The Role of Aryldiazonium Chemistry in Designing Electrochemical Aptasensors for the Detection of Food Contaminants
Abstract
:1. Introduction
2. The Role of Aptamers in Developing Biosensors for Food Contaminants
3. Strategies in Electrochemical Aptasensor Design: Aptamer Immobilization and Electrochemical Signal Generation
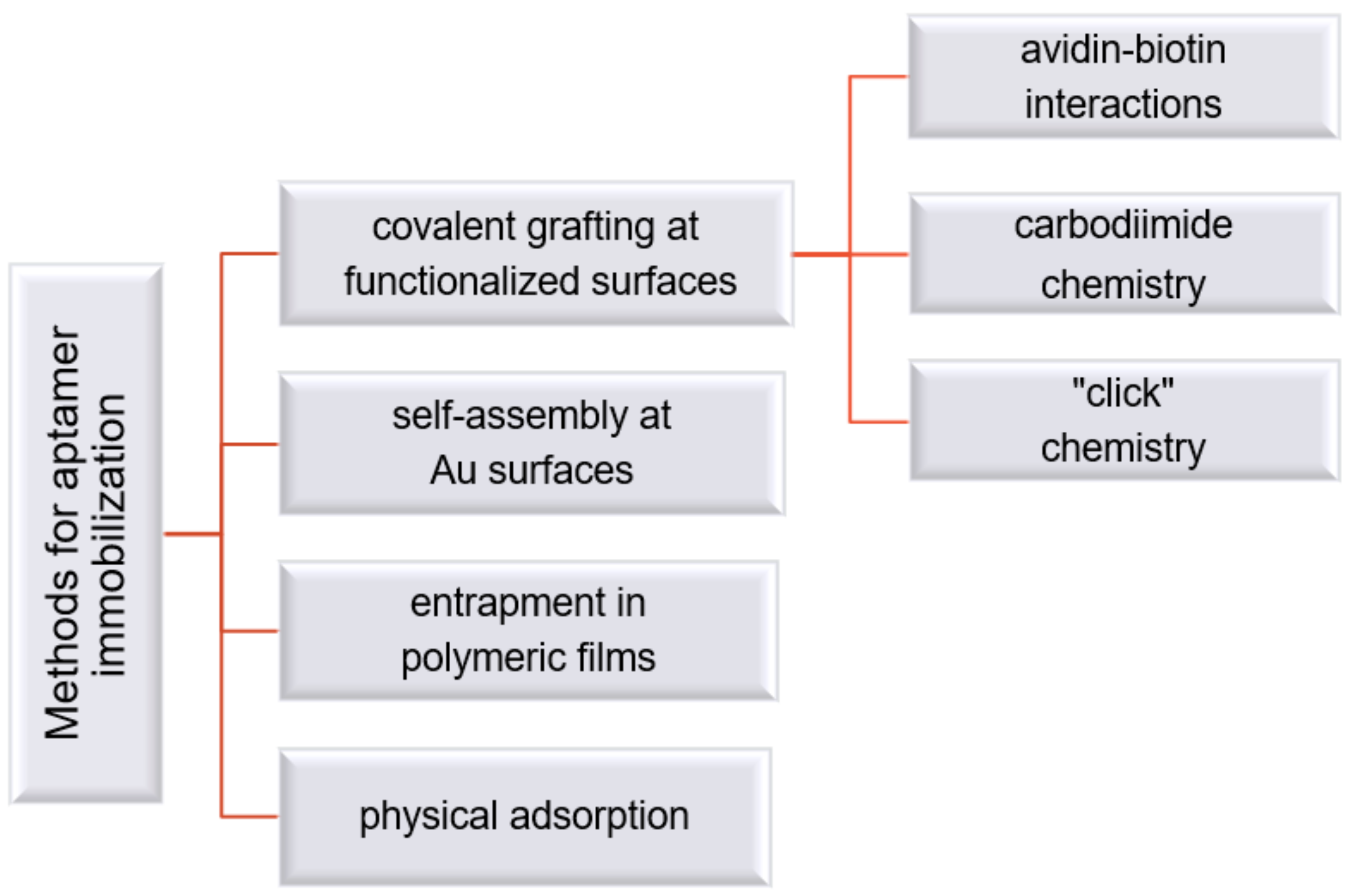
3.1. Strategies for Aptamer Modification and Immobilization at the Electrode Substrate
3.2. Strategies for Aptamer Modification and Immobilization at the Electrode Substrate
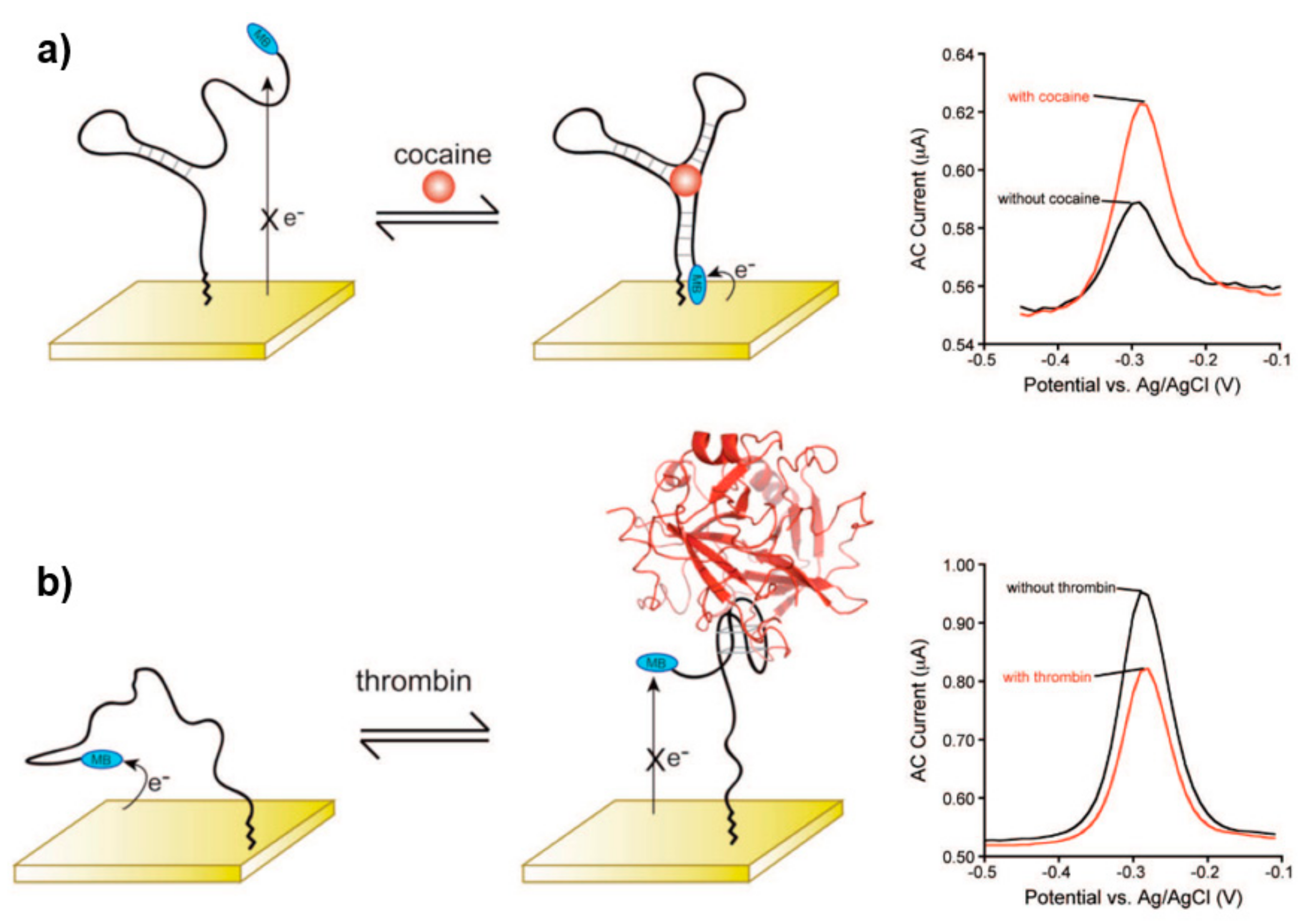
3.2.1. Strategies Based on Target Binding-Induced Aptamer Conformation Change
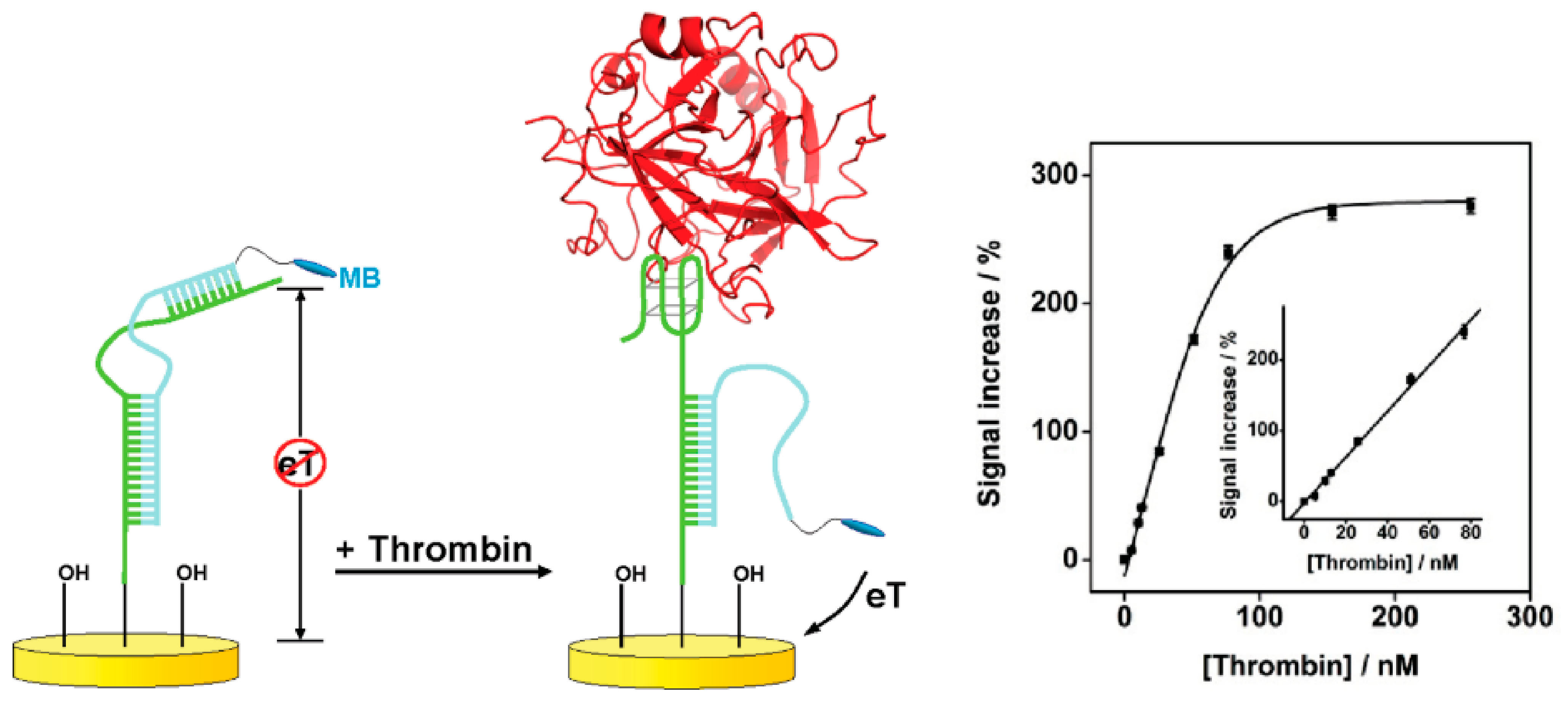
3.2.2. Strategies Based on Target Binding-Induced Aptamer-Complementary Sequence Dissociation
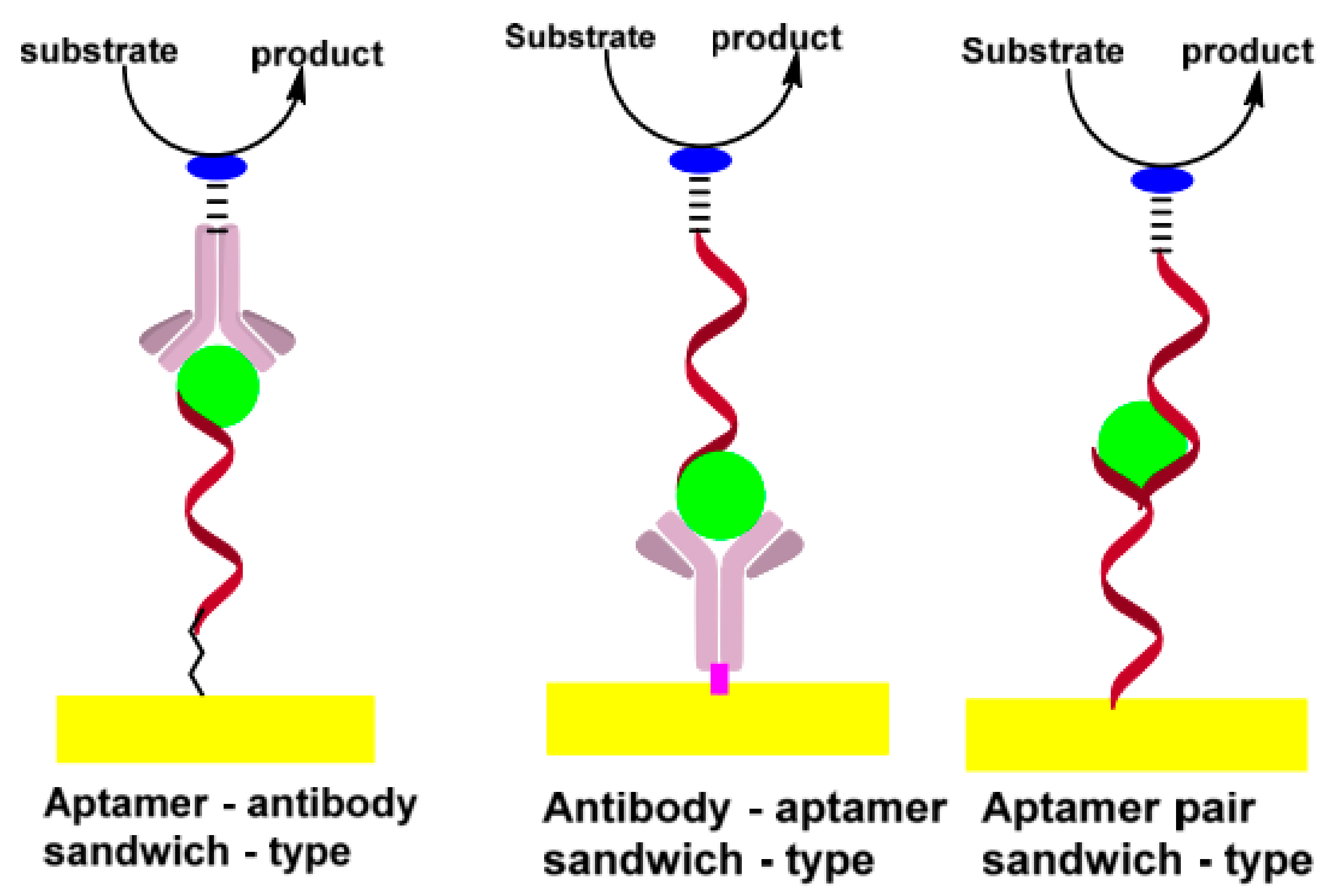
3.2.3. Sandwich Assays
3.2.4. Strategies Based on Label-Free Approaches
3.2.5. The Ratiometric Electrochemical Strategy
3.2.6. Other Amplification Strategies Employed in Electrochemical Aptasensors
4. The Role of Diazonium Electrochemistry for Aptasensors Development
4.1. Basic Considerations and Challenges Regarding the Aryldiazonium Grafting Strategy
4.2. The Role of Aryldiazonium Salts in Aptamer Immobilization on the Sensing Interface
4.3. The Role of Aryldiazonium Chemistry in Multiplexed Detection
4.4. The Role of Aryldiazonium Chemistry in Increasing the Selectivity of Aptasensors
4.5. The Role of Aryldiazonium Chemistry in Increasing the Stability of Aptasensors
4.6. The Role of Aryldiazonium Chemistry in the Assembly of Nanomaterials at the Sensing Interface
5. Electrochemical Aptasensors Developed Using Aryldiazonium Chemistry for Food Safety Monitoring Applications
5.1. Electrochemical Aptasensors for Antibiotics
5.1.1. Tetracyclines
5.1.2. Kanamycin
5.1.3. Penicillin
5.2. Electrochemical Biosensors for Hormonal Disruptors
Estradiol
5.3. Electrochemical Biosensors for Toxins
5.3.1. Ochratoxin A
5.3.2. Aflatoxin
5.3.3. Patulin
5.4. Electrochemical Biosensors for Lysozyme
5.5. Electrochemical Biosensors for Cadmium Ions
5.6. Electrochemical Aptasensors for Bacteria
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 12 April 2020).
- Li, F.; Yu, Z.; Han, X.; Lai, R.Y. Electrochemical aptamer-based sensors for food and water analysis: A review. Anal. Chim. Acta 2019, 1051, 1–23. [Google Scholar] [CrossRef]
- Mishra, G.K.; Barfidokht, A.; Tehrani, F.; Mishra, R.K. Food safety analysis using electrochemical biosensors. Foods 2018, 7, 141. [Google Scholar] [CrossRef] [Green Version]
- Sani, N.D.M.; Heng, L.Y.; Marugan, R.S.P.M.; Rajab, N.F. Electrochemical DNA biosensor for potential carcinogen detection in food sample. Food Chem. 2018, 269, 503–510. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Sun, L.; Liu, L.; Xu, C.; Kuang, H. Nanoparticle-based sensors for food contaminants. TrAC Trends Anal. Chem. 2019, 113, 74–83. [Google Scholar] [CrossRef]
- Mathisen, G.H.; Alexander, J.; Fæste, C.K.; Husøy, T.; Katrine Knutsen, H.; Ørnsrud, R.; Steffensen, I.L. A ranking method of chemical substances in foods for prioritisation of monitoring, based on health risk and knowledge gaps. Food Res. Int. 2020, 137, 109499. [Google Scholar] [CrossRef]
- Malik, A.K.; Blasco, C.; Picó, Y. Liquid chromatography-mass spectrometry in food safety. J. Chromatogr. A 2010, 1217, 4018–4040. [Google Scholar] [CrossRef] [PubMed]
- Escrivá, Ú.; Andrés-Costa, M.J.; Andreu, V.; Picó, Y. Analysis of cannabinoids by liquid chromatography–mass spectrometry in milk, liver and hemp seed to ensure food safety. Food Chem. 2017, 228, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Liu, Y.; Geng, J.; Kou, X.; Xin, Z.; Yang, D. Engineering nanomaterials-based biosensors for food safety detection. Biosens. Bioelectron. 2018, 106, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Leng, Y.; Li, X.; Huang, X.; Xiong, Y. Emerging strategies to enhance the sensitivity of competitive ELISA for detection of chemical contaminants in food samples. TrAC Trends Anal. Chem. 2020, 126, 115861. [Google Scholar] [CrossRef]
- Amiri, M.; Bezaatpour, A.; Jafari, H.; Boukherroub, R.; Szunerits, S. Electrochemical methodologies for the detection of pathogens. ACS Sens. 2018, 3, 1059–1086. [Google Scholar] [CrossRef] [PubMed]
- Dwidar, M.; Yokobayashi, Y. Development of a histamine aptasensor for food safety monitoring. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.A.; Halpern, J.M. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjug. Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; Chamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens. Bioelectron. 2016, 80, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Riu, J.; Giussani, B. Electrochemical biosensors for the detection of pathogenic bacteria in food. TrAC Trends Anal. Chem. 2020, 126, 115863. [Google Scholar] [CrossRef]
- Vlăsceanu, G.M.; Amărandi, R.M.; Ioniță, M.; Tite, T.; Iovu, H.; Pilan, L.; Burns, J.S. Versatile graphene biosensors for enhancing human cell therapy. Biosens. Bioelectron. 2018, 117, 283–302. [Google Scholar] [CrossRef] [PubMed]
- Azadbakht, A.; Roushani, M.; Abbasi, A.R.; Derikvand, Z. A novel impedimetric aptasensor, based on functionalized carbon nanotubes and prussian blue as labels. Anal. Biochem. 2016, 512, 58–69. [Google Scholar] [CrossRef]
- De Girolamo, A.; McKeague, M.; Pascale, M.; Cortese, M.; DeRosa, M.C. Immobilization of Aptamers on Substrates. In Aptamers for Analytical Applications; Dong, Y., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2018; pp. 85–116. ISBN 9783527342679. [Google Scholar]
- Vasilescu, A.; Hayat, A.; Gáspár, S.; Marty, J.L. Advantages of Carbon Nanomaterials in Electrochemical Aptasensors for Food Analysis. Electroanalysis 2018, 30, 2–19. [Google Scholar] [CrossRef]
- Hermann, C.A.; Duerkop, A.; Baeumner, A.J. Food Safety Analysis Enabled through Biological and Synthetic Materials: A Critical Review of Current Trends. Anal. Chem. 2019, 91, 569–587. [Google Scholar] [CrossRef] [PubMed]
- Khoshbin, Z.; Verdian, A.; Housaindokht, M.R.; Izadyar, M.; Rouhbakhsh, Z. Aptasensors as the future of antibiotics test kits-a case study of the aptamer application in the chloramphenicol detection. Biosens. Bioelectron. 2018, 122, 263–283. [Google Scholar] [CrossRef]
- Roushani, M.; Rahmati, Z.; Hoseini, S.J.; Hashemi Fath, R. Impedimetric ultrasensitive detection of chloramphenicol based on aptamer MIP using a glassy carbon electrode modified by 3-ampy-RGO and silver nanoparticle. Colloids Surf. B Biointerfaces 2019, 183, 110451. [Google Scholar] [CrossRef]
- Chen, Z.; Lai, G.; Liu, S.; Yu, A. Ultrasensitive electrochemical aptasensing of kanamycin antibiotic by enzymatic signal amplification with a horseradish peroxidase-functionalized gold nanoprobe. Sens. Actuators B Chem. 2018, 273, 1762–1767. [Google Scholar] [CrossRef]
- Alawad, A.; Istamboulié, G.; Calas-Blanchard, C.; Noguer, T. A reagentless aptasensor based on intrinsic aptamer redox activity for the detection of tetracycline in water. Sens. Actuators B Chem. 2019, 288, 141–146. [Google Scholar] [CrossRef]
- Jahanbani, S.; Benvidi, A. Comparison of two fabricated aptasensors based on modified carbon paste/oleic acid and magnetic bar carbon paste/Fe3O4@oleic acid nanoparticle electrodes for tetracycline detection. Biosens. Bioelectron. 2016, 85, 553–562. [Google Scholar] [CrossRef]
- Jahangiri–Dehaghani, F.; Zare, H.R.; Shekari, Z. Measurement of aflatoxin M1 in powder and pasteurized milk samples by using a label–free electrochemical aptasensor based on platinum nanoparticles loaded on Fe–based metal–organic frameworks. Food Chem. 2020, 310, 125820. [Google Scholar] [CrossRef]
- Deiminiat, B.; Rounaghi, G.H.; Arbab-Zavar, M.H.; Razavipanah, I. A novel electrochemical aptasensor based on f-MWCNTs/AuNPs nanocomposite for label-free detection of bisphenol A. Sens. Actuators B Chem. 2017, 242, 158–166. [Google Scholar] [CrossRef]
- Rajabnejad, S.H.; Badibostan, H.; Verdian, A.; Karimi, G.R.; Fooladi, E.; Feizy, J. Aptasensors as promising new tools in bisphenol A detection—An invisible pollution in food and environment. Microchem. J. 2020, 155, 104722. [Google Scholar] [CrossRef]
- Duffy, G.F.; Moore, E.J. Electrochemical Immunosensors for Food Analysis: A Review of Recent Developments. Anal. Lett. 2017, 50, 1–32. [Google Scholar] [CrossRef]
- Hetemi, D.; Noël, V.; Pinson, J. Grafting of diazonium salts on surfaces: Application to biosensors. Biosensors 2020, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- Prieto-Simon, B.; Campas, M.; Marty, J.-L. Biomolecule Immobilization in Biosensor Development: Tailored Strategies Based on Affinity Interactions. Protein Pept. Lett. 2008, 15, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, S.; Xiao, Y.; Wang, Y. Recent Advances in Immobilization Strategies for Biomolecules in Sensors Using Organic Field-Effect Transistors. Trans. Tianjin Univ. 2020, 26, 424–440. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Integrated Affinity Biosensing Platforms on Screen-Printed Electrodes Electrografted with Diazonium Salts. Sensors 2018, 18, 675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Moraes Silva, S.; Fan, S.; Wu, Y.; Alam, M.T.; Liu, G.; Justin Gooding, J. Aryldiazonium salt derived mixed organic layers: From surface chemistry to their applications. J. Electroanal. Chem. 2017, 785, 265–278. [Google Scholar] [CrossRef]
- Pilan, L. Tailoring the performance of electrochemical biosensors based on carbon nanomaterials via aryldiazonium electrografting. Bioelectrochemistry 2021, 138, 107697. [Google Scholar] [CrossRef] [PubMed]
- Raicopol, M.; Vlasceanu, I.; Lupulescu, I.; Brezoiu, A.M.; Pilan, L. Amperometric glucose biosensors based on functionalized electrochemically reduced graphene oxide. UPB Sci. Bull. Ser. B Chem. Mater. Sci. 2016, 78, 131–142. [Google Scholar]
- Levrie, K.; Jans, K.; Vos, R.; Ardakanian, N.; Verellen, N.; Van Hoof, C.; Lagae, L.; Stakenborg, T. Multiplexed site-specific electrode functionalization for multitarget biosensors. Bioelectrochemistry 2016, 112, 61–66. [Google Scholar] [CrossRef]
- Janegitz, B.C.; Silva, T.A.; Wong, A.; Ribovski, L.; Vicentini, F.C.; del Taboada Sotomayor, M.P.; Fatibello-Filho, O. The application of graphene for in vitro and in vivo electrochemical biosensing. Biosens. Bioelectron. 2017, 89, 224–233. [Google Scholar] [CrossRef] [Green Version]
- Hayat, A.; Sassolas, A.; Marty, J.L.; Radi, A.E. Highly sensitive ochratoxin A impedimetric aptasensor based on the immobilization of azido-apactamer onto electrografted binary film via click chemistry. Talanta 2013, 103, 14–19. [Google Scholar] [CrossRef]
- Villalonga, A.; Pérez-Calabuig, A.M.; Villalonga, R. Electrochemical biosensors based on nucleic acid aptamers. Anal. Bioanal. Chem. 2020, 412, 55–72. [Google Scholar] [CrossRef]
- Vasilescu, A.; Marty, J.L. Electrochemical aptasensors for the assessment of food quality and safety. TrAC Trends Anal. Chem. 2016, 79, 60–70. [Google Scholar] [CrossRef]
- Ebrahimi Vafaye, S.; Rahman, A.; Safaeian, S.; Adabi, M. An electrochemical aptasensor based on electrospun carbon nanofiber mat and gold nanoparticles for the sensitive detection of Penicillin in milk. J. Food Meas. Charact. 2021, 15, 876–882. [Google Scholar] [CrossRef]
- Malecka, K.; Mikuła, E.; Ferapontova, E.E. Design strategies for electrochemical aptasensors for cancer diagnostic devices. Sensors 2021, 21, 736. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labuda, J.; Oliveira Brett, A.M.; Evtugyn, G.; Fojta, M.; Mascini, M.; Ozsoz, M.; Palchetti, I.; Paleček, E.; Wang, J. Electrochemical nucleic acid-based biosensors: Concepts, terms, and methodology (IUPAC Technical Report). Pure Appl. Chem. 2010, 82, 1161–1187. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Wang, L.; Peng, X.; Fu, H.; Huang, C.; Li, Y.; Liu, Z. Recent advances in the development of electrochemical aptasensors for detection of heavy metals in food. Biosens. Bioelectron. 2020, 147, 111777. [Google Scholar] [CrossRef]
- Zhan, S.; Wu, Y.; Wang, L.; Zhan, X.; Zhou, P. A mini-review on functional nucleic acids-based heavy metal ion detection. Biosens. Bioelectron. 2016, 86, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Ștefan, G.; Hosu, O.; De Wael, K.; Lobo-Castañón, M.J.; Cristea, C. Aptamers in biomedicine: Selection strategies and recent advances. Electrochim. Acta 2021, 376, 137994. [Google Scholar] [CrossRef]
- Lyu, C.; Mahmood, I.; Wang, Z. Capture-SELEX for aptamer selection: A short review. Talanta 2021, 229, 122274. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Nikolaus, N.; Strehlitz, B. Capture-SELEX: Selection of DNA aptamers for aminoglycoside antibiotics. J. Anal. Methods Chem. 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Smart Carbon Nanomaterials in Electrochemical Biosensing for Clinical Analysis. Handb. Smart Mater. Anal. Chem. 2019, 859–894. [Google Scholar] [CrossRef]
- Ahirwar, R.; Nahar, S.; Aggarwal, S.; Ramachandran, S. In silico selection of an aptamer to estrogen receptor alpha using computational docking employing estrogen response elements as aptamer-alike molecules. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buglak, A.A.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. Methods and applications of in silico aptamer design and modeling. Int. J. Mol. Sci. 2020, 21, 8420. [Google Scholar] [CrossRef]
- Sharma, T.K.; Bruno, J.G.; Dhiman, A. ABCs of DNA aptamer and related assay development. Biotechnol. Adv. 2017, 35, 275–301. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Chumphukam, O.; Cass, A.E.G. Determination of minimal sequence for binding of an aptamer. A comparison of truncation and hybridization inhibition methods. RSC Adv. 2014, 4, 47227–47233. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, B.; Gao, S.X.; Liu, D.J.; Sun, M.J.; Jiao, B.H.; Wang, L.H. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 2015, 101, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alawad, A.; Latapie, L.; Evrard, D.; Gros, P.; Istamboulie, G.; Noguer, T.; Calas-Blanchard, C. SECM for Studying the Immobilization and Repartition of a Redox Anti-tetracycline Aptamer on Screen-printed Carbon Electrodes. Electroanalysis 2021, 33, 292–295. [Google Scholar] [CrossRef]
- Niazi, J.H.; Lee, S.J.; Gu, M.B. Single-stranded DNA aptamers specific for antibiotics tetracyclines. Bioorg. Med. Chem. 2008, 16, 7245–7253. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yan, W.; Guo, Y.; Wang, X.; Zhang, F.; Yu, L.; Guo, C.; Fang, G. Sensitive and selective electrochemical aptasensor via diazonium-coupling reaction for label-free determination of oxytetracycline in milk samples. Sens. Actuators Rep. 2020, 2, 100009. [Google Scholar] [CrossRef]
- Kim, Y.S.; Niazi, J.H.; Gu, M.B. Specific detection of oxytetracycline using DNA aptamer-immobilized interdigitated array electrode chip. Anal. Chim. Acta 2009, 634, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Istamboulie, G.; Hayat, A.; Catanante, G.; Bhand, S.; Marty, J.L. Disposable and portable aptamer functionalized impedimetric sensor for detection of kanamycin residue in milk sample. Sens. Actuators B Chem. 2017, 245, 507–515. [Google Scholar] [CrossRef]
- Song, K.M.; Cho, M.; Jo, H.; Min, K.; Jeon, S.H.; Kim, T.; Han, M.S.; Ku, J.K.; Ban, C. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer. Anal. Biochem. 2011, 415, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Paniel, N.; Istamboulié, G.; Triki, A.; Lozano, C.; Barthelmebs, L.; Noguer, T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta 2017, 162, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Rather, J.A.; Khudaish, E.A.; Kannan, P. Graphene-amplified femtosensitive aptasensing of estradiol, an endocrine disruptor. Analyst 2018, 143, 1835–1845. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Jung, H.S.; Matsuura, T.; Lee, H.Y.; Kawai, T.; Gu, M.B. Electrochemical detection of 17β-estradiol using DNA aptamer immobilized gold electrode chip. Biosens. Bioelectron. 2007, 22, 2525–2531. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Hayat, A.; Catanante, G.; Ocaña, C.; Marty, J.L. A label free aptasensor for Ochratoxin A detection in cocoa beans: An application to chocolate industries. Anal. Chim. Acta 2015, 889, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Hayat, A.; Catanante, G.; Istamboulie, G.; Marty, J.L. Sensitive quantitation of Ochratoxin A in cocoa beans using differential pulse voltammetry based aptasensor. Food Chem. 2016, 192, 799–804. [Google Scholar] [CrossRef]
- Gökçe, G.; Ben Aissa, S.; Nemčeková, K.; Catanante, G.; Raouafi, N.; Marty, J.L. Aptamer-modified pencil graphite electrodes for the impedimetric determination of ochratoxin A. Food Control 2020, 115, 4–10. [Google Scholar] [CrossRef]
- Cruz-Aguado, J.A.; Penner, G. Determination of ochratoxin A with a DNA aptamer. J. Agric. Food Chem. 2008, 56, 10456–10461. [Google Scholar] [CrossRef]
- Yugender Goud, K.; Catanante, G.; Hayat, A.; M., S.; Vengatajalabathy Gobi, K.; Marty, J.L. Disposable and portable electrochemical aptasensor for label free detection of aflatoxin B1 in alcoholic beverages. Sens. Actuators B Chem. 2016, 235, 466–473. [Google Scholar] [CrossRef]
- Le, L.C.; Allen, J.A.C.-A.; Penner, G.A. WO 2011/020198 Al 2011. Available online: https://patentimages.storage.googleapis.com/c3/c4/c5/abc0571addbb95/WO2011020198A1.pdf (accessed on 6 July 2021).
- Istamboulié, G.; Paniel, N.; Zara, L.; Granados, L.R.; Barthelmebs, L.; Noguer, T. Development of an impedimetric aptasensor for the determination of aflatoxin M1 in milk. Talanta 2016, 146, 464–469. [Google Scholar] [CrossRef]
- Nguyen, B.H.; Tran, L.D.; Do, Q.P.; Nguyen, H.L.; Tran, N.H.; Nguyen, P.X. Label-free detection of aflatoxin M1 with electrochemical Fe3O4/polyaniline-based aptasensor. Mater. Sci. Eng. C 2013, 33, 2229–2234. [Google Scholar] [CrossRef]
- Khan, R.; Aissa, S.B.; Sherazi, T.A.; Catanante, G.; Hayat, A.; Marty, J.L. Development of an impedimetric aptasensor for label free detection of patulin in apple juice. Molecules 2019, 24, 1017. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Duan, N.; Zhang, W.; Zhao, S.; Wang, Z. Screening and development of DNA aptamers as capture probes for colorimetric detection of patulin. Anal. Biochem. 2016, 508, 58–64. [Google Scholar] [CrossRef]
- Ocaña, C.; Hayat, A.; Mishra, R.K.; Vasilescu, A.; del Valle, M.; Marty, J.L. Label free aptasensor for Lysozyme detection: A comparison of the analytical performance of two aptamers. Bioelectrochemistry 2015, 105, 72–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ocaña, C.; Hayat, A.; Mishra, R.; Vasilescu, A.; Del Valle, M.; Marty, J.L. A novel electrochemical aptamer-antibody sandwich assay for lysozyme detection. Analyst 2015, 140, 4148–4153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, D.T.; Janssen, K.P.F.; Pollet, J.; Lammertyn, E.; Anné, J.; Van Schepdael, A.; Lammertyn, J. Selection and characterization of DNA aptamers for egg white lysozyme. Molecules 2010, 15, 1127. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Aguayo, D.; del Valle, M. Label-free aptasensor for lysozyme detection using electrochemical impedance spectroscopy. Sensors 2018, 18, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Subramanian, P.; Schechter, A.; Teblum, E.; Yemini, R.; Nessim, G.D.; Vasilescu, A.; Li, M.; Boukherroub, R.; Szunerits, S. Vertically Aligned Nitrogen-Doped Carbon Nanotube Carpet Electrodes: Highly Sensitive Interfaces for the Analysis of Serum from Patients with Inflammatory Bowel Disease. ACS Appl. Mater. Interfaces 2016, 8, 9600–9609. [Google Scholar] [CrossRef]
- Cox, J.C.; Ellington, A.D. Automated selection of anti-protein aptamers. Bioorg. Med. Chem. 2001, 9, 2525–2531. [Google Scholar] [CrossRef]
- Wang, Q.; Vasilescu, A.M.; Wang, Q.; Li, M.; Boukherroub, R.; Szunerits, S.; Coffinier, Y.; Li, M.; Boukherroub, R.; Szunerits, S.; et al. Electrophoretic approach for the simultaneous deposition and functionalization of reduced graphene oxide nanosheets with diazonium compounds: Application for sensing in serum. ACS Appl. Mater. Interfaces 2017, 9, 12823–12831. [Google Scholar] [CrossRef] [PubMed]
- Kirby, R.; Cho, E.J.; Gehrke, B.; Bayer, T.; Park, Y.S.; Neikirk, D.P.; McDevitt, J.T.; Ellington, A.D. Aptamer-based sensor arrays for the detection and quantitation of proteins. Anal. Chem. 2004, 76, 4066–4075. [Google Scholar] [CrossRef] [PubMed]
- Rabai, S.; Benounis, M.; Baraket, A.; Errachid, A.; Jaffrezic, N.; Louis, J.; Rhouati, A. Development of a label-free electrochemical aptasensor based on diazonium electrodeposition: Application to cadmium detection in water. Anal. Biochem. 2021, 612, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhan, S.; Wang, L.; Zhou, P. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst 2014, 139, 1550–1561. [Google Scholar] [CrossRef]
- Bagheryan, Z.; Raoof, J.B.; Golabi, M.; Turner, A.P.F.; Beni, V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens. Bioelectron. 2016, 80, 566–573. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Janagama, H.; Dwivedi, H.P.; Senthil Kumar, T.M.A.; Jaykus, L.A.; Schefers, J.; Sreevatsan, S. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Mol. Cell. Probes 2009, 23, 20–28. [Google Scholar] [CrossRef]
- Meirinho, G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Voltammetric aptasensors for protein disease biomarkers detection: A review. Biotechnol. Adv. 2016, 34, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Catanante, G.; Mishra, R.K.; Hayat, A.; Marty, J.L. Sensitive analytical performance of folding based biosensor using methylene blue tagged aptamers. Talanta 2016, 153, 138–144. [Google Scholar] [CrossRef]
- Evtugyn, G.; Porfireva, A.; Shamagsumova, R.; Hianik, T. Advances in electrochemical aptasensors based on carbon nanomaterials. Chemosensors 2020, 8, 96. [Google Scholar] [CrossRef]
- Simon, L.; Bognár, Z.; Gyurcsányi, R.E. Finding the Optimal Surface Density of Aptamer Monolayers by SPR Imaging Detection-Based Aptamer. Electroanalysis 2020, 32, 1–9. [Google Scholar] [CrossRef]
- White, R.J.; Phares, N.; Lubin, A.A.; Xiao, Y.; Plaxco, K.W. Optimization of electrochemical aptamer-based sensors via optimization of probe packing density and surface chemistry. Langmuir 2008, 24, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Qiao, X.; Zhao, H.; He, Y.; Sheng, Q.; Yue, T. Ultrasensitive and label-free electrochemical aptasensor based on carbon dots-black phosphorus nanohybrid for the detection of Ochratoxins A. Microchem. J. 2021, 106378. [Google Scholar] [CrossRef]
- Liu, M.; Ke, H.; Sun, C.; Wang, G.; Wang, Y.; Zhao, G. A simple, supersensitive and highly selective electrochemical label-free aptasensor of 17β-estradiol based on signal amplification of bi-functional graphene. Talanta 2018, 194, 266–272. [Google Scholar] [CrossRef]
- Yu, P.; Liu, Y.; Zhang, X.; Zhou, J.; Xiong, E.; Li, X.; Chen, J. A novel electrochemical aptasensor for bisphenol A assay based on triple-signaling strategy. Biosens. Bioelectron. 2016, 79, 22–28. [Google Scholar] [CrossRef]
- Yan, L.; Luo, C.; Cheng, W.; Mao, W.; Zhang, D.; Ding, S. A simple and sensitive electrochemical aptasensor for determination of Chloramphenicol in honey based on target-induced strand release. J. Electroanal. Chem. 2012, 687, 89–94. [Google Scholar] [CrossRef]
- Guarisco, M.; Gandolfi, D.; Guider, R.; Vanzetti, L.; Bartali, R.; Ghulinyan, M.; Cretich, M.; Chiari, M.; Bettotti, P.; Pavesi, L.; et al. A new aptamer immobilization strategy for protein recognition. Sens. Actuators B Chem. 2017, 252, 222–231. [Google Scholar] [CrossRef]
- Ocaña, C.; Lukic, S.; del Valle, M.; Valle, M. Aptamer-antibody sandwich assay for cytochrome c employing an MWCNT platform and electrochemical impedance. Microchim. Acta 2015, 182, 2045–2053. [Google Scholar] [CrossRef] [Green Version]
- Ni, S.; Qiao, L.; Shen, Z.; Gao, Y.; Liu, G. Physical absorption vs covalent binding of graphene oxide on glassy carbon electrode towards a robust aptasensor for ratiometric electrochemical detection of vascular endothelial growth factor (VEGF) in serum. Electrochim. Acta 2020, 331, 135321. [Google Scholar] [CrossRef]
- Acquah, C.; Danquah, M.K.; Yon, J.L.S.; Sidhu, A.; Ongkudon, C.M. A review on immobilised aptamers for high throughput biomolecular detection and screening. Anal. Chim. Acta 2015, 888, 10–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paleček, E.; Bartošík, M. Electrochemistry of nucleic acids. Chem. Rev. 2012, 112, 3427–3481. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Disney, M.D.; Miller, B.L.; Krauss, T.D. Hybridization-based unquenching of DNA hairpins on Au surfaces: Prototypical “molecular beacon” biosensors. J. Am. Chem. Soc. 2003, 125, 4012–4013. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, I.; Mascini, M. Electrochemical nanomaterial-based nucleic acid aptasensors. Anal. Bioanal. Chem. 2012, 402, 3103–3114. [Google Scholar] [CrossRef]
- Cheng, A.K.H.; Sen, D.; Yu, H. Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry 2009, 77, 1–12. [Google Scholar] [CrossRef]
- Sassolas, A.; Blum, L.J.; Béatrice, D.L.B. Electrochemical aptasensors. Electroanalysis 2009, 21, 1237–1250. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, J.A.; Wang, J.; Kawde, A.; Xiang, Y.; Gothelf, K.V.; Collins, G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J. Am. Chem. Soc. 2006, 2228–2229. [Google Scholar] [CrossRef]
- Xiao, Y.; Lubin, A.A.; Heeger, A.J.; Plaxco, K.W. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem. Int. Ed. 2005, 44, 5456–5459. [Google Scholar] [CrossRef]
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heegert, A.J. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991. [Google Scholar] [CrossRef] [PubMed]
- Lubin, A.A.; Plaxco, K.W. Folding-based electrochemical biosensors: The case for responsive nucleic acid architectures. Acc. Chem. Res. 2010, 43, 496–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palchetti, I.; Mascini, M. Electrochemical Nucleic Acid Aptamer–Based Biosensors. In ELECTROCHEMİCAL Biosensors; Cosnier, S., Ed.; Taylor & Francis Group, LLC: Abingdon, UK, 2013; pp. 75–87. ISBN 9789814303613. [Google Scholar]
- Abd-Ellatief, R.; Abd-Ellatief, M.R. Electrochemical Aptasensors: Current Status and Future Perspectives. Diagnostics 2021, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Chrouda, A.; Sbartai, A.; Baraket, A.; Renaud, L.; Maaref, A.; Jaffrezic-Renault, N. An aptasensor for ochratoxin A based on grafting of polyethylene glycol on a boron-doped diamond microcell. Anal. Biochem. 2015, 488, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Steel, A.B.; Herne, T.M.; Tarlov, M.J. Electrochemical quantitation of DNA immobilized on gold. Anal. Chem. 1998, 70, 4670–4677. [Google Scholar] [CrossRef]
- Shen, L.; Chen, Z.; Li, Y.; Jing, P.; Xie, S.; He, S.; He, P.; Shao, Y. A chronocoulometric aptamer sensor for adenosine monophosphate. Chem. Commun. 2007, 2169–2171. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.K.H.; Ge, B.; Yu, H.Z. Aptamer-based biosensors for label-free voltammetric detection of lysozyme. Anal. Chem. 2007, 79, 5158–5164. [Google Scholar] [CrossRef] [PubMed]
- Vis, P.; Lemmers, J. An Introduction to the Use of Fluorescent Probes in Ratiometric Fluorescence Microscopy BT—Signal Transduction—Single Cell Techniques; Van Duijn, B., Wiltink, A., Eds.; Springer: Berlin/Heidelberg, Germany, 1998; pp. 285–305. ISBN 978-3-642-80368-0. [Google Scholar]
- Spring, S.A.; Goggins, S.; Frost, C.G. Ratiometric Electrochemistry: Improving the Robustness, Reproducibility and Reliability of Biosensors. Molecules 2021, 26, 2130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Liu, D.; Li, Y.; Shen, X.; Ma, S.; Liu, Y.; You, T. Ratiometric electrochemical aptasensor for ultrasensitive detection of Ochratoxin A based on a dual signal amplification strategy: Engineering the binding of methylene blue to DNA. Biosens. Bioelectron. 2020, 150, 111814. [Google Scholar] [CrossRef]
- Yang, T.; Yu, R.; Yan, Y.; Zeng, H.; Luo, S.; Liu, N.; Morrin, A.; Luo, X.; Li, W. A review of ratiometric electrochemical sensors: From design schemes to future prospects. Sens. Actuators B Chem. 2018, 274, 501–516. [Google Scholar] [CrossRef]
- Yu, P.; Zhou, J.; Wu, L.; Xiong, E.; Zhang, X.; Chen, J. A ratiometric electrochemical aptasensor for sensitive detection of protein based on aptamer—Target—Aptamer sandwich structure. J. Electroanal. Chem. 2014, 732, 61–65. [Google Scholar] [CrossRef]
- Lu, M.; Cao, C.; Wang, F.; Liu, G. A polyethyleneimine reduced graphene oxide/gold nanocubes based electrochemical aptasensor for chloramphenicol detection using single-stranded DNA-binding protein. Mater. Des. 2021, 199, 109409. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Xu, W.; Leng, X.; Wang, H.; Guo, Y.; Huang, J. A Novel Sandwich-type Electrochemical Aptasensor Based on GR-3D Au and Aptamer-AuNPs-HRP for Sensitive Detection of Oxytetracycline. Procedia Technol. 2017, 27, 151–152. [Google Scholar] [CrossRef]
- Yao, J.; Li, Y.; Xie, M.; Yang, Q.; Liu, T. The electrochemical behaviors and kinetics of AuNPs/N, S-GQDs composite electrode: A novel label-free amplified BPA aptasensor with extreme sensitivity and selectivity. J. Mol. Liq. 2020, 320, 114384. [Google Scholar] [CrossRef]
- Hui, Y.; Wang, B.; Ren, R.; Zhao, A.; Zhang, F.; Song, S.; He, Y. An electrochemical aptasensor based on DNA-AuNPs-HRP nanoprobes and exonuclease-assisted signal amplification for detection of aflatoxin B1. Food Control 2020, 109. [Google Scholar] [CrossRef]
- Yan, M.; Bai, W.; Zhu, C.; Huang, Y.; Yan, J.; Chen, A. Design of nuclease-based target recycling signal amplification in aptasensors. Biosens. Bioelectron. 2016, 77, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Liu, D.; Dong, N.; Li, Y.; Meng, S.; You, T. Interaction between the functionalized probes: The depressed efficiency of dual-amplification strategy on ratiometric electrochemical aptasensor for aflatoxin B1. Biosens. Bioelectron. 2021, 182, 113169. [Google Scholar] [CrossRef] [PubMed]
- Delamar, M.; Hitmi, R.; Pinson, J.; Saveant, J.M. Covalent modification of carbon surfaces by grafting of functionalized aryl radicals produced from electrochemical reduction of diazonium salts. J. Am. Chem. Soc. 1992, 114, 5883–5884. [Google Scholar] [CrossRef]
- Kirkman, P.M.; Güell, A.G.; Cuharuc, A.S.; Unwin, P.R. Spatial and temporal control of the diazonium modification of sp 2 carbon surfaces. J. Am. Chem. Soc. 2014, 136, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Adenier, A.; Barré, N.; Cabet-Deliry, E.; Chaussé, A.; Griveau, S.; Mercier, F.; Pinson, J.; Vautrin-Ul, C. Study of the spontaneous formation of organic layers on carbon and metal surfaces from diazonium salts. Surf. Sci. 2006, 600, 4801–4812. [Google Scholar] [CrossRef]
- Bourdillon, C.; Delamar, M.; Demaille, C.; Hitmi, R.; Moiroux, J.; Pinson, J. Immobilization of glucose oxidase on a carbon surface derivatized by electrochemical reduction of diazonium salts. J. Electroanal. Chem. 1992, 336, 113–123. [Google Scholar] [CrossRef]
- Ungureanu, E.-M.; Pilan, L.; Meghea, A.; Le Floch, F.; Simonato, J.-P.; Bidan, G. Preliminary tests for grafting p-nitrophenyl on carbon nanotubes. Rev. Chim. 2008, 59. [Google Scholar] [CrossRef]
- Ungureanu, E.-M.; Pilan, L.; Meghea, A.; Le Floch, F.; Simonato, J.-P.; Bidan, G. Preliminary Tests for N,N-Diethylaniline Grafting on Carbon Nanotubes. Rev. Chim. 2007, 58, 866–870. [Google Scholar]
- Le Floch, F.; Bidan, G.; Pilan, L.; Ungureanu, E.-M.; Simonato, J.-P. Carbon substrate functionalization with diazonium salts toward sensor applications. Mol. Cryst. Liq. Cryst. 2008, 486, 37–41. [Google Scholar] [CrossRef]
- Mohamed, A.A.; Salmi, Z.; Dahoumane, S.A.; Mekki, A.; Carbonnier, B.; Chehimi, M.M. Functionalization of nanomaterials with aryldiazonium salts. Adv. Colloid Interface Sci. 2016, 225, 16–36. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Tailoring Sensitivity in Electrochemical Nucleic Acid Hybridization Biosensing: Role of Surface Chemistry and Labeling Strategies. ChemElectroChem 2018, 6, 60–72. [Google Scholar] [CrossRef]
- Laure, W.; De Bruycker, K.; Espeel, P.; Fournier, D.; Woisel, P.; Du Prez, F.E.; Lyskawa, J. Ultrafast Tailoring of Carbon Surfaces via Electrochemically Attached Triazolinediones. Langmuir 2018. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Alshehri, N.; Rahman, A.M.A.; Dasouki, M.; Salah, K.M.A.; Zourob, M. Electrochemical immunosensors for the detection of survival motor neuron (SMN) protein using different carbon nanomaterials-modified electrodes. Biosens. Bioelectron. 2018, 101, 282–289. [Google Scholar] [CrossRef]
- Branzoi, V.; Branzoi, F.; Raicopol, M.; Pilan, L. Blocking behaviour of covalently attached p-substituted phenyls towards solution-based redox probes. Rev. Chim. 2011, 62, 436–442. [Google Scholar]
- Baranton, S.; Belanger, D. Electrochemical derivatization of carbon surface by reduction of in situ generated diazonium cations. J. Phys. Chem. B 2005, 109, 24401–24410. [Google Scholar] [CrossRef]
- Ghilane, J.; Martin, P.; Lacaze, P.-C.; Randriamahazaka, H.; Lacroix, J.; Santos, L. Host-Guest Complexation: A Convenient Route for the Electroreduction of Diazonium Salts in Aqueous Media and the Formation of Composite Materials. J. Am. Chem. Soc. 2010, 132, 1690–1698. [Google Scholar]
- Bahr, J.L.; Yang, J.; Kosynkin, D.V.; Bronikowski, M.J.; Smalley, R.E.; Tour, J.M. Functionalization of Carbon Nanotubes by Electrochemical Reduction of Aryl Diazonium Salts: A Bucky Paper Electrode. J. Am. Chem. Soc. 2001, 123, 6536–6542. [Google Scholar] [CrossRef]
- Baranton, S.; Bélanger, D. In situ generation of diazonium cations in organic electrolyte for electrochemical modification of electrode surface. Electrochim. Acta 2008, 53, 6961–6967. [Google Scholar] [CrossRef]
- Tour, J.M.; Bahr, J.L. Highly functionalized carbon nanotubes using in situ generated diazonium compounds. Chem. Mater. 2001, 13, 3823–3824. [Google Scholar]
- Flavin, K.; Chaur, M.N.; Echegoyen, L.; Giordani, S. Functionalization of Multilayer Fullerenes (Carbon Nano-Onions) using Diazonium Compounds and “Click” Chemistry. Org. Lett. 2010, 12, 840–843. [Google Scholar] [CrossRef]
- Wang, A.; Yu, W.; Huang, Z.; Zhou, F.; Song, J.; Song, Y. Covalent functionalization of reduced graphene oxide with porphyrin by means of diazonium chemistry for nonlinear optical performance. Sci. Rep. 2016, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCreery, R.; Bergren, A.J. Diazonium Compounds in Molecular Electronics. In Aryl Diazonium Salts New Coupling Agents in Polymer and Surface Science; Chehimi, M.M., Ed.; Wiley-VCH Verlag & Co. KGaA: Weinheim, Germany, 2012; pp. 219–239. ISBN 978-3-527-65047-7. [Google Scholar]
- Mévellec, V.; Roussel, S.; Tessier, L.; Chancolon, J.; Mayne-L’Hermite, M.; Deniau, G.; Viel, P.; Palacin, S. Grafting polymers on surfaces: A new powerful and versatile diazonium salt-based one-step process in aqueous media. Chem. Mater. 2007, 19, 6323–6330. [Google Scholar] [CrossRef]
- Chamoulaud, G.; Bélanger, D. Spontaneous derivatization of a copper electrode with in situ generated diazonium cations in aprotic and aqueous media. J. Phys. Chem. C 2007, 111, 7501–7507. [Google Scholar] [CrossRef]
- Garrett, D.J.; Lehr, J.; Miskelly, G.M.; Downard, A.J. Microcontact printing using the spontaneous reduction of aryldiazonium salts. J. Am. Chem. Soc. 2007, 129, 15456–15457. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.P.; Maya, F.; Kosynkin, D.V.; Dirk, S.M.; Stapleton, J.J.; McGuiness, C.L.; Allara, D.L.; Tour, J.M. Direct Covalent Grafting of Conjugated Molecules onto Si, GaAs, and Pd Surfaces from Aryldiazonium Salts. J. Am. Chem. Soc. 2004, 126, 370–378. [Google Scholar] [CrossRef]
- Lehr, J.; Williamson, B.E.; Flavel, B.S.; Downard, A.J. Reaction of gold substrates with diazonium salts in acidic solution at open-circuit potential. Langmuir 2009, 25, 13503–13509. [Google Scholar] [CrossRef]
- Hurley, B.L.; McCreery, R.L. Covalent bonding of organic molecules to Cu and Al alloy 2024 T3 surfaces via diazonium ion reduction. J. Electrochem. Soc. 2004, 151, 252–259. [Google Scholar] [CrossRef] [Green Version]
- Podvorica, F.I.; Combellas, C.; Delamar, M.; Kanoufi, F.A.A.; Pinson, J. Spontaneous grafting of iron surfaces by reduction of aryldiazonium salts in acidic water: Applications to the inhibition of iron corrosion. In Passivation of Metals and Semiconductors, and Properties of Thin Oxide Layers; Elsevier Science: Amsterdam, The Netherlands, 2006; pp. 697–702. [Google Scholar] [CrossRef]
- Adenier, A.; Cabet-Deliry, E.; Chaussé, A.; Griveau, S.; Mercier, F.; Pinson, J.; Vautrin-Ul, C. Grafting of nitrophenyl groups on carbon and metallic surfaces without electrochemical induction. Chem. Mater. 2005, 17, 491–501. [Google Scholar] [CrossRef]
- Toupin, M.; Bélanger, D. Spontaneous Functionalization of Carbon Black by Reaction with 4-Nitrophenyldiazonium Cations. Langmuir 2008, 24, 1910–1917. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Lee, J.S.; Shin, H.-J.; Shin, H.S.; Choi, H.C. Spatially Resolved Spontaneous Reactivity of Diazonium Salt on Edge and Basal Plane of Graphene without Surfactant and Its Doping Effect. Langmuir 2010, 26, 12278–12284. [Google Scholar] [CrossRef] [PubMed]
- Bekyarova, E.; Itkis, M.E.; Ramesh, P.; Berger, C.; Sprinkle, M.; de Heer, W.A.; Haddon, R.C. Chemical Modification of Epitaxial Graphene: Spontaneous Grafting of Aryl Groups. J. Am. Chem. Soc. 2009, 131, 1336–1337. [Google Scholar] [CrossRef]
- Le Floch, F.; Simonato, J.P.; Bidan, G. Electrochemical signature of the grafting of diazonium salts: A probing parameter for monitoring the electro-addressed functionalization of devices. Electrochim. Acta 2009, 54, 3078–3085. [Google Scholar] [CrossRef]
- Raicopol, M.; Andronescu, C.; Atasiei, R.; Hanganu, A.; Vasile, E.; Brezoiu, A.M.; Pilan, L. Organic layers via aryl diazonium electrochemistry: Towards modifying platinum electrodes for interference free glucose biosensors. Electrochim. Acta 2016, 206, 226–237. [Google Scholar] [CrossRef]
- Raicopol, M.; Necula, L.; Ionita, M.; Pilan, L. Electrochemical reduction of aryl diazonium salts: A versatile way for carbon nanotubes functionalisation. Surf. Interface Anal. 2012, 44, 1081–1085. [Google Scholar] [CrossRef]
- Pilan, L.; Ungureanu, E.-M.; Bidan, G. Electrochemical grafting: A modern tool to achieve new materials used for recognition. Nonlinear Opt. Quantum Opt. 2009, 39. [Google Scholar]
- Raicopol, M.; Andronescu, C.; Atasiei, R.; Hanganu, A.; Pilan, L. Post-Polymerization Electrochemical Functionalization of a Conducting Polymer: Diazonium Salt Electroreduction at Polypyrrole Electrodes. J. Electrochem. Soc. 2014, 161, G103–G113. [Google Scholar] [CrossRef]
- Pinson, J.; Delamar, M.; Desbat, B.; Hitmi, R.; Savéant, J.-M.; Allongue, P.; Fagebaume, O. Covalent Modification of Carbon Surfaces by Aryl Radicals Generated from the Electrochemical Reduction of Diazonium Salts. J. Am. Chem. Soc. 1997, 119, 201–207. [Google Scholar] [CrossRef]
- Tahara, K.; Kubo, Y.; Lindner, B.; Hashimoto, S.; Hirose, S.; Brown, A.; Hirsch, B.; Daukiya, L.; De Feyter, S.; Tobe, Y. Steric and electronic effects of electrochemically generated aryl radicals on grafting of the graphite surface. Langmuir 2019, 35, 2089–2098. [Google Scholar] [CrossRef]
- Radtke, M.; Ignaszak, A. A Surface Grafting of Carbon Allotropes with in-situ Generated 3-aryl Diazonium Chlorides: Electrochemical Kinetic Studies. Electroanalysis 2016, 28, 2900–2909. [Google Scholar] [CrossRef]
- López, I.; Cesbron, M.; Levillain, E.; Breton, T. Diazonium Grafting Control through a Redox Cross-Reaction: Elucidation of the Mechanism Involved when using 2,2-Diphenylpicrylhydrazyl as an Inhibitor. ChemElectroChem 2018, 5, 1197–1202. [Google Scholar] [CrossRef]
- Nielsen, L.T.; Vase, K.H.; Dong, M.; Besenbacher, F.; Pedersen, S.U.; Daasbjerg, K. Electrochemical approach for constructing a monolayer of thiophenolates from grafted multilayers of diaryl disulfides. J. Am. Chem. Soc. 2007, 129, 1888–1889. [Google Scholar] [CrossRef]
- Peng, Z.; Holm, A.H.; Nielsen, L.T.; Pedersen, S.U.; Daasbjerg, K. Covalent sidewall functionalization of carbon nanotubes by a “Formation—Degradation” approach. Chem. Mater. 2008, 20, 6068–6075. [Google Scholar] [CrossRef]
- Leroux, Y.R.; Hapiot, P. Nanostructured monolayers on carbon substrates prepared by electrografting of protected aryldiazonium salts. Chem. Mater. 2013, 25, 489–495. [Google Scholar] [CrossRef]
- Wang, R.-C.C.; Yin, T.-L.L.; Wei, P.-J.J.; Liu, J.-G.G. A copper complex covalently grafted on carbon nanotubes and reduced graphene oxide promotes oxygen reduction reaction activity and catalyst stability. RSC Adv. 2015, 5, 66487–66493. [Google Scholar] [CrossRef]
- Mishyn, V.; Aspermair, P.; Leroux, Y.; Happy, H.; Knoll, W.; Boukherroub, R.; Szunerits, S. “Click” Chemistry on Gold Electrodes Modified with Reduced Graphene Oxide by Electrophoretic Deposition. Surfaces 2019, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Menanteau, T.; Levillain, E.; Breton, T. Electrografting via diazonium chemistry: From multilayer to monolayer using radical scavenger. Chem. Mater. 2013, 25, 2905–2909. [Google Scholar] [CrossRef] [Green Version]
- Menanteau, T.; Levillain, E.; Downard, A.J.; Breton, T. Evidence of monolayer formation via diazonium grafting with a radical scavenger: Electrochemical, AFM and XPS monitoring. Phys. Chem. Chem. Phys. 2015, 17, 13137–13142. [Google Scholar] [CrossRef]
- Leroux, Y.R.; Hui, F.; Noel, J.-M.; Roux, C.; Downard, A.J.; Hapiot, P. Design of Robust Binary Film onto Carbon Surface Using Diazonium Electrochemistry. Langmuir 2011, 27, 11222–11228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Vilà, N.; Walcarius, A.; Etienne, M. Molecular and Biological Catalysts Coimmobilization on Electrode by Combining Diazonium Electrografting and Sequential Click Chemistry. ChemElectroChem 2018, 5, 2208–2217. [Google Scholar] [CrossRef]
- Louault, C.; D’Amours, M.; Bélanger, D. The electrochemical grafting of a mixture of substituted phenyl groups at a glassy carbon electrode surface. ChemPhysChem 2008, 9, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Zhang, Y.; Jiang, C.; Qi, M.; Liu, G. Advances on Aryldiazonium Salt Chemistry Based Interfacial Fabrication for Sensing Applications. ACS Appl. Mater. Interfaces 2017, 9, 5031–5049. [Google Scholar] [CrossRef] [PubMed]
- Flavel, B.S.; Gross, A.J.; Garrett, D.J.; Nock, V.; Downard, A.J. A simple approach to patterned protein immobilization on silicon via electrografting from diazonium salt solutions. ACS Appl. Mater. Interfaces 2010, 2, 1184–1190. [Google Scholar] [CrossRef]
- Kuo, T.M.; Shen, M.Y.; Huang, S.Y.; Li, Y.K.; Chuang, M.C. Facile Fabrication of a Sensor with a Bifunctional Interface for Logic Analysis of the New Delhi Metallo-β-Lactamase (NDM)-Coding Gene. ACS Sens. 2016, 1, 124–130. [Google Scholar] [CrossRef]
- Santos, L.; Ghilane, J.; Lacroix, J.C. Surface patterning based on nanosphere lithography and electroreduction of in situ generated diazonium cation. Electrochem. Commun. 2012, 18, 20–23. [Google Scholar] [CrossRef]
- Ruffien, A.; Dequaire, M.; Brossier, P. Covalent immobilization of oligonucleotides on p-aminophenyl-modified carbon screen-printed electrodes for viral DNA sensing. Chem. Commun. 2003, 912–913. [Google Scholar] [CrossRef]
- Revenga-Parra, M.; García-Mendiola, T.; González-Costas, J.; González-Romero, E.; Marín, A.G.; Pau, J.L.; Pariente, F.; Lorenzo, E. Simple diazonium chemistry to develop specific gene sensing platforms. Anal. Chim. Acta 2014, 813, 41–47. [Google Scholar] [CrossRef]
- Bartolome, J.P.; Echegoyen, L.; Fragoso, A. Reactive Carbon Nano-Onion Modified Glassy Carbon Surfaces as DNA Sensors for Human Papillomavirus Oncogene Detection with Enhanced Sensitivity. Anal. Chem. 2015, 87, 6744–6751. [Google Scholar] [CrossRef]
- Perréard, C.; Ladner, Y.; D’Orlyé, F.; Descroix, S.; Taniga, V.; Varenne, A.; Kanoufi, F.; Slim, C.; Griveau, S.; Bedioui, F. Electrochemically assisted micro localized grafting of aptamers in a microchannel engraved in fluorinated thermoplastic polymer Dyneon THV. RSC Adv. 2015, 5, 11128–11131. [Google Scholar] [CrossRef]
- Liu, Y.; Tuleouva, N.; Ramanculov, E.; Revzin, A. Aptamer-based electrochemical biosensor for interferon gamma detection. Anal. Chem. 2010, 82, 8131–8136. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Li, H.; Wang, W.; Cao, J.; Gan, N.; Han, H. Application of Multiplexed Aptasensors in Food Contaminants Detection. ACS Sens. 2020, 5, 3721–3738. [Google Scholar] [CrossRef]
- Shen, Z.; Huang, J.; Wei, H.; Niu, H.; Li, B.; Li, R.; Liu, G. Validation of an in vivo electrochemical immunosensing platform for simultaneous detection of multiple cytokines in Parkinson’s disease mice model. Bioelectrochemistry 2020, 134, 107532. [Google Scholar] [CrossRef]
- Martínez-García, G.; Agüí, L.; Yáñez-Sedeño, P.; Pingarrón, J.M. Multiplexed electrochemical immunosensing of obesity-related hormones at grafted graphene-modified electrodes. Electrochim. Acta 2016, 202, 209–215. [Google Scholar] [CrossRef]
- Gautier, C.; López, I.; Breton, T. A post-functionalization toolbox for diazonium (electro)-grafted surfaces: Review of the coupling methods. Mater. Adv. 2021. [Google Scholar] [CrossRef]
- Harper, J.C.; Polsky, R.; Wheeler, D.R.; Dirk, S.M.; Brozik, S.M. Selective immobilization of DNA and antibody probes on electrode arrays: Simultaneous electrochemical detection of DNA and protein on a single platform. Langmuir 2007, 23, 8285–8287. [Google Scholar] [CrossRef] [PubMed]
- Ott, C.; Raicopol, M.; Andronescu, C.; Vasile, E.; Hanganu, A.; Pruna, A.; Pilan, L. Functionalized polypyrrole/sulfonated graphene nanocomposites: Improved biosensing platforms through aryl diazonium electrochemistry. Synth. Met. 2018, 235, 20–28. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, G.; Hein, R.; Liu, N.; Luo, X.; Davis, J.J. Antifouling Strategies for Selective in Vitro and in Vivo Sensing. Chem. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gui, A.L.; Luais, E.; Peterson, J.R.; Gooding, J.J. Zwitterionic phenyl layers: Finally, stable, anti-biofouling coatings that do not passivate electrodes. ACS Appl. Mater. Interfaces 2013, 5, 4827–4835. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Alam, M.T.; Silva, S.M.; Taufik, S.; Fan, S.; Gooding, J.J. Unique Sensing Interface That Allows the Development of an Electrochemical Immunosensor for the Detection of Tumor Necrosis Factor α in Whole Blood. ACS Sens. 2016, 1, 1432–1438. [Google Scholar] [CrossRef]
- Guo, L.; Ma, L.; Zhang, Y.; Cheng, X.; Xu, Y.; Wang, J.; Wang, E.; Peng, Z. Spectroscopic Identification of the Au-C Bond Formation upon Electroreduction of an Aryl Diazonium Salt on Gold. Langmuir 2016, 32, 11514–11519. [Google Scholar] [CrossRef]
- Liu, G.; Böcking, T.; Gooding, J.J. Diazonium salts: Stable monolayers on gold electrodes for sensing applications. J. Electroanal. Chem. 2007, 600, 335–344. [Google Scholar] [CrossRef]
- Civit, L.; Fragoso, A.; O’Sullivan, C.K. Thermal stability of diazonium derived and thiol-derived layers on gold for application in genosensors. Electrochem. Commun. 2010, 12, 1045–1048. [Google Scholar] [CrossRef]
- Jia, X.; Dong, S.; Wang, E. Engineering the bioelectrochemical interface using functional nanomaterials and microchip technique toward sensitive and portable electrochemical biosensors. Biosens. Bioelectron. 2016, 76, 80–90. [Google Scholar] [CrossRef]
- Wu, L.; Xiong, E.; Zhang, X.; Zhang, X.; Chen, J. Nanomaterials as signal amplification elements in DNA-based electrochemical sensing. Nano Today 2014, 9, 197–211. [Google Scholar] [CrossRef]
- Arias De Fuentes, O.; Ferri, T.; Frasconi, M.; Paolini, V.; Santucci, R. Highly-Ordered Covalent Anchoring of Carbon Nanotubes on Electrode Surfaces by Diazonium Salt Reactions. Angew. Chem. Int. Ed. 2011, 50, 3457–3461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayanan, J.; Sharma, M.K.; Ponmariappan, S.; Shaik, M.; Upadhyay, S. Electrochemical immunosensor for botulinum neurotoxin type-E using covalently ordered graphene nanosheets modified electrodes and gold nanoparticles-enzyme conjugate. Biosens. Bioelectron. 2015, 69, 249–256. [Google Scholar] [CrossRef]
- Liu, G.; Qi, M.; Zhang, Y.; Cao, C.; Goldys, E.M. Nanocomposites of gold nanoparticles and graphene oxide towards an stable label-free electrochemical immunosensor for detection of cardiac marker troponin-I. Anal. Chim. Acta 2016, 909, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kesavan, S.; Gowthaman, N.S.K.; Alwarappan, S.; John, S.A. Real time detection of adenosine and theophylline in urine and blood samples using graphene modified electrode. Sens. Actuators B Chem. 2019, 278, 46–54. [Google Scholar] [CrossRef]
- Hicks, J.M.; Wong, Z.Y.; Scurr, D.J.; Silman, N.; Jackson, S.K.; Mendes, P.M.; Aylott, J.W.; Rawson, F.J. Tailoring the Electrochemical Properties of Carbon Nanotube Modified Indium Tin Oxide via in Situ Grafting of Aryl Diazonium. Langmuir 2017, 33, 4924–4933. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ge, L.; Sun, X.; Hou, T.; Li, F. Graphene-Assisted Label-Free Homogeneous Electrochemical Biosensing Strategy based on Aptamer-Switched Bidirectional DNA Polymerization. ACS Appl. Mater. Interfaces 2015, 7, 28566–28575. [Google Scholar] [CrossRef]
- Lian, Y.; He, F.; Wang, H.; Tong, F. A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus. Biosens. Bioelectron. 2015, 65, 314–319. [Google Scholar] [CrossRef]
- Yang, L.; Xu, Y.; Wang, X.; Zhu, J.; Zhang, R.; He, P.; Fang, Y. The application of beta-cyclodextrin derivative functionalized aligned carbon nanotubes for electrochemically DNA sensing via host – guest recognition. Anal. Chim. Acta 2011, 689, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rawson, F.J.; Yeung, C.L.; Jackson, S.K.; Mendes, P.M. Tailoring 3D single-walled carbon nanotubes anchored to indium tin oxide for natural cellular uptake and intracellular sensing. Nano Lett. 2013, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Shadman, S.M.; Daneshi, M.; Shafiei, F.; Azimimehr, M.; Khorasgani, M.R.; Sadeghian, M.; Motaghi, H.; Mehrgardi, M.A. Aptamer-Based Electrochemical Biosensors; Elsevier Inc.: Amsterdam, The Netherlands, 2019; ISBN 9780128164914. [Google Scholar]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-based biosensors for antibiotic detection: A review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- El Alami El Hassani, N.; Baraket, A.; Boudjaoui, S.; Taveira Tenório Neto, E.; Bausells, J.; El Bari, N.; Bouchikhi, B.; Elaissari, A.; Errachid, A.; Zine, N. Development and application of a novel electrochemical immunosensor for tetracycline screening in honey using a fully integrated electrochemical Bio-MEMS. Biosens. Bioelectron. 2019, 130, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Vega, D.; Agüí, L.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Voltammetry and amperometric detection of tetracyclines at multi-wall carbon nanotube modified electrodes. Anal. Bioanal. Chem. 2007, 389, 951–958. [Google Scholar] [CrossRef]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 1–22. [Google Scholar] [CrossRef]
- Jampasa, S.; Pummoree, J.; Siangproh, W.; Khongchareonporn, N.; Ngamrojanavanich, N.; Chailapakul, O.; Chaiyo, S. “Signal-On” electrochemical biosensor based on a competitive immunoassay format for the sensitive determination of oxytetracycline. Sens. Actuators B Chem. 2020, 320, 128389. [Google Scholar] [CrossRef]
- Niazi, J.H.; Lee, S.J.; Kim, Y.S.; Gu, M.B. ssDNA aptamers that selectively bind oxytetracycline. Bioorg. Med. Chem. 2008, 16, 1254–1261. [Google Scholar] [CrossRef]
- Kulikova, T.; Gorbatchuk, V.; Stoikov, I.; Rogov, A.; Evtugyn, G.; Hianik, T. Impedimetric determination of kanamycin in milk with aptasensor based on carbon black-oligolactide composite. Sensors 2020, 20, 4738. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, X.; Sun, X.; Guo, Y.; Zhao, W. A dual-signal amplification strategy for kanamycin based on ordered mesoporous carbon-chitosan/gold nanoparticles-streptavidin and ferrocene labelled DNA. Anal. Chim. Acta 2018, 1033, 185–192. [Google Scholar] [CrossRef]
- Yu, Z.; Cui, P.; Xiang, Y.; Li, B.; Han, X.; Shi, W.; Yan, H.; Zhang, G. Developing a fast electrochemical aptasensor method for the quantitative detection of penicillin G residue in milk with high sensitivity and good anti-fouling ability. Microchem. J. 2020, 157, 105077. [Google Scholar] [CrossRef]
- Jalili, R.; Khataee, A.; Rashidi, M.R.; Razmjou, A. Detection of penicillin G residues in milk based on dual-emission carbon dots and molecularly imprinted polymers. Food Chem. 2020, 314, 126172. [Google Scholar] [CrossRef]
- Wen, T.; Wang, M.; Luo, M.; Yu, N.; Xiong, H.; Peng, H. A nanowell-based molecularly imprinted electrochemical sensor for highly sensitive and selective detection of 17β-estradiol in food samples. Food Chem. 2019, 297, 124968. [Google Scholar] [CrossRef]
- Qiao, L.; Wang, H.; He, J.; Yang, S.; Chen, A. Truncated affinity-improved aptamers for 17β-estradiol determination by AuNPs-based colorimetric aptasensor. Food Chem. 2021, 340, 128181. [Google Scholar] [CrossRef]
- Anfossi, L.; Giovannoli, C.; Baggiani, C. Mycotoxin detection. Curr. Opin. Biotechnol. 2016, 37, 120–126. [Google Scholar] [CrossRef]
- Evtugyn, G.; Hianik, T. Electrochemical immuno- and aptasensors for mycotoxin determination. Chemosensors 2019, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Tang, D. Recent advances in photoelectrochemical biosensors for analysis of mycotoxins in food. TrAC Trends Anal. Chem. 2020, 124, 115814. [Google Scholar] [CrossRef]
- Nan, M.N.; Bi, Y.; Xue, H.L.; Long, H.T.; Xue, S.L.; Pu, L.M.; Prusky, D. Modification performance and electrochemical characteristics of different groups of modified aptamers applied for label-free electrochemical impedimetric sensors. Food Chem. 2021, 337. [Google Scholar] [CrossRef]
- Li, Z.; Mohamed, M.A.; Vinu Mohan, A.M.; Zhu, Z.; Sharma, V.; Mishra, G.K.; Mishra, R.K. Application of electrochemical aptasensors toward clinical diagnostics, food, and environmental monitoring: Review. Sensors 2019, 19, 5435. [Google Scholar] [CrossRef] [Green Version]
- De Girolamo, A.; McKeague, M.; Miller, J.D.; Derosa, M.C.; Visconti, A. Determination of ochratoxin A in wheat after clean-up through a DNA aptamer-based solid phase extraction column. Food Chem. 2011, 127, 1378–1384. [Google Scholar] [CrossRef]
- Williams, C.D.; Jaeschke, H. Liver Toxicology. In Encyclopedia of Environmental Health; Nriagu, J.O., Ed.; Elsevier: Burlington, VT, USA, 2011; pp. 509–514. ISBN 978-0-444-52272-6. [Google Scholar]
- Zhang, Y.; Huang, Y.; Yue, Z.; Fan, H.; Wu, S. Preparation and application of aptamer-functionalized sorbent for the analysis of ultra-trace Aflatoxin M1 and analogues in milk. Microchem. J. 2021, 106179. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, E.; Jin, Z.; Irudayaraj, J. An ultrasensitive aptasensor based on fluorescent resonant energy transfer and exonuclease-assisted target recycling for patulin detection. Food Chem. 2018, 249, 136–142. [Google Scholar] [CrossRef]
- He, B.; Lu, X. An electrochemical aptasensor based on tetrahedral DNA nanostructures as a signal probe carrier platform for sensitive detection of patulin. Anal. Chim. Acta 2020, 1138, 123–131. [Google Scholar] [CrossRef]
- Liburdi, K.; Benucci, I.; Esti, M. Lysozyme in Wine: An Overview of Current and Future Applications. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1062–1073. [Google Scholar] [CrossRef]
- Chen, K.; Han, S.Y.; Zhang, B.; Li, M.; Sheng, W.J. Development of lysozyme-combined antibacterial system to reduce sulfur dioxide and to stabilize italian Riesling ice wine during aging process. Food Sci. Nutr. 2015, 3, 453–465. [Google Scholar] [CrossRef]
- Panel, E.; Nda, A. Scientific Opinion related to a notification from the Oenological Products and Practices International Association (OENOPPIA) on lysozyme from hen’s egg to be used in the manufacture of wine as an anti-microbial stabilizer/additive pursuant to Article 6. EFSA J. 2011, 9. [Google Scholar] [CrossRef] [Green Version]
- Luan, Y.; Lu, A.; Chen, J.; Fu, H.; Xu, L. A label-free aptamer-based fluorescent assay for cadmium detection. Appl. Sci. 2016, 6, 432. [Google Scholar] [CrossRef] [Green Version]
- Food Safety Authority of Ireland Mercury, Lead, Cadmium, Tin and Arsenic in Food. Toxicol. Factsheet Ser. 2009, 1, 1–13.
- Air Quality Guidelines—Second Edition, Chapter 6.3 Cadmium; Environment Health Criteria World Health Organization Regional Office Europe: Copenhagen, Denmark, 2000; pp. 1–11.
- Tang, C.X.; Zhao, Y.; He, X.W.; Yin, X.B. A “turn-on” electrochemiluminescent biosensor for detecting Hg2+ at femtomole level based on the intercalation of Ru(phen)32+ into ds-DNA. Chem. Commun. 2010, 46, 9022–9024. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, N.; Kumari, A.; Lee, H.J.; Kim, T.Y.; Tripathi, K.M. Nano-carbon based sensors for bacterial detection and discrimination in clinical diagnosis: A junction between material science and biology. Appl. Mater. Today 2020, 18, 100467. [Google Scholar] [CrossRef]
- Majdinasab, M.; Hayat, A.; Marty, J.L. Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. TrAC Trends Anal. Chem. 2018, 107, 60–77. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, D.; Zhou, X.; Yu, Y.; Liu, J.; Hu, N.; Wang, H.; Li, G.; Wu, Y. Recent progress in the construction of nanozyme-based biosensors and their applications to food safety assay. TrAC Trends Anal. Chem. 2019, 121, 1–22. [Google Scholar] [CrossRef]
- Phopin, K.; Tantimongcolwat, T. Pesticide aptasensors—State of the art and perspectives. Sensors 2020, 20, 6809. [Google Scholar] [CrossRef]
- Malekzad, H.; Jouyban, A.; Hasanzadeh, M.; Shadjou, N.; de la Guardia, M. Ensuring food safety using aptamer based assays: Electroanalytical approach. TrAC Trends Anal. Chem. 2017, 94, 77–94. [Google Scholar] [CrossRef]
- Song, S.H.; Gao, Z.F.; Guo, X.; Chen, G.H. Aptamer-Based Detection Methodology Studies in Food Safety. Food Anal. Methods 2019, 2019, 966–990. [Google Scholar] [CrossRef]
- Mishra, G.K.; Sharma, V.; Mishra, R.K. Electrochemical aptasensors for food and environmental safeguarding: A review. Biosensors 2018, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Rapini, R.; Marrazza, G. Electrochemical aptasensors for contaminants detection in food and environment: Recent advances. Bioelectrochemistry 2017, 118, 47–61. [Google Scholar] [CrossRef]
- Liu, M.; Khan, A.; Wang, Z.; Liu, Y.; Yang, G.; Deng, Y.; He, N. Aptasensors for pesticide detection. Biosens. Bioelectron. 2019, 130, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Farré, M.; Barceló, D. Microfluidic devices: Biosensors. In Chemical Analysis of Food; Elsevier Inc.: Burlington, VT, USA, 2020; pp. 287–351. ISBN 9780128132661. [Google Scholar]
- Moro, G.; De Wael, K.; Moretto, L.M. Challenges in the electrochemical (bio)sensing of nonelectroactive food and environmental contaminants. Curr. Opin. Electrochem. 2019, 16, 57–65. [Google Scholar] [CrossRef]
- Han, K.; Liu, T.; Wang, Y.; Miao, P. Electrochemical aptasensors for detection of small molecules, macromolecules, and cells. Rev. Anal. Chem. 2016, 35, 201–211. [Google Scholar] [CrossRef]

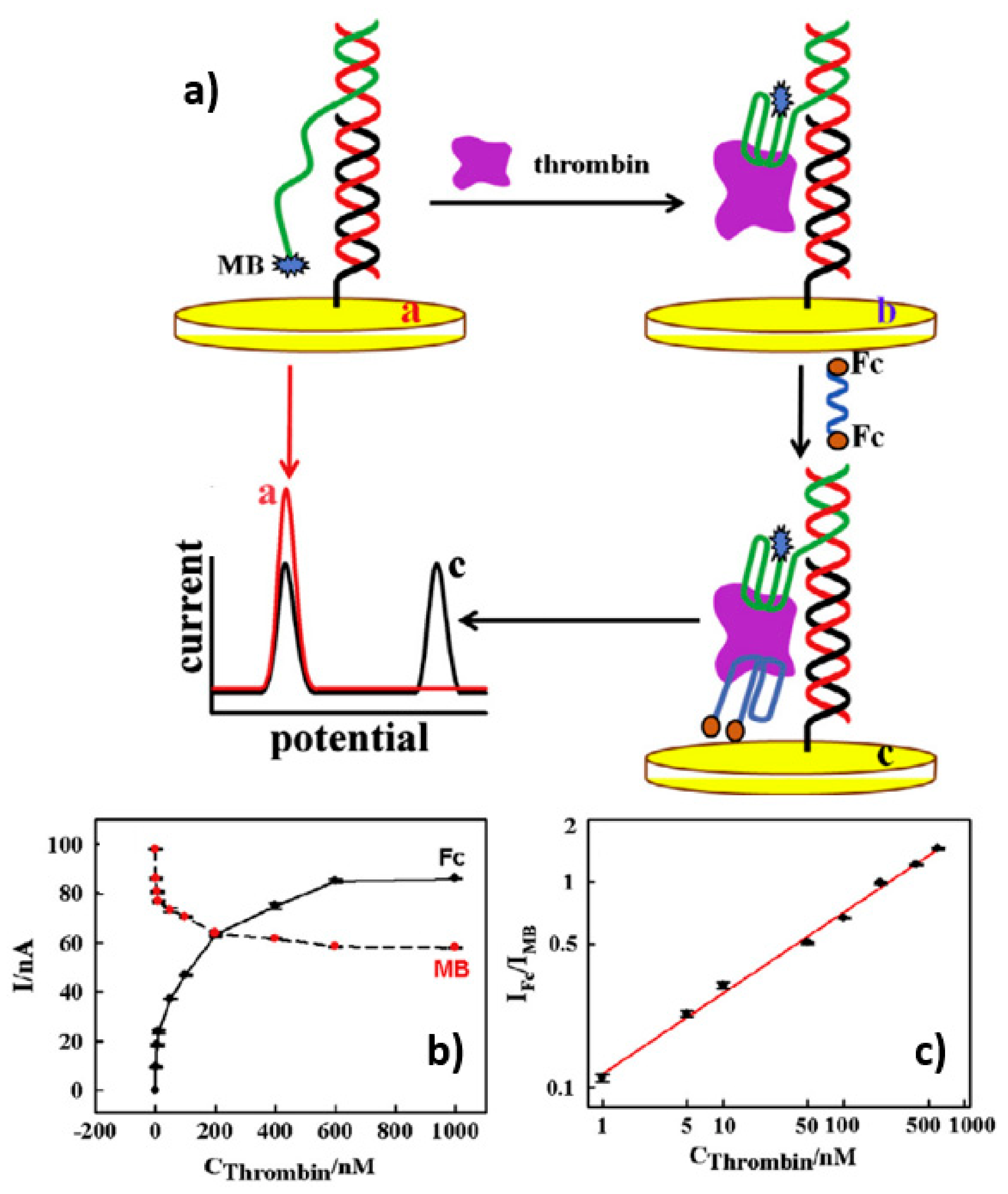

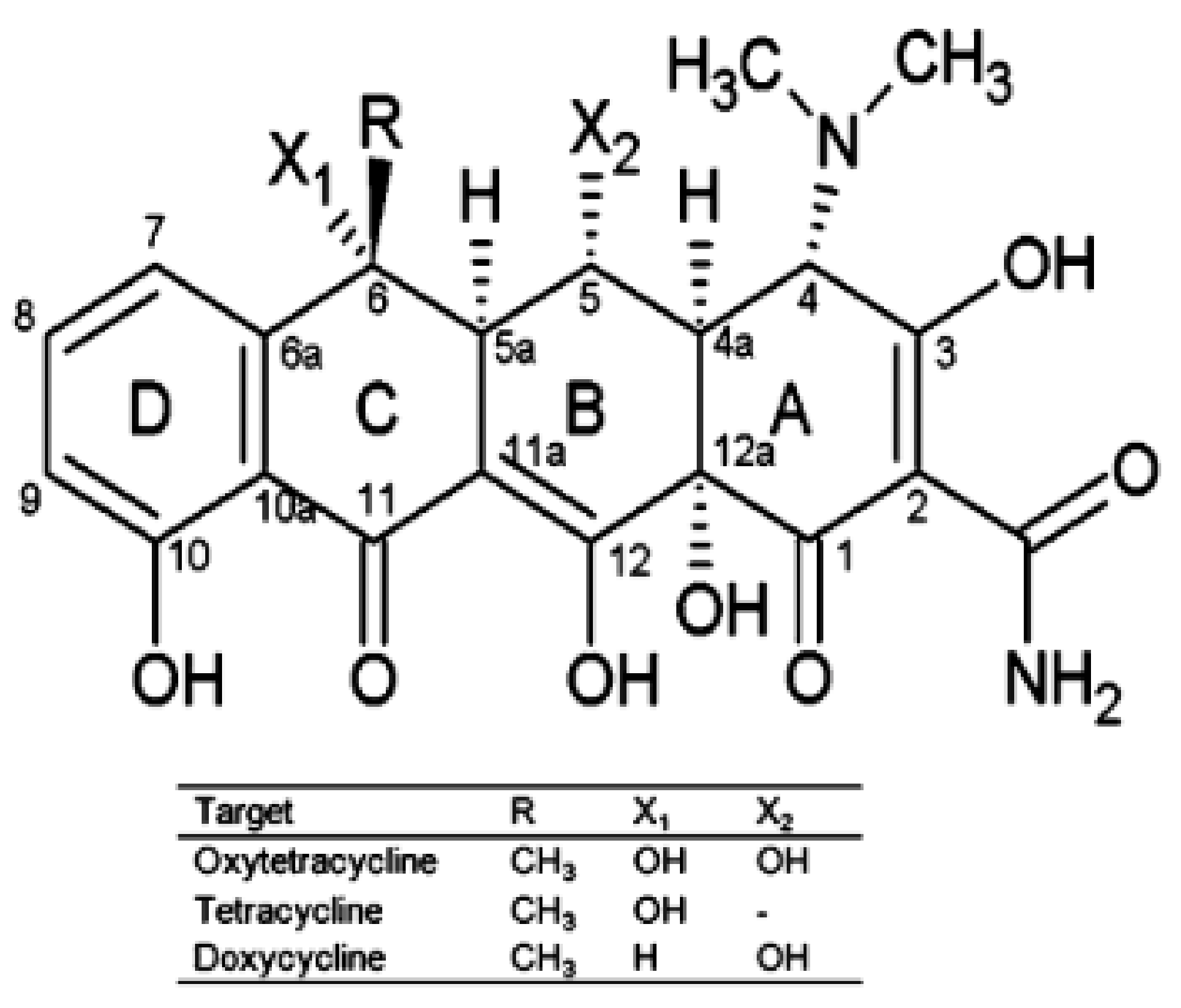
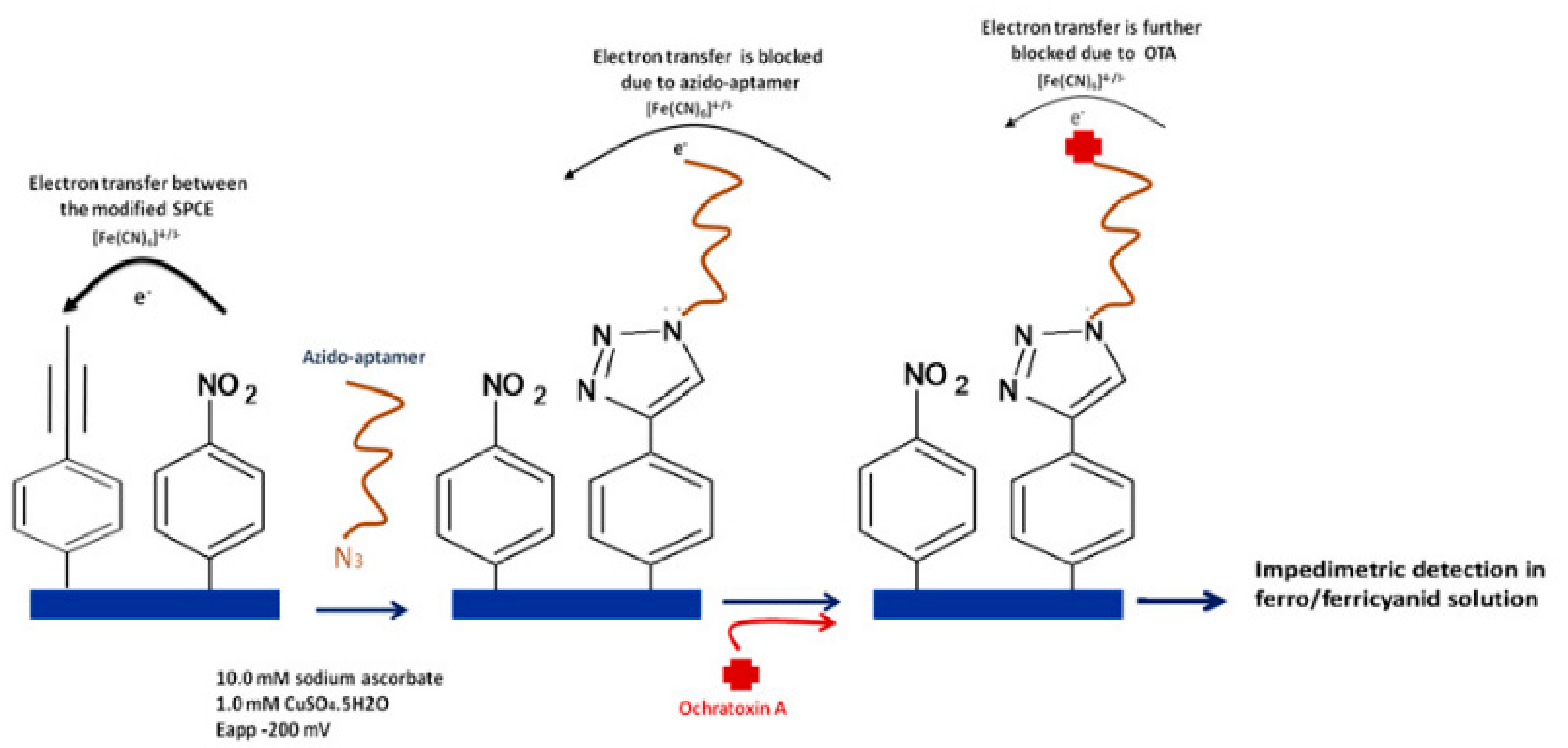

| Detected Analyte | Reference (Aptasensor) | Aptamer Sequence | Kd | Reference (SELEX Aptamer) |
|---|---|---|---|---|
| TET | [25,59] | 5′- CGT ACG GAA TTC GCT AGC CCC CCG GCA GGC CAC GGC TTG GGT TGG TCC CAC TGC GCG TGG ATC CGA GCT CCA CGT G-3′ | 63 nM | [60] |
| OTC | [61] | 5′-GGA ATT CGC TAG CAC GTT GAC GCT GGT GCC CGG TTG TGG TGC GAG TGT TGT GTG GAT CCG AGC TCC ACG TG-3′ | 11.13 nM | [62] |
| Kan | [63] | 5′-TGG GGG TTG AGG CTA AGC CGA-3′ | 78.8 nM | [64] |
| Pen-G | [65] | 5′-GGG AGG ACG AAG CGG AAC GAG ATG TAG ATG AGG CTC GAT CCG AAT GCG TGA CGT CTA TCG GAA TAC TCG TTT TTA CGC CTC AGA AGA CAC GCC CGA CA-3′ | - | [65] |
| E2 | [66] | 5′-GCT TCC AGC TTA TTG AAT TAC ACG CAG AGG GTA GCG GCT CTG CGC ATT CAA TTG CTG CGC GCT GAA GCG CGG AAG C-3′ (truncated to 38 mer) | 2.7 nM | [67] |
| OTA | [40,68,69,70] | 5′-GAT CGG GTG TGG GTG GCG TAA AGG GAG CAT CGG ACA-3′ | 0.2 μM | [71] |
| AFB1 | [72] | 5′-GTT GGG CAC GTG TTG TCT CTC TGT GTC TCG TGC CCT TCG CTA GGC CCA CA-3′ | 75 nM | [73] |
| AFM1 | [74] | 5′-ACT GCT AGA GAT TTT CCA CAT-3′ | - | [75] |
| Pat | [76] | 5′-GGC CCG CCA ACC CGC ATC ATC TAC ACT GAT ATT TTA CCT T-3′ | 21.83 nM | [77] |
| LYS | [78,79] | 5′-NH2-GCA GCT AAG CAG GCG GCT CAC AAA ACC ATT CGC ATG CGG C-3′ | 2.8 nM | [80] |
| LYS | [78,81,82] | 5′-NH2-ATC AGG GCT AAA GAG TGC AGA GTT ACT TAG-3′ | 31 nM | [83] |
| LYS | [84] | 5′-GGG AAT GCA TCC ACA TCT ACG AAT TCA TCA GGG CTA AAG AG-3′ | 29 ± 5 nM | [85] |
| Cd2+ | [86] | 5′-ACC GAC CGT GCT GGA CTC TGG ACT GTT GTG GTA TTA TTT TTG GTT GTG CAG TAT GAG CGA GCG TTG CG-3′ | 34.5 nM | [87] |
| S. typhimurium | [88] | 5′-TTT GGT CCT TGT CTT ATG TCC AGA ATG CGA GGA AAG TCT ATA GCA GAG GAG ATG TGT GAA CCG AGT AAA TTT CTC CTA CTG GGA TAG GTG GAT TAT-3′ | - | [89] |
| Analyte | Substrate | Diazonium Salt/Antifouling Layer; Apt Coupling Method | Apt Selection Reference | Detection Method; Principle | LOD; Linearity Range | Real Sample; Recovery Rate | Reference |
|---|---|---|---|---|---|---|---|
| TET | SPCE | 4-carboxyphenyl; EDC-NHS; 5′-amino-Apt | [60] | CV; Apt intrinsic redox activity | 0.035 μg L−1; 0.05 μg L−1–20 μg L−1 | Natural waters; 87.8–97% | [25] |
| OTC | GC | 4-carboxyphenyl/ BSA; EDC-NHS; 5′-amino-Apt | [62] | DPV; [Fe(CN)6]3−/4− redox probe | 0.229 μg L−1; 1.0 μg L−1–1.0 × 105 μg L−1 | Spiked milk; 87.0–99.3% | [61] |
| TET | SPCE | 4-carboxyphenyl; EDC-NHS; 5′-amino-Apt | [60] | SECM; Apt intrinsic redox activity | - | - | [59] |
| Kan | SPCE | 4-carboxyphenyl/Casein; EDC-NHS; 3′-amino-Apt | [64] | EIS; [Fe(CN)6]3−/4− redox probe | 0.11 μg L−1; 1.2 μg L−1–75 μg L−1 | Spiked milk; 96.88–100.5% | [63] |
| Pen-G | SPCE | 4-nitrophenyl, electrochemically reduced to 4-aminophenyl/Casein; GA crosslinking; 5′-amino-Apt | [65] | EIS; [Fe(CN)6]3−/4− redox probe | 0.17 µg L−1; 0.4–1000 µg L−1 | Spiked milk; 83–100% | [65] |
| E2 | ERGO/GC | 4-carboxyphenyl; EDC-NHS; 5′-amino- Apt | [67] | SWV; E2 redox activity | 0.5 × 10−15 mol L−1; 1.0 10−15 mol L−1–9.0 10−12 mol L−1 and 1.2 10−11 mol L−1–2.3 × 10−10 mol L−1 | Waste waters: 94.05–100.47%; pharmaceutical samples: 96.87%–100.22% | [66] |
| OTA | SPCE | 4-ethynyl phenyl (protected), 4-nitrophenyl; “click” chemistry; 5′-azido- Apt | [71] | EIS; [Fe(CN)6]3−/4− redox probe | 0.25 μg L−1; 1.25 μg L−1–500 μg L−1 | Spiked beer samples | [40] |
| OTA | SPCE | 4-carboxyphenyl/ BSA; EDC-NHS; 5′-amino-Apt | [71] | EIS; [Fe(CN)6]3−/4− redox probe | 0.15 μg L−1; 0.15–2.5 μg L−1 | Spiked cocoa samples; 91–95% | [68] |
| OTA | SPCE | 4-carboxyphenyl/ BSA; EDC-NHS; 5′-amino-Apt | [71] | DPV; oxidation current of 1-naphthol (ALP enzyme, 1-NPP substrate) | 0.07 μg L−1; 0.15–5 μg L−1 | Spiked cocoa samples; 82.1–85% | [69] |
| OTA | PGE | 4-carboxyphenyl; EDC-NHS; 5′-amino- Apt | [71] | EIS; [Fe(CN)6]3−/4− redox probe | 0.1 μg L−1; 0.1–2 μg L−1 | Spiked beer samples; 93.4 ± 6.6% | [70] |
| AFB1 | SPCE | 4-carboxyphenyl/ BSA; EDC-NHS; 5′-amino- Apt | [73] (seq B) | EIS; [Fe(CN)6]3−/4− redox probe | Seq. A: 0.125 μg L−1; 0.25–16.0 μg L−1 and Seq. B: 0.25 μg L−1; 0.1–16.0 μg L−1 | Spiked beer and wine samples; 92–102% | [72] |
| AFM1 | GCE | 4-carboxyphenyl; EDC-NHS; -5′-amino- Apt or 5′-HEG-amino-Apt | [75] | EIS; [Fe(CN)6]3−/4− redox probe | 1.15 ng L−1, 0.02 μg L−1–1 μg L−1(for 5′-HEG-amino-Apt) | Spiked milk; 98%–114% | [74] |
| PAT | SPCE | 4-carboxyphenyl/ BSA; EDC-NHS + carboxy-amino PEG; 3′-amino-Apt | [77] | EIS; [Fe(CN)6]3−/4− redox probe | 2.8 ng L−1; 1–25 ng L−1 | Spiked fresh apple juice; up to 99% | [76] |
| LYS | SPCE | 4-carboxyphenyl/ BSA; EDC-NHS; 5′-amino-Apt | [80,83] | EIS; [Fe(CN)6]3−/4− redox probe | 25 nM; 0.025–0.8 μM for Apt [80] and 100 nM; 0.1–0.8. μM for Apt [83] | Spiked wine; 94.2–102% | [78] |
| LYS | GECE | 4-carboxyphenyl/ PEG; EDC-NHS; 5′-amino-C6-Apt | [83] | EIS; [Fe(CN)6]3−/4− redox probe | 1.67 µM; 1.67 µM–5 µM | Spiked wine; 77% | [81] |
| LYS | SPCE | 4-carboxyphenyl/ BSA; EDC-NHS; 5′-amino-Apt + LYS-Ab-Bio/Av-ALP (sandwich assay) | [80] | DPV; oxidation current of 1-naphthol (ALP enzyme, 1-NPP substrate) | 4.3 fmol L−1; 5 fmol L−1–5 nmol L−1 | Spiked wine; 95.2–102.0% | [79] |
| LYS | VA-NCNT | 4-carboxyphenyl; EDC-NHS + NeutrAv; 5′-Bio-T24-Apt | [83] | DPV; [Fe(CN)6]4− oxidation signal | 100 fM; 0.1–7 pM | Serum samples | [82] |
| LYS | RGO/PEI | 4-ethynylphenyl/ BSA; “click” chemistry; 5′-azido-Apt | [85] | DPV; [Fe(CN)6]4− oxidation signal | 1 pM; 1–15 pM | Serum samples | [84] |
| Cadmium | Au SPE | 4-carboxyphenyl; EDC-NHS; 5′-amino-Apt | [87] | EIS; [Fe(CN)6]3−/4− redox probe | 2.75 × 10−10 mol L−1; 10−3–10−9 mol L−1 | Real water samples | [86] |
| S. typhimurium | 4-carboxyphenyl; EDC-NHS; 5′-amino-Apt | [89] | EIS; [Fe(CN)6]3−/4− redox probe | 6 CFU mL−1; 1 × 101–1 × 108 CFU mL−1 | Spiked apple juice | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raicopol, M.; Pilan, L. The Role of Aryldiazonium Chemistry in Designing Electrochemical Aptasensors for the Detection of Food Contaminants. Materials 2021, 14, 3857. https://doi.org/10.3390/ma14143857
Raicopol M, Pilan L. The Role of Aryldiazonium Chemistry in Designing Electrochemical Aptasensors for the Detection of Food Contaminants. Materials. 2021; 14(14):3857. https://doi.org/10.3390/ma14143857
Chicago/Turabian StyleRaicopol, Matei, and Luisa Pilan. 2021. "The Role of Aryldiazonium Chemistry in Designing Electrochemical Aptasensors for the Detection of Food Contaminants" Materials 14, no. 14: 3857. https://doi.org/10.3390/ma14143857
APA StyleRaicopol, M., & Pilan, L. (2021). The Role of Aryldiazonium Chemistry in Designing Electrochemical Aptasensors for the Detection of Food Contaminants. Materials, 14(14), 3857. https://doi.org/10.3390/ma14143857






