- Mazumdar, D.; Liu, J.; Lu, G.; Zhou, J.; Lu, Y. Easy-to-use dipstick tests for detection of lead in paints using non-cross-linked gold nanoparticle–DNAzyme conjugates. Chem. Commun. 2010, 46, 1416–1418. [Google Scholar] [CrossRef]
- Miao, X.; Ling, L.; Shuai, X. Ultrasensitive detection of lead (II) with DNAzyme and gold nanoparticles probes by using a dynamic light scattering technique. Chem. Commun. 2011, 47, 4192–4194. [Google Scholar] [CrossRef]
- Zhang, Z.; Balogh, D.; Wang, F.; Willner, I. Smart mesoporous SiO2 nanoparticles for the DNAzyme-induced multiplexed release of substrates. J. Am. Chem. Soc. 2013, 135, 1934–1940. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhou, Y.; Tang, Z. Chiral inorganic nanoparticles, origin, optical properties and bioapplications. Nanoscale 2011, 3, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Liu, W.; Li, Z.; Han, B.; Zhou, Y.; Gao, Y.; Tang, Z. Manipulation of collective optical activity in one-dimensional plasmonic assembly. ACS Nano 2012, 6, 2326–2332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, J.; Wang, Y.; Chen, H. Emerging chirality in nanoscience. Chem. Soc. Rev. 2013, 42, 2930–2962. [Google Scholar] [CrossRef] [PubMed]
- Mastroianni, A.J.; Claridge, S.A.; Alivisatos, A.P. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J. Am. Chem. Soc. 2009, 131, 8455–8459. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Schreiber, R.; Fan, Z.; Pardatscher, G.; Roller, E.M.; Högele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhu, Z.; Liu, W.; Zhou, Y.; Han, B.; Gao, Y.; Tang, Z. Reversible plasmonic circular dichroism of Au nanorod and DNA assemblies. J. Am. Chem. Soc. 2012, 134, 3322–3325. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Xu, L.; Xu, C.; Ma, W.; Kuang, H.; Wang, L.; Kotov, N.A. Self-assembly of chiral nanoparticle pyramids with strong R/S optical activity. J. Am. Chem. Soc. 2012, 134, 15114–15121. [Google Scholar] [PubMed]
- Zhao, Y.; Xu, L.; Kuang, H.; Wang, L.B.; Xu, C.L. Asymmetric and symmetric PCR of gold nanoparticles, a pathway to scaled-up self-assembly with tunable chirality. J. Mater. Chem. 2012, 22, 5574–5580. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, L.; Liz-Marzán, L.M.; Kuang, H.; Ma, W.; Asenjo-García, A.; Garcia de Abajo, J.; Kotov, N.A.; Wang, L.; Xu, C. Alternating plasmonic nanoparticle heterochains made by polymerase chain reaction and their optical properties. J. Phys. Chem. Lett. 2013, 4, 641–647. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, L.; Zhu, Y.; Ma, W.; Kuang, H.; Wang, L.; Xu, C. Chirality based sensor for bisphenol A detection. Chem. Commun. 2012, 48, 5760–5762. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, L.; Ma, W.; Xu, Z.; Kuang, H.; Wang, L.B.; Xu, C.L. A one-step homogeneous plasmonic circular dichroism detection of aqueous mercury ions using nucleic acid functionalized gold nanorods. Chem. Commun. 2012, 48, 11889–11891. [Google Scholar] [CrossRef]
- Kim, H.; Ren, W.; Kim, J.; Yoon, J. Fluorescent and colorimetric sensors for detection of lead, cadmium, and mercury ions. Chem. Soc. Rev. 2012, 41, 3210–3244. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Xing, C.R.; Hao, C.L.; Liu, L.Q.; Wang, L.B.; Xu, C.L. Rapid and highly sensitive detection of lead ions in drinking water based on a strip immunosensor. Sensors 2013, 13, 4214–4224. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xia, Y. Triangular nanoplates of silver, synthesis, characterization, and use as sacrificial templates for generating triangular nanorings of gold. Adv. Mater. 2003, 15, 695–699. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kuang, H.; Yin, H.; Xing, C.; Xu, C. A Sensitive DNAzyme-Based Chiral Sensor for Lead Detection. Materials 2013, 6, 5038-5046. https://doi.org/10.3390/ma6115038
Kuang H, Yin H, Xing C, Xu C. A Sensitive DNAzyme-Based Chiral Sensor for Lead Detection. Materials. 2013; 6(11):5038-5046. https://doi.org/10.3390/ma6115038
Chicago/Turabian StyleKuang, Hua, Honghong Yin, Changrui Xing, and Chuanlai Xu. 2013. "A Sensitive DNAzyme-Based Chiral Sensor for Lead Detection" Materials 6, no. 11: 5038-5046. https://doi.org/10.3390/ma6115038
APA StyleKuang, H., Yin, H., Xing, C., & Xu, C. (2013). A Sensitive DNAzyme-Based Chiral Sensor for Lead Detection. Materials, 6(11), 5038-5046. https://doi.org/10.3390/ma6115038




