Biological Synthesis of Copper Nanoparticles Using Edible Plant Allium monanthum: Characterization of Antibacterial, Antioxidant, and Anti-Inflammatory Properties Using In Silico Molecular Docking Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Methods
2.2. Preparation of A. monanthum Plant Extract
2.3. Synthesis of Copper Nanoparticles Using A. monanthum Extract
2.4. Characterization of the AM-CuNPs
2.5. Antibacterial Activity of AM-CuNPs
2.6. Antioxidant Activity of Synthesized AM-CuNPs
2.7. In Silico Analysis of Anti-Inflammatory Effect of AM Synthesized CuNPs
2.7.1. Ligand Preparation
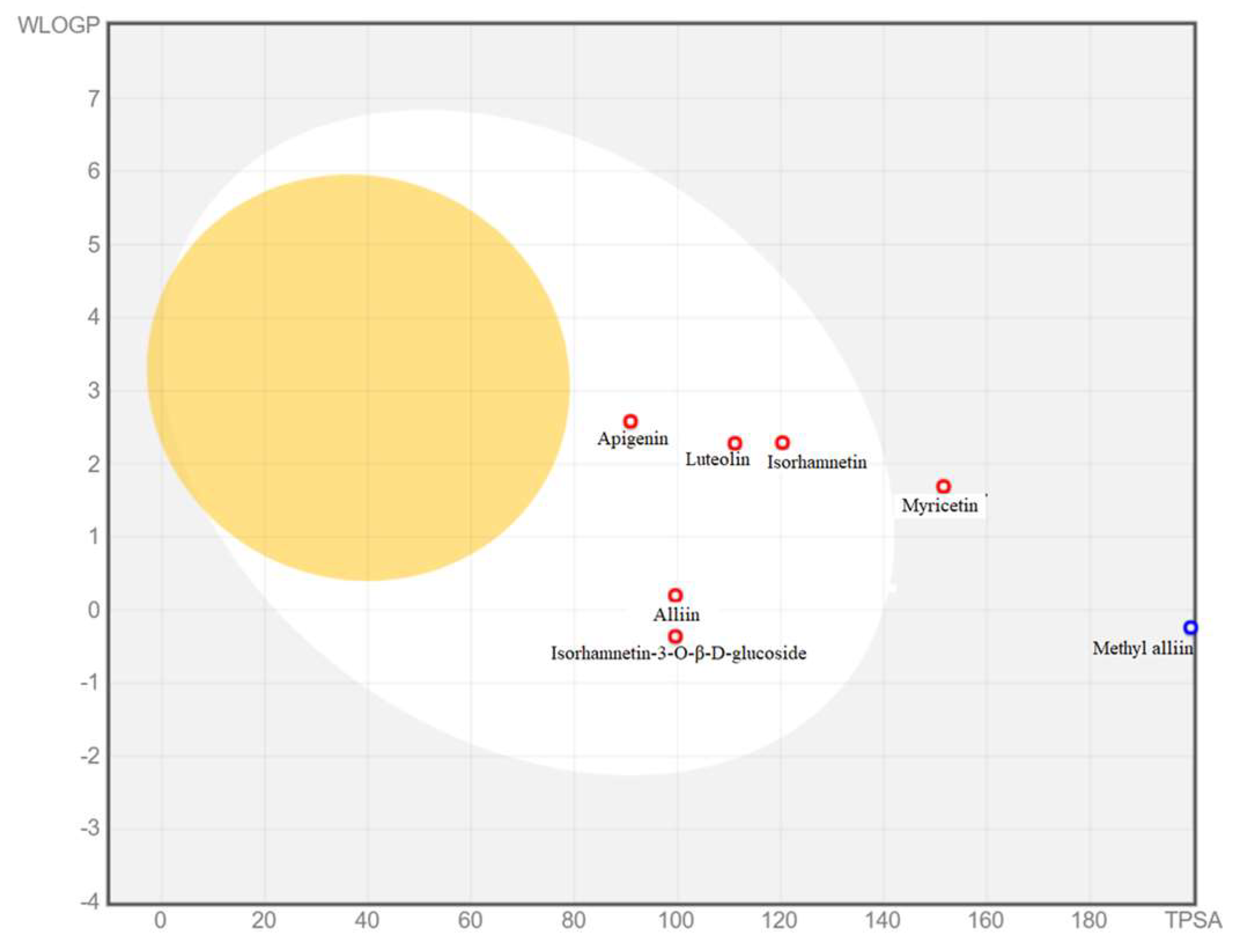
2.7.2. ADME Pharmacokinetics Analysis
2.7.3. In Silico Target Prediction
2.7.4. Molecular Docking Studies
3. Results
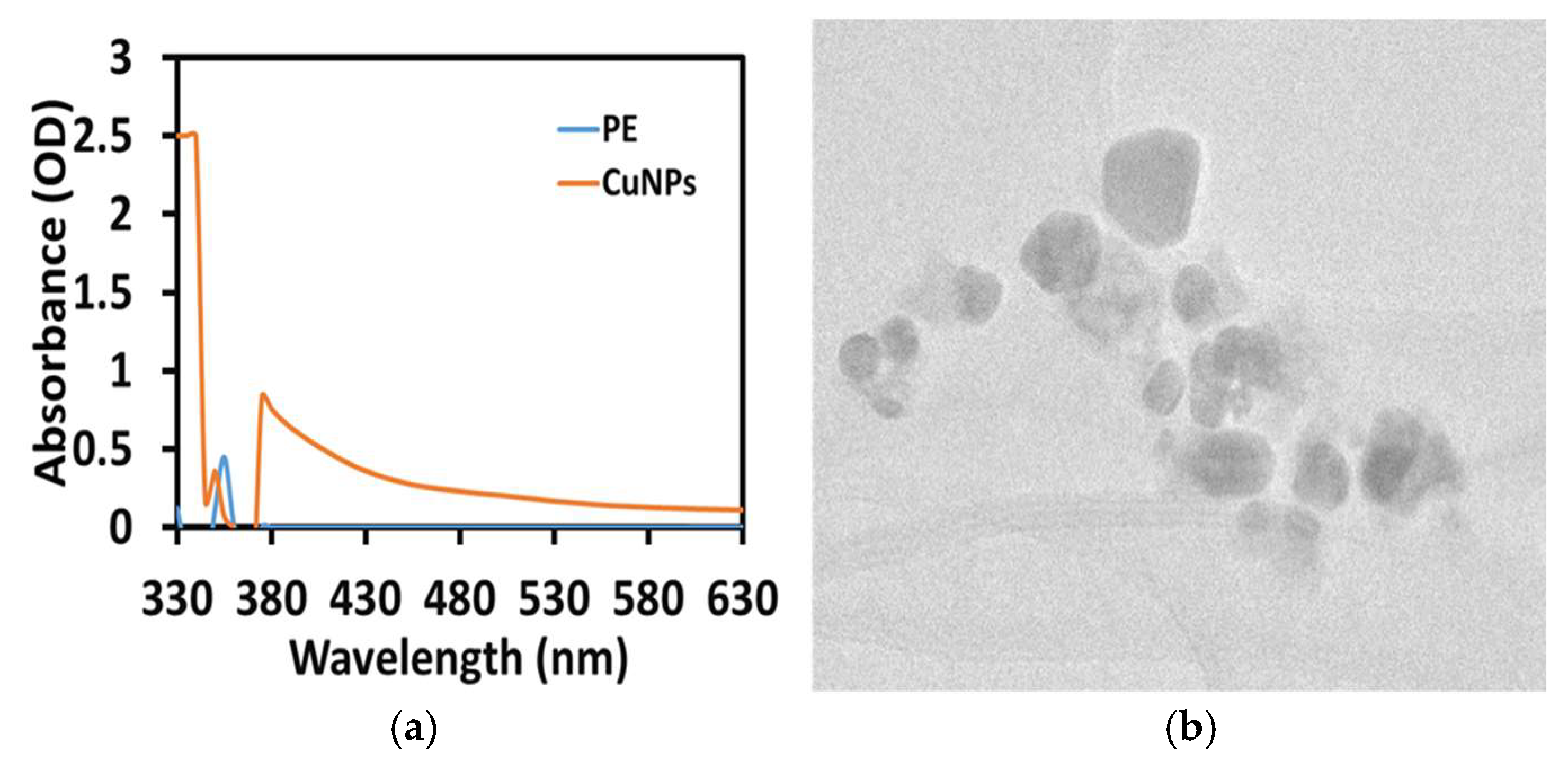
3.1. Preliminary Confirmation of CuNP Synthesis Using UV-Vis Spectra
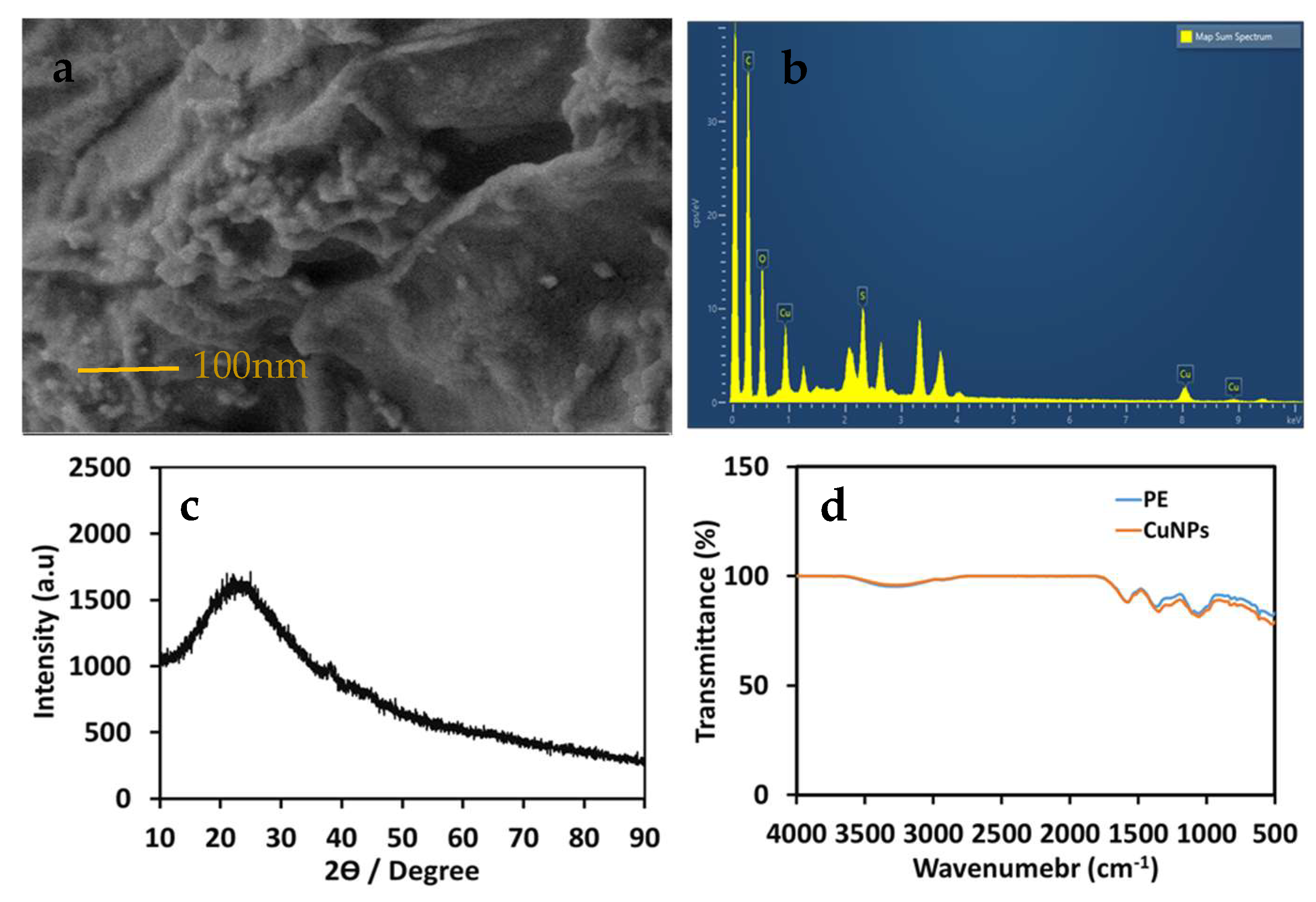
3.2. Morphology and Size Characterization of AM-CuNPs by Electron Microscopy
3.3. XRD Analysis of Biosynthesized AM-CuNPs
3.4. FT-IR Spectra of Biosynthesized AM-CuNPs
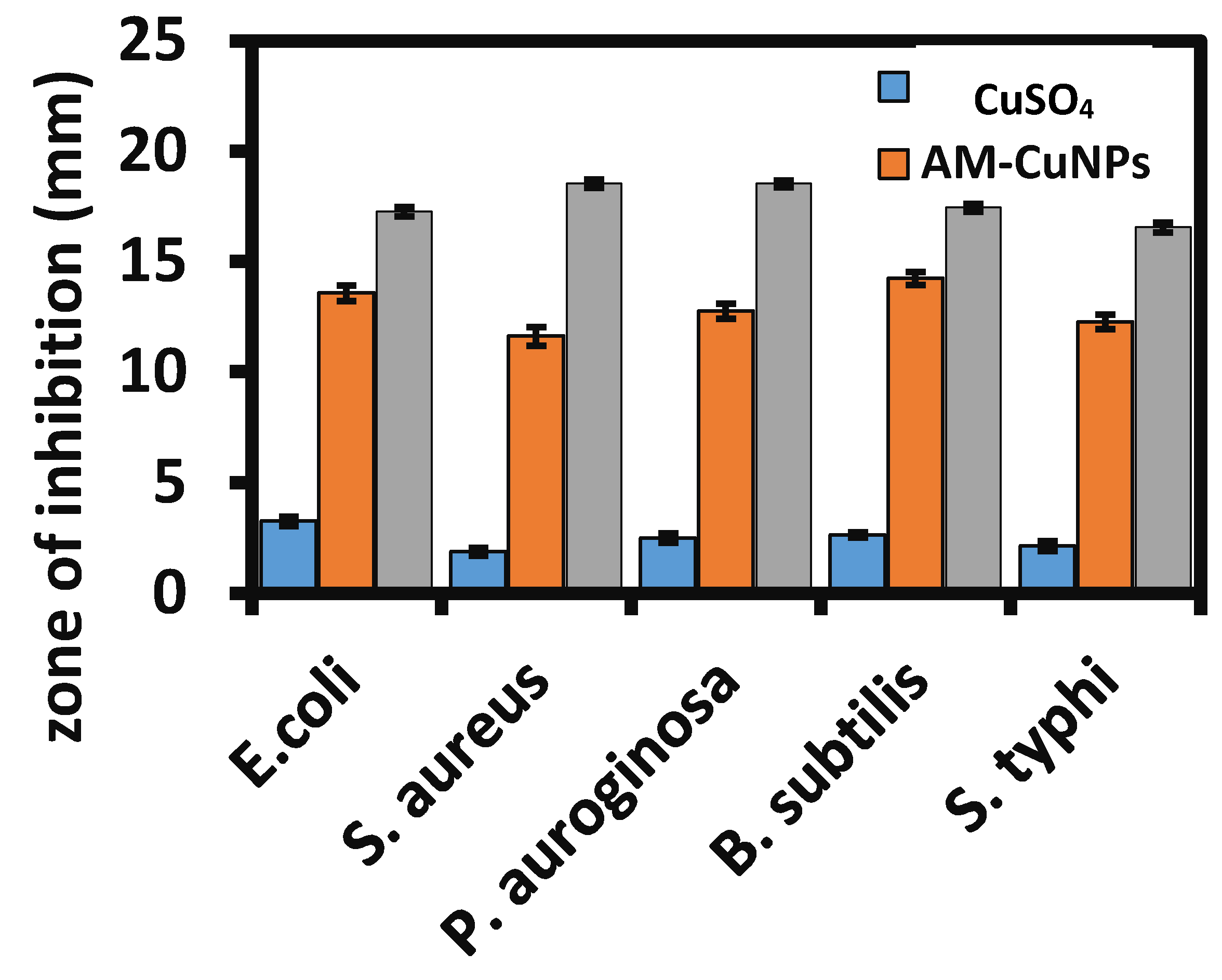
3.5. Assessment of Antibacterial Activity of AM-CuNPs
3.6. Antioxidant Activity of AM-CuNPs
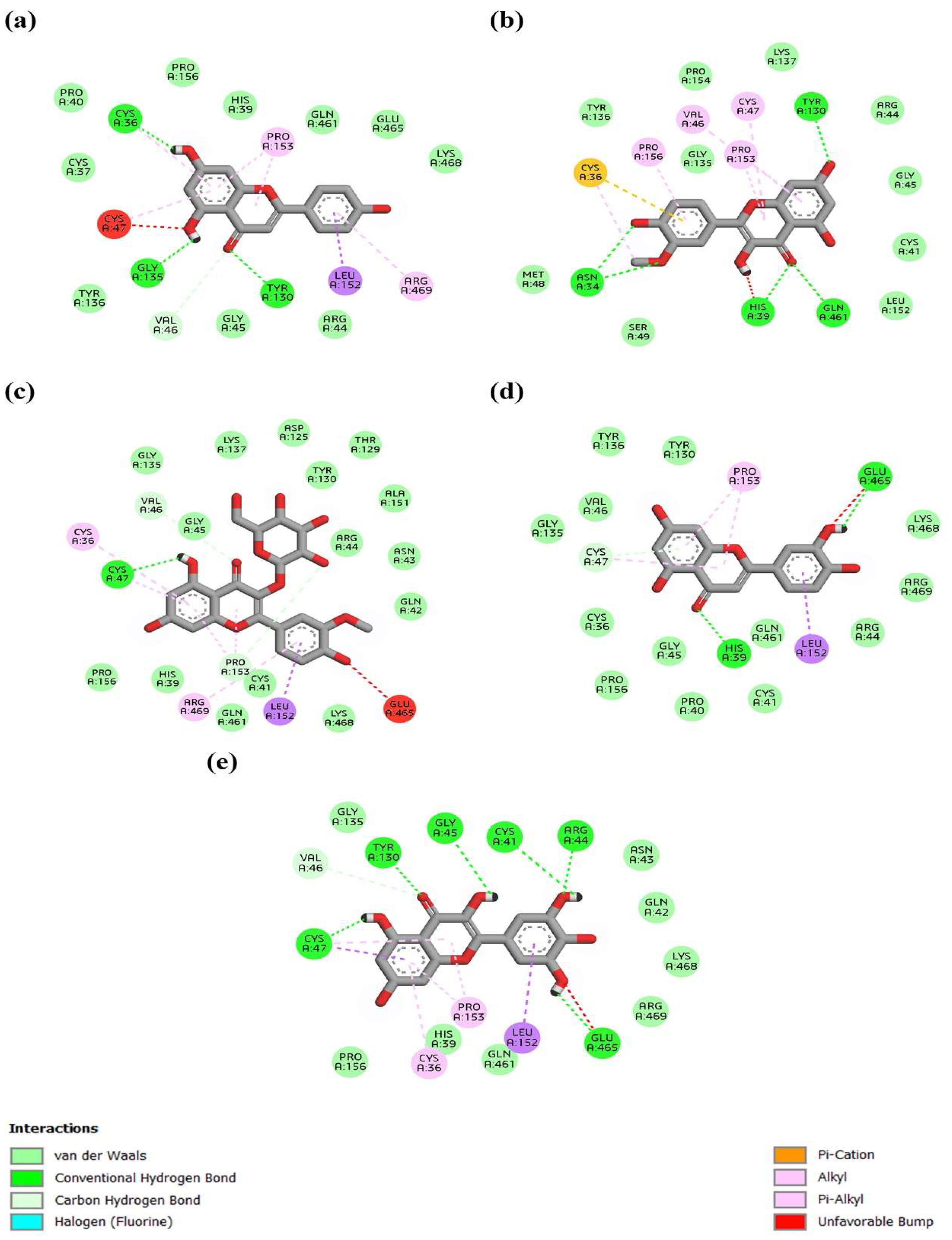
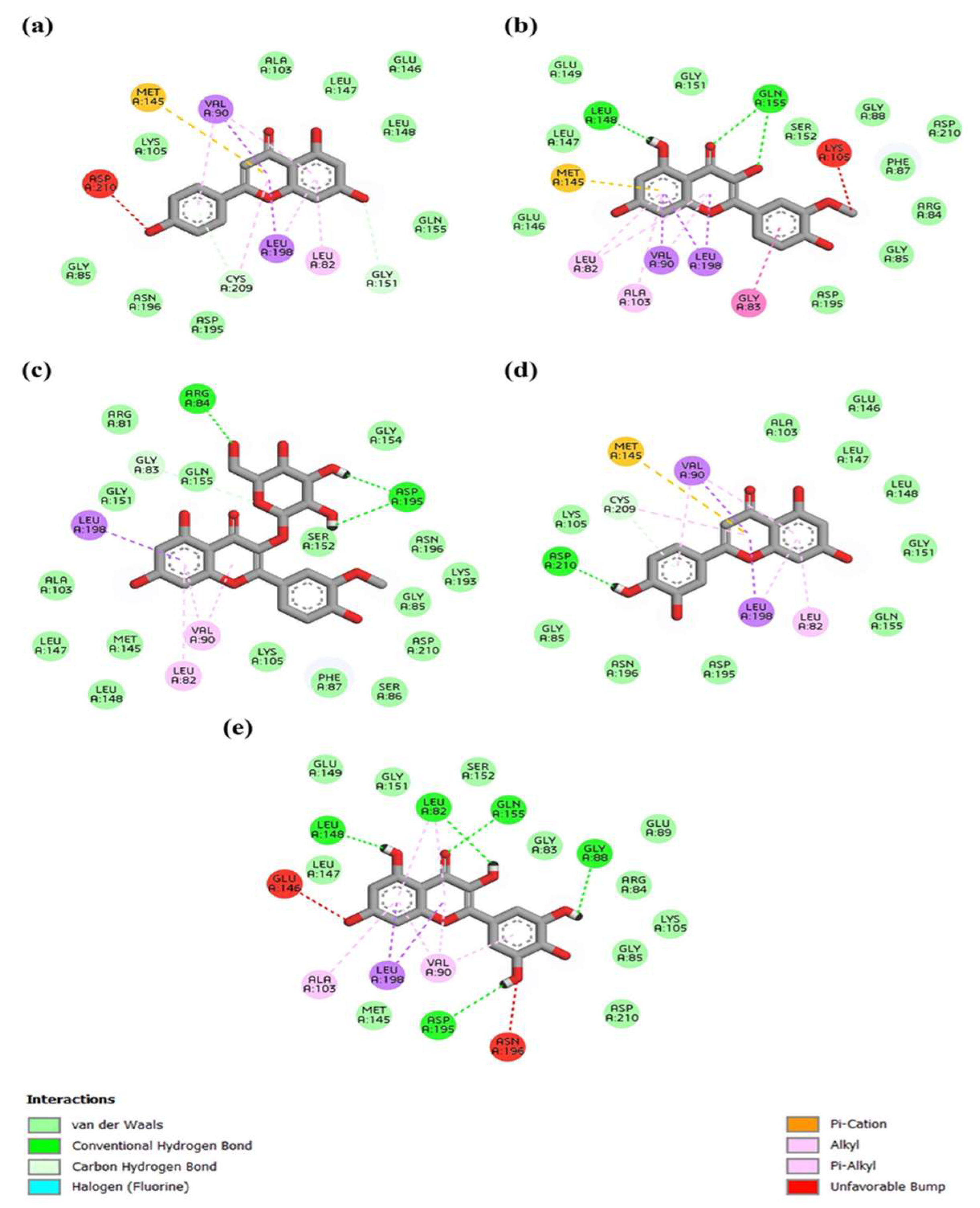
3.7. In Silico Analysis of Anti-Inflammatory Properties of A. monanthum Extract and CuNPs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, M.A.; Ahmed, T.; Wu, W.; Hossain, A.; Hafeez, R.; Masum, M.M.I.; Wang, Y.; An, Q.; Sun, G.; Li, B. Advancements in Plant and Microbe-Based Synthesis of Metallic Nanoparticles and Their Antimicrobial Activity against Plant Pathogens. Nanomaterials 2020, 10, 1146. [Google Scholar] [CrossRef] [PubMed]
- Das, R.K.; Pachapur, V.L.; Lonappan, L.; Naghdi, M.; Pulicharla, R.; Maiti, S.; Cledon, M.; Dalila, L.M.A.; Sarma, S.J.; Brar, S.K. Biological Synthesis of Metallic Nanoparticles: Plants, Animals and Microbial Aspects. Nanotechnol. Environ. Eng. 2017, 2, 18. [Google Scholar] [CrossRef]
- Ghosh, M.K.; Sahu, S.; Gupta, I.; Ghorai, T.K. Green Synthesis of Copper Nanoparticles from an Extract of Jatropha curcasleaves: Characterization, Optical Properties, CT-DNA Binding and Photocatalytic Activity. RSC Adv. 2020, 10, 22027–22035. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Song, J.Y.; Kim, B.S. Biological Synthesis of Copper Nanoparticles Using Magnolia kobus Leaf Extract and Their Antibacterial Activity. J. Chem. Technol. Biotechnol. 2013, 88, 1971–1977. [Google Scholar] [CrossRef]
- Abboud, Y.; Saffaj, T.; Chagraoui, A.; El Bouari, A.; Brouzi, K.; Tanane, O.; Ihssane, B. Biosynthesis, Characterization and Antimicrobial Activity of Copper Oxide Nanoparticles (CONPs) Produced Using Brown Alga Extract (Bifurcaria bifurcata). Appl. Nanosci. 2014, 4, 571–576. [Google Scholar] [CrossRef]
- Sivaraj, R.; Rahman, P.K.S.M.; Rajiv, P.; Salam, H.A.; Venckatesh, R. Biogenic Copper Oxide Nanoparticles Synthesis Using Tabernaemontana divaricate Leaf Extract and Its Antibacterial Activity against Urinary Tract Pathogen. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2014, 133, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Ramyadevi, J.; Jeyasubramanian, K.; Marikani, A.; Rajakumar, G.; Rahuman, A.A.; Santhoshkumar, T.; Kirthi, A.V.; Jayaseelan, C.; Marimuthu, S. Copper Nanoparticles Synthesized by Polyol Process Used to Control Hematophagous Parasites. Parasitol. Res. 2011, 109, 1403–1415. [Google Scholar] [CrossRef]
- Rajasekar, A.; Rajeshkumar, S. Antioxidant and Anti-Inflammatory Property of Copper Nanoparticles (Cunps) Synthesised Using Blue Tea. J. Complement. Med. Res. 2021, 12, 81. [Google Scholar] [CrossRef]
- Yaqoob, A.A.; Ahmad, H.; Parveen, T.; Ahmad, A.; Oves, M.; Ismail, I.M.I.; Qari, H.A.; Umar, K.; Mohamad Ibrahim, M.N. Recent Advances in Metal Decorated Nanomaterials and Their Various Biological Applications: A Review. Front. Chem. 2020, 8, 341. [Google Scholar] [CrossRef]
- Alshammari, S.O.; Mahmoud, S.Y.; Farrag, E.S. Synthesis of Green Copper Nanoparticles Using Medicinal Plant Krameria sp. Root Extract and Its Applications. Molecules 2023, 28, 4629. [Google Scholar] [CrossRef]
- Ermini, M.L.; Voliani, V. Antimicrobial Nano-Agents: The Copper Age. ACS Nano 2021, 15, 6008–6029. [Google Scholar] [CrossRef] [PubMed]
- Seyyed Hajizadeh, Y.; Babapour, E.; Harzandi, N.; Yazdanian, M.; Ranjbar, R. The Effect of Cytotoxicity and Antimicrobial of Synthesized CuO NPs from Propolis on HEK-293 Cells and Lactobacillus acidophilus. Evid.-Based Complement. Altern. Med. 2023, 2023, 1430839. [Google Scholar] [CrossRef] [PubMed]
- Huq, M.A.; Ashrafudoulla, M.; Rahman, M.M.; Balusamy, S.R.; Akter, S. Green Synthesis and Potential Antibacterial Applications of Bioactive Silver Nanoparticles: A Review. Polymers 2022, 14, 742. [Google Scholar] [CrossRef]
- Singh, H.; Desimone, M.F.; Pandya, S.; Jasani, S.; George, N.; Adnan, M.; Aldarhami, A.; Bazaid, A.S.; Alderhami, S.A. Revisiting the Green Synthesis of Nanoparticles: Uncovering Influences of Plant Extracts as Reducing Agents for Enhanced Synthesis Efficiency and Its Biomedical Applications. Int. J. Nanomed. 2023, 18, 4727–4750. [Google Scholar] [CrossRef]
- Shenhar, B.R.; Norsten, T.B.; Rotello, V.M. Polymer-Mediated Nanoparticle Assembly: Structural Control and Applications. Adv. Mater. 2005, 17, 657–669. [Google Scholar] [CrossRef]
- Moon, H.I. Larvicidal Activity of Major Essential Oils from Stems of Allium monanthum Maxim. against Aedes aegypti L. J. Enzyme Inhib. Med. Chem. 2011, 26, 827–830. [Google Scholar] [CrossRef]
- Yi, G.; Kim, K.-M. Comparison of Major Agricultural Traits and Genetic Diversity in Indigenous Korean Rocambole (Allium monanthum Max.) Based on Collecting Sites. J. Korean Soc. Int. Agric. 2015, 27, 393–397. [Google Scholar] [CrossRef]
- Chung, H.S. Antiproliferative Activity of Allium monanthum in HT-29 Human Colorectal Adenocarcinoma Cells. Korean Tea Soc. 2022, 28, 22–32. [Google Scholar] [CrossRef]
- Yoon, K.R.; Ryu, J.K.; Lee, E. Biological Effects of Allium monanthum Extracts on Lipid Metabolism, Anti-Oxidation and the Production of Pro-Inflammatory Cytokines in Rats Fed a High-Fat Diet. Korean J. Plant Resour. 2013, 26, 337–346. [Google Scholar] [CrossRef]
- Maham, M.; Sajadi, S.M.; Kharimkhani, M.M.; Nasrollahzadeh, M. Biosynthesis of the CuO Nanoparticles Using Euphorbia chamaesyce Leaf Extract and Investigation of Their Catalytic Activity for the Reduction of 4-Nitrophenol. IET Nanobiotechnol. 2017, 11, 766–772. [Google Scholar] [CrossRef]
- Bogdanović, U.; Lazić, V.; Vodnik, V.; Budimir, M.; Marković, Z.; Dimitrijević, S. Copper Nanoparticles with High Antimicrobial Activity. Mater. Lett. 2014, 128, 75–78. [Google Scholar] [CrossRef]
- Rajeshkumar, S.; Menon, S.; Venkat Kumar, S.; Ponnanikajamideen, M.; Ali, D.; Arunachalam, K. Anti-Inflammatory and Antimicrobial Potential of Cissus Quadrangularis-Assisted Copper Oxide Nanoparticles. J. Nanomater. 2021, 2021, 5742981. [Google Scholar] [CrossRef]
- Bojarska, J.; Remko, M.; Breza, M.; Madura, I.D.; Kaczmarek, K.; Zabrocki, J.; Wolf, W.M. A Supramolecular Approach to Structure-Based Design with a Focus on Synthons Hierarchy in Ornithine-Derived Ligands: Review, Synthesis, Experimental and in Silico Studies. Molecules 2020, 25, 1135. [Google Scholar] [CrossRef] [PubMed]
- Fatoki, T.H.; Ajiboye, B.O.; Aremu, A.O. In Silico Evaluation of the Antioxidant, Anti-Inflammatory, and Dermatocosmetic Activities of Phytoconstituents in Licorice (Glycyrrhiza glabra L.). Cosmetics 2023, 10, 69. [Google Scholar] [CrossRef]
- Khairy, A.; Ghareeb, D.A.; Celik, I.; Hammoda, H.M.; Zaatout, H.H.; Ibrahim, R.S. Forecasting of Potential Anti-Inflammatory Targets of Some Immunomodulatory Plants and Their Constituents Using in Vitro, Molecular Docking and Network Pharmacology-Based Analysis. Sci. Rep. 2023, 13, 9539. [Google Scholar] [CrossRef]
- Mali, S.C.; Dhaka, A.; Githala, C.K.; Trivedi, R. Green Synthesis of Copper Nanoparticles Using Celastrus paniculatus Willd. Leaf Extract and Their Photocatalytic and Antifungal Properties. Biotechnol. Rep. 2020, 27, e00518. [Google Scholar] [CrossRef]
- Amaliyah, S.; Pangesti, D.P.; Masruri, M.; Sabarudin, A.; Sumitro, S.B. Green Synthesis and Characterization of Copper Nanoparticles Using Piper retrofractum Vahl Extract as Bioreductor and Capping Agent. Heliyon 2020, 6, e04636. [Google Scholar] [CrossRef]
- Khashan, K.S.; Jabir, M.S.; Abdulameer, F.A. Preparation and Characterization of Copper Oxide Nanoparticles Decorated Carbon Nanoparticles Using Laser Ablation in Liquid. J. Phys. Conf. Ser. 2018, 1003, 012100. [Google Scholar] [CrossRef]
- Sankara Narayanan, V.P.; Kathirason, S.G.; Elango, P.; Subramanian, R.; Sivagurusundar, R.; Gurusamy, A. Emilia sonchifolia Leaf Extract-Mediated Green Synthesis, Characterization, in Vitro Biological Activities, Photocatalytic Degradation and in Vivo Danio Rerio Embryo Toxicity of Copper Nanoparticles. RSC Adv. 2023, 13, 16724–16740. [Google Scholar] [CrossRef]
- Sadia, B.O.; Cherutoi, J.K.; Achisa, C.M. Optimization, Characterization, and Antibacterial Activity of Copper Nanoparticles Synthesized Using Senna didymobotrya Root Extract. J. Nanotechnol. 2021, 2021, 5611434. [Google Scholar] [CrossRef]
- Vijayakumar, G.; Kesavan, H.; Kannan, A.; Arulanandam, D.; Kim, J.H.; Kim, K.J.; Song, H.J.; Kim, H.J.; Rangarajulu, S.K. Phytosynthesis of Copper Nanoparticles Using Extracts of Spices and Their Antibacterial Properties. Processes 2021, 9, 1341. [Google Scholar] [CrossRef]
- Murthy, H.C.A.; Desalegn, T.; Kassa, M.; Abebe, B.; Assefa, T. Synthesis of Green Copper Nanoparticles Using Medicinal Plant Hagenia abyssinica (Brace) JF. Gmel. Leaf Extract: Antimicrobial Properties. J. Nanomater. 2020, 2020, 3924081. [Google Scholar] [CrossRef]
- Ramasubbu, K.; Padmanabhan, S.; Al-Ghanim, K.A.; Nicoletti, M.; Govindarajan, M.; Sachivkina, N.; Rajeswari, V.D. Green Synthesis of Copper Oxide Nanoparticles Using Sesbania grandiflora Leaf Extract and Their Evaluation of Anti-Diabetic, Cytotoxic, Anti-Microbial, and Anti-Inflammatory Properties in an In-Vitro Approach. Fermentation 2023, 9, 332. [Google Scholar] [CrossRef]
- Apak, R.; Özyürek, M.; Güçlü, K.; Çapanoğlu, E. Antioxidant Activity/Capacity Measurement. 1. Classification, Physicochemical Principles, Mechanisms, and Electron Transfer (ET)-Based Assays. J. Agric. Food Chem. 2016, 64, 997–1027. [Google Scholar] [CrossRef]
- Rehana, D.; Mahendiran, D.; Kumar, R.S.; Rahiman, A.K. Evaluation of Antioxidant and Anticancer Activity of Copper Oxide Nanoparticles Synthesized Using Medicinally Important Plant Extracts. Biomed. Pharmacother. 2017, 89, 1067–1077. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Sultana, M.J.; Nibir, A.I.S.; Ahmed, F.R.S. Biosensing and Anti-Inflammatory Effects of Silver, Copper and Iron Nanoparticles from the Leaf Extract of Catharanthus roseus. Beni-Suef Univ. J. Basic Appl. Sci. 2023, 12, 26. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, A.L.; Huang, S.K.H.; Sevilla, U.T.A.; Hsieh, C.Y.; Tsai, P.W. In Silico Analysis of Anti-Inflammatory and Antioxidant Properties of Bioactive Compounds from Crescentia cujete L. Molecules 2023, 28, 3547. [Google Scholar] [CrossRef] [PubMed]
- Utami, W.; Aziz, H.A.; Fitriani, I.N.; Zikri, A.T.; Mayasri, A.; Nasrudin, D. In Silico Anti-Inflammatory Activity Evaluation of Some Bioactive Compound from Ficus religiosa through Molecular Docking Approach. J. Phys. Conf. Ser. 2020, 1563, 012024. [Google Scholar] [CrossRef]

| Microorganisms | Zone of Inhibition (mm) | ||
|---|---|---|---|
| CuSO4 | AM-CuNPs | Standard | |
| 1. E. coli | 3.2 ± 0.2 d | 13.58 ± 0.35 c | 18.27 ± 0.21 b |
| 2. S. aureus | 1.8 ± 0.18 a | 11.63 ± 0.42 a | 19.54 ± 0.18 c |
| 3. P. aureginosa | 2.4 ± 0.22 c | 12.77 ± 0.34 b | 19.53 ± 0.16 c |
| 4. B. subtilis | 2.6 ± 0.11 c | 14.25 ± 0.29 d | 17.45 ± 0.18 a |
| 5. S. typhi | 2.1 ± 0.22 b | 12.28 ± 0.33 a | 18.56 ± 0.2 b |
| Sample Name | Anti-Oxidant Assay | ||
|---|---|---|---|
| DPPH (%) | ABTS (%) | H2O2 (%) | |
| AM-CuNPs | 68.46 ± 0.85 c | 62.55 ± 0.68 c | 66.17 ± 0.73 c |
| CuSO4 1 mM | 22.35 ± 0.58 a | 19.26 ± 0.75 a | 17.58 ± 0.66 a |
| A. monanthum extract | 63.28 ± 0.63 b | 61.54 ± 0.82 b | 65.29 ± 0.68 b |
| Vitamin C-standard | 92.56 ± 0.45 d | 83.66 ± 0.38 d | 85.23 ± 0.35 d |
| Molecule | MW (g/mol) | Water Solubility | GI Absorption | BBB Perm-Eant | P-glyco -Protein Substrate | Lipinski |
|---|---|---|---|---|---|---|
| Alliin | 177.22 | Highly soluble | High | No | No | Yes |
| Apigenin | 270.24 | Moderately Soluble | High | No | No | Yes |
| Isorhamnetin-3-O-β-D-glucoside | 478.4 | Soluble | Low | No | Yes | No |
| Isorhamnetin | 316.26 | Soluble | High | No | No | Yes |
| Luteolin | 286.24 | Soluble | High | No | No | Yes |
| Methyl-alliin | 151.18 | Highly soluble | High | No | No | Yes |
| Myricetin | 318.24 | Soluble | Low | No | No | Yes |
| Ligand | Average Binding Energy | Top Binding Energy |
|---|---|---|
| Myricetin_5281672 | −8.17 | −9.8 |
| Luteolin_5280445 | −8 | −9.4 |
| Isorhamnetin-glucoside_5318645 | −7.9 | −9.7 |
| Apigenin_5280443 | −7.58 | −9 |
| Isorhamnetin_5281654 | −7.22 | −8.9 |
| Alliin_87310 | −4.78 | −5.2 |
| Methyl_alliin_99483 | −4.43 | −4.8 |
| Ligand | Average Binding Energy | Top Binding Energy |
|---|---|---|
| Isorhamnetin-glucoside_5318645 | −7.78 | −9.1 |
| Isorhamnetin_5281654 | −7.6 | −8.9 |
| Myricetin_5281672 | −7.5 | −8.9 |
| Luteolin_5280445 | −7.47 | −9.3 |
| Apigenin_5280443 | −7.46 | −9 |
| Alliin_87310 | −4 | −4.6 |
| Methyl_alliin_99483 | −3.52 | −4.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, H.S.; Jung, J.S.; Jeong, Y.-I.; Choi, K.C. Biological Synthesis of Copper Nanoparticles Using Edible Plant Allium monanthum: Characterization of Antibacterial, Antioxidant, and Anti-Inflammatory Properties Using In Silico Molecular Docking Analysis. Materials 2023, 16, 6669. https://doi.org/10.3390/ma16206669
Han HS, Jung JS, Jeong Y-I, Choi KC. Biological Synthesis of Copper Nanoparticles Using Edible Plant Allium monanthum: Characterization of Antibacterial, Antioxidant, and Anti-Inflammatory Properties Using In Silico Molecular Docking Analysis. Materials. 2023; 16(20):6669. https://doi.org/10.3390/ma16206669
Chicago/Turabian StyleHan, Hyo Shim, Jeong Sung Jung, Young-Il Jeong, and Ki Choon Choi. 2023. "Biological Synthesis of Copper Nanoparticles Using Edible Plant Allium monanthum: Characterization of Antibacterial, Antioxidant, and Anti-Inflammatory Properties Using In Silico Molecular Docking Analysis" Materials 16, no. 20: 6669. https://doi.org/10.3390/ma16206669
APA StyleHan, H. S., Jung, J. S., Jeong, Y.-I., & Choi, K. C. (2023). Biological Synthesis of Copper Nanoparticles Using Edible Plant Allium monanthum: Characterization of Antibacterial, Antioxidant, and Anti-Inflammatory Properties Using In Silico Molecular Docking Analysis. Materials, 16(20), 6669. https://doi.org/10.3390/ma16206669









