Evaluation of the Antitumour and Antiproliferative Effect of Xanthohumol-Loaded PLGA Nanoparticles on Melanoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Production of XN-Loaded PLGA Nanoparticles
2.3. Particle Size, Polydispersity Index and Zeta Potential Characterization of XN-Loaded PLGA Nanoparticles
2.4. Association Efficiency (AE) and Loading Capacity (LC) of XN
2.5. Attenuated Total Reflectance Fourier Transform Infrared Spectroscopy (ATR-FTIR)
2.6. Scanning Electron Microscopy (SEM)
2.7. Cell Culture
2.8. Cell Culture Studies
2.9. Cell Viability Assay
2.10. Wound Healing Experiment
2.11. Fluorescence Immunocytochemistry for Detection of Membrane Markers
2.12. Statistical Analysis
3. Results and Discussion
3.1. Particle Size, Polydispersity Index, Zeta Potential, AE and LC
3.2. Interaction of XN and PLGA Nanoparticles Assessed by ATR-FTIR
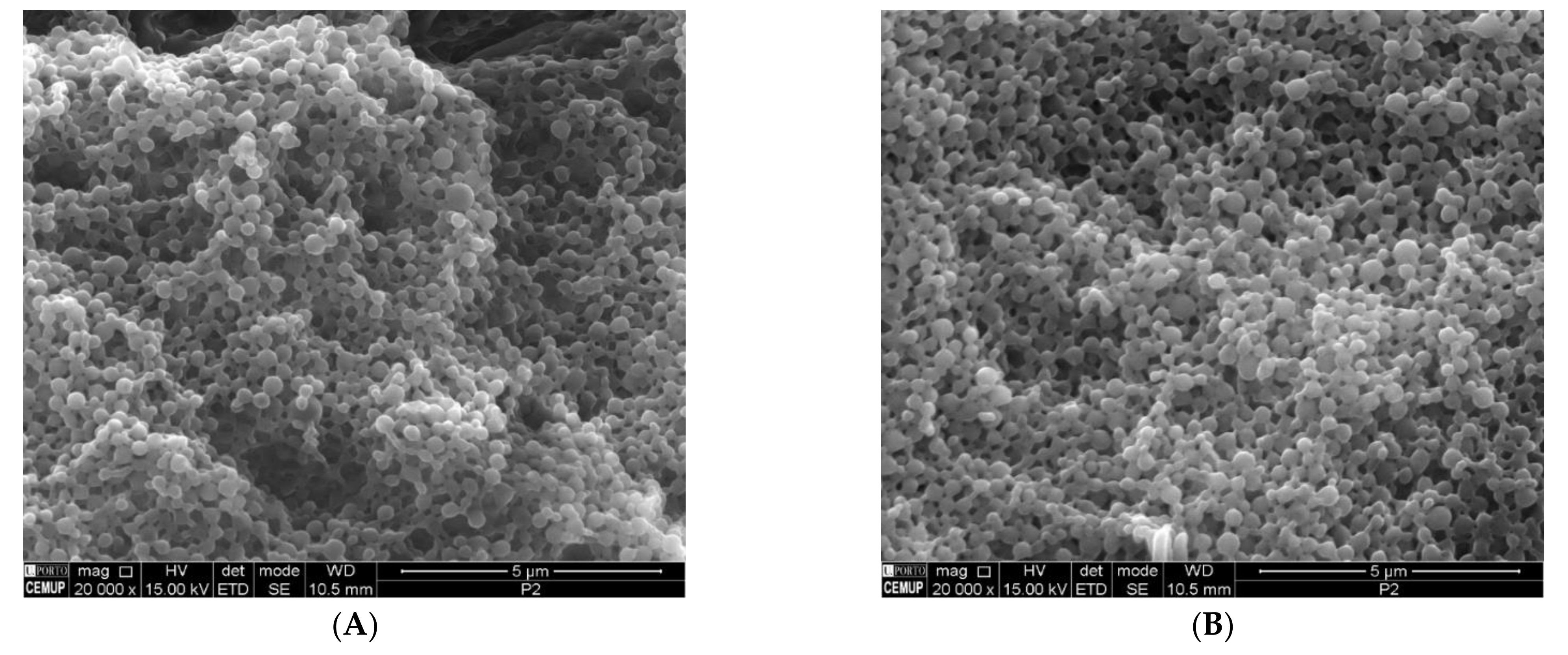
3.3. SEM Analysis
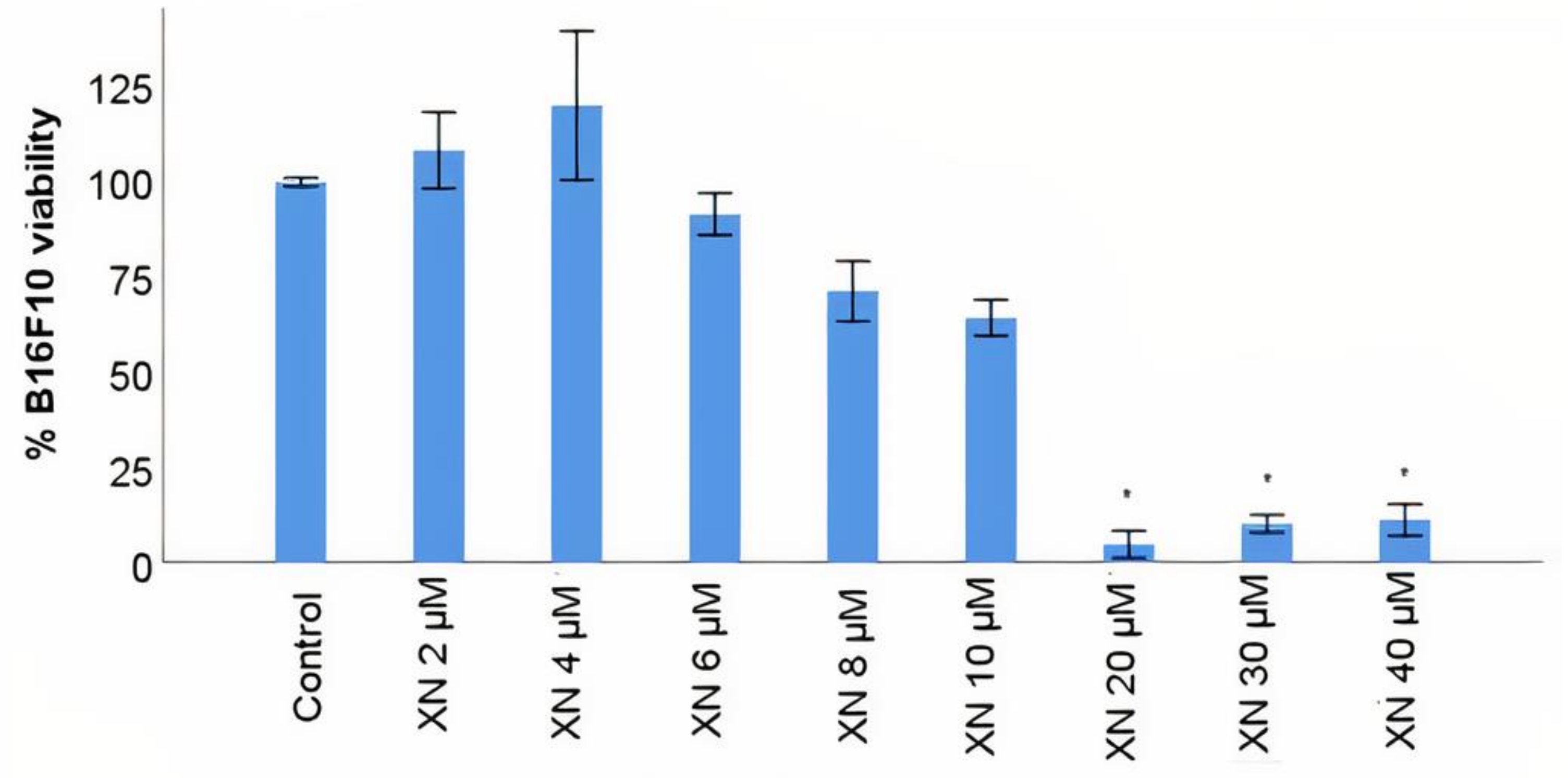
3.4. B16F10 Viability Study with XN Solutions
3.5. B16F10 Viability Study with XN-Loaded PLGA Nanoparticles Compared to XN Solubilized Form
3.6. B16F10 Migration Study with XN-Loaded PLGA Nanoparticles Compared to XN Solubilized Form
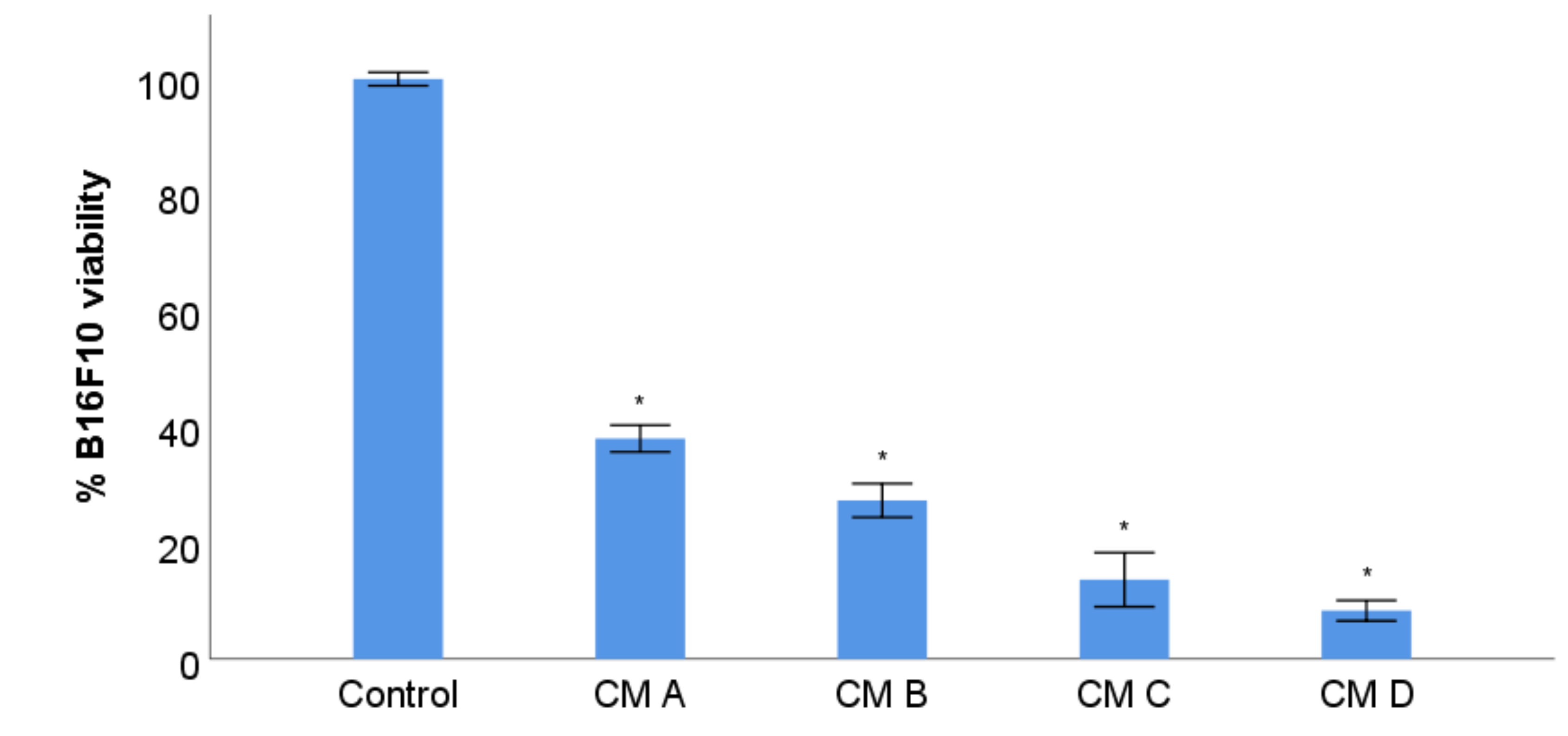
3.7. B16F10 Viability Study with Conditioned Media from Stimulation of RAW 264.7 with XN-Loaded PLGA Nanoparticles and XN Solubilized Form
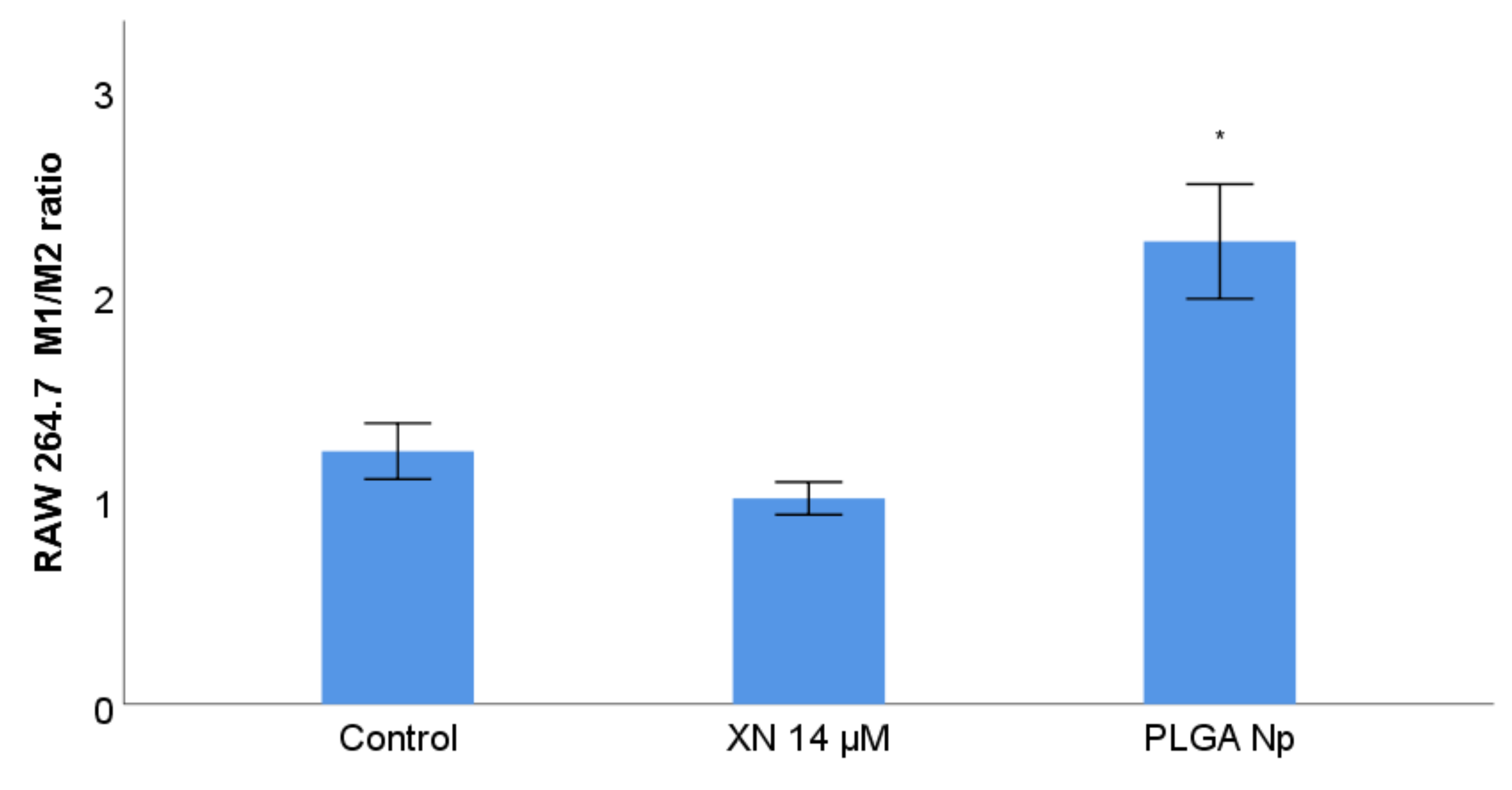
3.8. RAW 264.7 Phenotype Membrane Markers Study with Previous Stimulation with XN Solubilized Form and Non-Encapsulated PLGA Nanoparticles
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prado, G.; Svoboda, R.M.; Rigel, D.R. What’s New in Melanoma. Dermatol. Clin. 2019, 37, 159–168. [Google Scholar]
- Apalla, Z.; Nashan, D.; Weller, R.; Castellsagué, X. Skin cancer: Epidemiology, disease burden, pathophysiology, diagnosis, and therapeutic approaches. Dermatol. Ther. 2017, 7, 5–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, S.; Smith, C.; Wernberg, J. Epidemiology and risk factors of melanoma. Surg. Clin. North Am. 2020, 100, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Che, G.; Huang, B.; Xie, Z.; Zhao, J.; Yan, Y.; Wu, J.; Sun, H.; Ma, H. Trends in incidence and survival in patients with melanoma, 1974–2013. Am. J. Cancer Res. 2019, 9, 1396–1414. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [Green Version]
- Miller, K.; Siegel, R.; Lin, C.; Mariotto, A.; Kramer, J.; Rowland, J.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Steen, C.; Newman, A. Computational approaches for characterizing the tumor immune microenvironment. Immunology 2019, 158, 70–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuccitto, A.; Shahaj, E.; Vergani, E.; Ferro, S.; Huber, V.; Rodolfo, M.; Castelli, C.; Rivoltini, L.; Vallacchi, V. Immunosuppressive circuits in tumor microenvironment and their influence on cancer treatment efficacy. Virchows Arch. 2019, 474, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Butturini, E.; Carcereri de Prati, A.; Boriero, D.; Mariotto, S. Tumor Dormancy and Interplay with Hypoxic Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 4305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinshaw, D.S.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, D.; Cang, H.; Guo, B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019, 8, 4709–4721. [Google Scholar] [CrossRef]
- Larionova, I.; Cherdyntseva, N.; Liu, T.; Patysheva, M.; Rakina, M.; Kzhyshkowska, J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology 2019, 8, 1596004. [Google Scholar] [CrossRef] [Green Version]
- Salmaninejad, A.; Valilou, S.; Soltani, A.; Ahmadi, S.; Abarghan, Y.; Rosengren, R.; Sahebkar, A. Tumor-associated macrophages: Role in cancer development and therapeutic implications. Cell. Oncol. 2019, 42, 591–608. [Google Scholar] [CrossRef]
- Wang, H.; Yang, L.; Wang, D.; Zhang, Q.; Zhang, L. Pro-tumor activities of macrophages in the progression of melanoma. Hum. Vaccin. Immunother. 2017, 13, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Ley, K.; Pramod, A.B.; Croft, M.; Ravichandran, K.S.; Ting, J.P. How mouse macrophages sense what is going on. Front. Immunol. 2016, 7, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, R.; Li, S.; Geng, H.; Wang, X.; Guan, Q.; Li, X.; Ren, C.; Yuan, X. Reversing the polarization of tumor-associated macrophages inhibits tumor metastasis. Int. Immunopharmacol. 2017, 49, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zitelli, J.A.; Mohs, F.E.; Larson, P.; Snow, S. Mohs micrographic surgery for melanoma. Dermatol. Clin. 1989, 7, 833–843. [Google Scholar] [CrossRef]
- Boyle, G. Therapy for metastatic melanoma: An overview and update. Expert. Rev. Anticancer. Ther. 2011, 11, 725–737. [Google Scholar] [CrossRef]
- Institute Report. Global Oncology Trends 2018-Innovation, Expansion and Disruption. The IQVIA Institute. Available online: https://www.iqvia.com/insights/the-iqvia-institute/reports/global-oncology-trends-2018 (accessed on 1 October 2021).
- Broman, K.K.; Dossett, L.A.; Sun, J.; Eroglu, Z.; Zager, J.S. Update on BRAF and MEK inhibition for treatment of melanoma in metastatic, unresectable, and adjuvant settings. Expert Opin. Drug. Saf. 2019, 18, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Rose, A. Encorafenib and binimetinib for the treatment of BRAF V600E/K-mutated melanoma. Drugs Today 2019, 55, 247–264. [Google Scholar] [CrossRef]
- Arozarena, I.; Wellbrock, C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Sileni, V.C.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef]
- Singh, V.K.; Arora, D.; Ansari, M.I.; Sharma, P.K. Phytochemicals based chemopreventive and chemotherapeutic strategies and modern technologies to overcome limitations for better clinical applications. Phytother. Res. 2019, 33, 3064–3089. [Google Scholar]
- Arora, I.; Sharma, M.; Tollefsbol, T.O. Combinatorial Epigenetics Impact of Polyphenols and Phytochemicals in Cancer Prevention and Therapy. Int. J. Mol. Sci. 2019, 20, 4567. [Google Scholar] [CrossRef] [Green Version]
- Heenatigala Palliyage, G.; Singh, S.; Ashby, C.R.; Tiwari, A.K.; Chauhan, H. Pharmaceutical Topical Delivery of Poorly Soluble Polyphenols: Potential Role in Prevention and Treatment of Melanoma. AAPS PharmSciTech 2019, 20, 250. [Google Scholar] [CrossRef] [PubMed]
- Sajadimajd, S.; Bahramsoltani, R.; Iranpanah, A.; Patra, J.K.; Das, G.; Gouda, S.; Rahimi, R.; Rezaeiamiri, E.; Cao, H.; Giampieri, F.; et al. Advances on natural polyphenols as anticancer agents for skin cancer. Pharmacol. Res. 2020, 151, 104584. [Google Scholar] [CrossRef]
- Ravikumar, P.; Katariya, M.; Patil, S.; Tatke, P.; Pillai, R. Skin delivery of resveratrol encapsulated lipidic formulation for melanoma chemoprevention. J. Microencapsul. 2019, 36, 535–551. [Google Scholar] [CrossRef]
- Prieto, K.; Cao, Y.; Mohamed, E.; Trillo-Tinoco, J.; Sierra, R.A.; Urueña, C.; Sandoval, T.A.; Fiorentino, S.; Rodriguez, P.C.; Barreto, A. Polyphenol-rich extract induces apoptosis with immunogenic markers in melanoma cells through the ER stress-associated kinase PERK. Cell Death Discov. 2019, 5, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afaq, F.; Katiyar, S. Polyphenols: Skin photoprotection and inhibition of photocarcinogenesis. Mini Rev. Med. Chem. 2011, 11, 1200–1215. [Google Scholar] [PubMed] [Green Version]
- Strickland, L.R.; Pal, H.C.; Elmets, C.A.; Afaq, F. Targeting drivers of melanoma with synthetic small molecules and phytochemicals. Cancer Lett. 2015, 359, 20–35. [Google Scholar] [CrossRef] [Green Version]
- Leonida, M.D.; Kumar, I. Bionanomaterials for Skin Regeneration; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Nair, H.B.; Sung, B.; Yadav, V.R.; Kannappan, R.; Chaturvedi, M.M.; Aggarwal, B.B. Delivery of antiinflammatory nutraceuticals by nanoparticles for the prevention and treatment of cancer. Biochem. Pharmacol. 2010, 80, 1833–1843. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, I.A.; Mukhtar, H. Nanochemoprevention by bioactive food components: A perspective. Pharm. Res. 2010, 27, 1054–1060. [Google Scholar] [CrossRef] [Green Version]
- Fortina, P.; Kricka, L.J.; Surrey, S.; Grodzinski, P. Nanobiotechnology: The promise and reality of new approaches to molecular recognition. Trends Biotechnol. 2005, 23, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, H.; Gorain, B.; Pandey, M.; Khurana, R.K.; Kesharwani, P. Strategizing biodegradable polymeric nanoparticles to cross the biological barriers for cancer targeting. Int. J. Pharm. 2019, 565, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Clemente, N.; Boggio, E.; Gigliotti, L.C.; Raineri, D.; Ferrara, B.; Miglio, G.; Argenziano, M.; Chiocchetti, A.; Cappellano, G.; Trotta, F.; et al. Immunotherapy of experimental melanoma with ICOS-Fc loaded in biocompatible and biodegradable nanoparticles. J. Control. Release 2020, 320, 112–124. [Google Scholar] [CrossRef]
- Lee, S.Y.; Koo, J.S.; Yang, M.; Cho, H.-J. Application of temporary agglomeration of chitosan-coated nanoparticles for the treatment of lung metastasis of melanoma. J. Colloid. Interface Sci. 2019, 544, 266–275. [Google Scholar] [CrossRef]
- Jain, R. The manufacturing techniques of various drug loaded biodegradable poly(lactide-co-glycolide) (PLGA) devices. Biomaterials 2000, 21, 2475–2490. [Google Scholar] [CrossRef]
- Sousa, F.; Cruz, A.; Fonte, P.; Pinto, I.M.; Neves-Petersen, M.T.; Sarmento, B. A new paradigm for antiangiogenic therapy through controlled release of bevacizumab from PLGA nanoparticles. Sci. Rep. 2017, 7, 3736. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Ríos, R.; Sánchez-Ramírez, D.R.; Ruiz-Saray, K.; Oceguera-Basurto, P.E.; Almada, M.; Juárez, J.; Zepeda-Moreno, A.; Del Toro-Arreola, A.; Topete, A.; Daneri-Navarro, A. Cisplatin-loaded PLGA nanoparticles for HER2 targeted ovarian cancer therapy. Colloids Surf. B Biointerfaces 2019, 178, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Sims, L.B.; Curry, K.C.; Parupalli, S.; Horner, G.; Frieboes, H.B.; Steinbach-Rankins, J.M. Efficacy of Surface-Modified PLGA Nanoparticles as a Function of Cervical Cancer Type. Pharm. Res. 2019, 36, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fonte, P.; Andrade, F.; Azevedo, C.; Pinto, J.; Seabra, V.; van de Weert, M.; Reis, S.; Sarmento, B. Effect of the Freezing Step in the Stability and Bioactivity of Protein-Loaded PLGA Nanoparticles Upon Lyophilization. Pharm. Res. 2016, 33, 2777–2793. [Google Scholar] [CrossRef]
- Fonte, P.; Lino, P.R.; Seabra, V.; Almeida, A.J.; Reis, S.; Sarmento, B. Annealing as a tool for the optimization of lyophilization and ensuring of the stability of protein-loaded PLGA nanoparticles. Int. J. Pharm. 2016, 503, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Liu, T.; Yang, C. Development of PLGA-lipid nanoparticles with covalently conjugated indocyanine green as a versatile nanoplatform for tumor-targeted imaging and drug delivery. Int. J. Nanomed. 2016, 11, 5807–5821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nassir, A.M.; Shahzad, N.; Ibrahim, I.A.A.; Ahmad, I.; Md, S.; Ain, M.R. Resveratrol-loaded PLGA nanoparticles mediated programmed cell death in prostate cancer cells. Saudi Pharm. J. 2018, 26, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huan, M.-l.; Liu, M.; Cheng, Y.; Sun, Y.; Cui, H.; Liu, D.-Z.; Mei, Q.-B.; Zhou, S.-Y. Doxorubicin and resveratrol co-delivery nanoparticle to overcome doxorubicin resistance. Sci. Rep. 2016, 6, 35267. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei Mirakabad, F.S.; Akbarzadeh, A.; Milani, M.; Zarghami, N.; Taheri-Anganeh, M.; Zeighamian, V.; Badrzadeh, F.; Rahmati-Yamchi, M. A Comparison between the cytotoxic effects of pure curcumin and curcumin-loaded PLGA-PEG nanoparticles on the MCF-7 human breast cancer cell line. Artif. Cells Nanomed. Biotechnol. 2016, 44, 423–430. [Google Scholar] [CrossRef]
- Singh, M.; Bhatnagar, P.; Mishra, S.; Kumar, P.; Shukla, Y.; Gupta, K.C. PLGA-encapsulated tea polyphenols enhance the chemotherapeutic efficacy of cisplatin against human cancer cells and mice bearing Ehrlich ascites carcinoma. Int. J. Nanomed. 2015, 10, 6789–6809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Sharma, S.; Sharma, V.; Sharma, K.; Yadav, S.K.; Dwivedi, P.; Agrawal, S.; Paliwal, S.K.; Dwivedi, A.K.; Maikhuri, J.P.; et al. Oleanolic–bioenhancer coloaded chitosan modified nanocarriers attenuate breast cancer cells by multimode mechanism and preserve female fertility. Int. J. Biol. Macromol. 2017, 104, 1345–1358. [Google Scholar] [CrossRef]
- Das, S.; Das, J.; Samadder, A.; Paul, A.; Khuda-Bukhsh, A.R. Strategic formulation of apigenin-loaded PLGA nanoparticles for intracellular trafficking, DNA targeting and improved therapeutic effects in skin melanoma in vitro. Toxicol. Lett. 2013, 223, 124–138. [Google Scholar] [CrossRef]
- Chiu, H.I.; Samad, N.A.; Fang, L.; Lim, V. Cytotoxicity of targeted PLGA nanoparticles: A systematic review. RSC Adv. 2021, 11, 9433–9449. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Thapa, R.; Hariani, H.N.; Volyanyuk, M.; Ogle, S.D.; Orloff, K.A.; Ankireddy, S.; Lai, K.; Žiniauskaitė, A.; Stubbs, E.B.; et al. Poly(lactic-co-glycolic acid) nanoparticles encapsulating the prenylated flavonoid, xanthohumol, protect corneal epithelial cells from dry eye disease-associated oxidative stress. Pharmaceutics 2021, 13, 1362. [Google Scholar] [CrossRef] [PubMed]
- Barichello, J.M.; Morishita, M.; Takayama, K.; Nagai, T. Encapsulation of hydrophilic and lipophilic drugs in PLGA nanoparticles by the nanoprecipitation method. Drug Dev. Ind. Pharm. 1999, 25, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Walden, D.; Kunnimalaiyaan, S.; Sokolowski, K.; Clark, T.G.; Kunnimalaiyaan, M. Antiproliferative and apoptotic effects of xanthohumol in cholangiocarcinoma. Oncotarget 2017, 8, 88069–88078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, D.O.; Freitas, J.; Nogueira, P.; Henriques, S.N.; Carmo, A.M.; Castro, M.A.; Guido, L.F. Xanthohumol inhibits cell proliferation and induces apoptosis in human thyroid cells. Food Chem. Toxicol. 2018, 121, 450–457. [Google Scholar]
- Koo, J.-H.; Kim, H.T.; Yoon, H.-Y.; Kwon, K.-B.; Choi, I.-W.; Jung, S.H.; Kim, H.-U.; Park, B.-H.; Park, J.-W. Effect of xanthohumol on melanogenesis in B16 melanoma cells. Exp. Mol. Med. 2008, 40, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Shi, X.B.; Xu, B.; Yuan, C.S.; Zheng, W.; Li, G.; Li, J.; Wang, Z.H. Endoplasmic reticulum stress mediated the xanthohumol induced murine melanoma B16-F10 cell death. J. Asian Nat. Prod. Res. 2020, 22, 850–863. [Google Scholar] [CrossRef]
- Dorn, C.; Weiss, T.S.; Heilmann, J.; Hellerbrand, C. Xanthohumol, a prenylated chalcone derived from hops, inhibits proliferation, migration and interleukin-8 expression of hepatocellular carcinoma cells. Int. J. Oncol. 2010, 36, 435–441. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Yin, H.; Qian, X.; Dong, J.; Qian, Z.; Miao, J. Xanthohumol, a prenylated chalcone from hops, inhibits the viability and stemness of doxorubicin-resistant MCF-7/ADR Cells. Molecules 2016, 22, 36. [Google Scholar] [CrossRef] [Green Version]
- Denton, A.E.; Roberts, E.W.; Fearon, D.T. Stromal Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2018, 1060, 99–114. [Google Scholar]
- Shaul, M.E.; Fridlender, Z.G. Cancer-related circulating and tumor-associated neutrophils—Subtypes, sources and function. FEBS J. 2018, 285, 4316–4342. [Google Scholar] [CrossRef]
- Moura, V.; Lacerda, M.; Figueiredo, P.; Corvo, M.L.; Cruz, M.E.; Soares, R.; de Lima, M.C.; Simões, S.; Moreira, J.N. Targeted and intracellular triggered delivery of therapeutics to cancer cells and the tumor microenvironment: Impact on the treatment of breast cancer. Breast Cancer Res. Treat. 2012, 133, 61–73. [Google Scholar] [CrossRef]
- Ostuni, R.; Kratochvill, F.; Murray, P.J.; Natoli, G. Macrophages and cancer: From mechanisms to therapeutic implications. Trends Immunol. 2015, 36, 229–239. [Google Scholar] [CrossRef]
- Suh, K.S.; Rhee, S.Y.; Kim, Y.S.; Lee, Y.S.; Choi, E.M. Xanthohumol modulates the expression of osteoclast-specific genes during osteoclastogenesis in RAW264.7 cells. Food Chem. Toxicol. 2013, 62, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.C.; You, S.K.; Kim, H.J.; Cho, C.W.; Lee, I.S.; Kang, B.Y. Xanthohumol inhibits IL-12 production and reduces chronic allergic contact dermatitis. Int. Immunopharmacol. 2010, 10, 556–561. [Google Scholar] [CrossRef]
- Strathmann, J.; Klimo, K.; Sauer, S.W.; Okun, J.G.; Prehn, J.H.; Gerhäuser, C. Xanthohumol-induced transient superoxide anion radical formation triggers cancer cells into apoptosis via a mitochondria-mediated mechanism. FASEB J. 2010, 24, 2938–2950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Song, Z.; Bai, J.; Nauwynck, H.; Zhao, Y.; Jiang, P. Xanthohumol inhibits PRRSV proliferation and alleviates oxidative stress induced by PRRSV via the Nrf2-HMOX1 axis. Vet. Res. 2019, 50, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima-Fontes, M.; Costa, R.; Rodrigues, I.; Soares, R. Xanthohumol Restores Hepatic Glucolipid Metabolism Balance in Type 1 Diabetic Wistar Rats. J. Agric Food Chem. 2017, 65, 7433–7439. [Google Scholar] [CrossRef]
- Costa, R.; Rodrigues, I.; Guardão, L.; Lima, J.Q.; Sousa, E.; Soares, R.; Negrão, R. Modulation of VEGF signaling in a mouse model of diabetes by xanthohumol and 8-prenylnaringenin: Unveiling the angiogenic paradox and metabolism interplay. Mol. Nutr. Food Res. 2017, 61, 1–12. [Google Scholar] [CrossRef]
- Tukulula, M.; Hayeshi, R.; Fonteh, P.; Meyer, D.; Ndamase, A.; Madziva, M.T.; Khumalo, V.; Lubuschagne, P.; Naicker, B.; Swai, H.; et al. Curdlan-Conjugated PLGA Nanoparticles Possess Macrophage Stimulant Activity and Drug Delivery Capabilities. Pharm. Res. 2015, 32, 2713–2726. [Google Scholar] [CrossRef] [Green Version]



| Formulation | Size (nm) | PdI | ZP (mV) | AE (%) | LC (%) |
|---|---|---|---|---|---|
| PLGA Np | 273 ± 18 | 0.285 ± 0.015 | −15.4 ± 2.1 | - | - |
| XN-PLGA Np | 312 ± 49 | 0.259 ± 0.015 | −18.2 ± 1.4 | 88.7 ± 4.3 | 15.9 ± 1.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, M.; Macedo, A.S.; Lima, S.A.C.; Reis, S.; Soares, R.; Fonte, P. Evaluation of the Antitumour and Antiproliferative Effect of Xanthohumol-Loaded PLGA Nanoparticles on Melanoma. Materials 2021, 14, 6421. https://doi.org/10.3390/ma14216421
Fonseca M, Macedo AS, Lima SAC, Reis S, Soares R, Fonte P. Evaluation of the Antitumour and Antiproliferative Effect of Xanthohumol-Loaded PLGA Nanoparticles on Melanoma. Materials. 2021; 14(21):6421. https://doi.org/10.3390/ma14216421
Chicago/Turabian StyleFonseca, Magda, Ana S. Macedo, Sofia A. Costa Lima, Salette Reis, Raquel Soares, and Pedro Fonte. 2021. "Evaluation of the Antitumour and Antiproliferative Effect of Xanthohumol-Loaded PLGA Nanoparticles on Melanoma" Materials 14, no. 21: 6421. https://doi.org/10.3390/ma14216421
APA StyleFonseca, M., Macedo, A. S., Lima, S. A. C., Reis, S., Soares, R., & Fonte, P. (2021). Evaluation of the Antitumour and Antiproliferative Effect of Xanthohumol-Loaded PLGA Nanoparticles on Melanoma. Materials, 14(21), 6421. https://doi.org/10.3390/ma14216421









