Pillared Graphene Structures Supported by Vertically Aligned Carbon Nanotubes as the Potential Recognition Element for DNA Biosensors
Abstract
:1. Introduction
2. Methods and Results
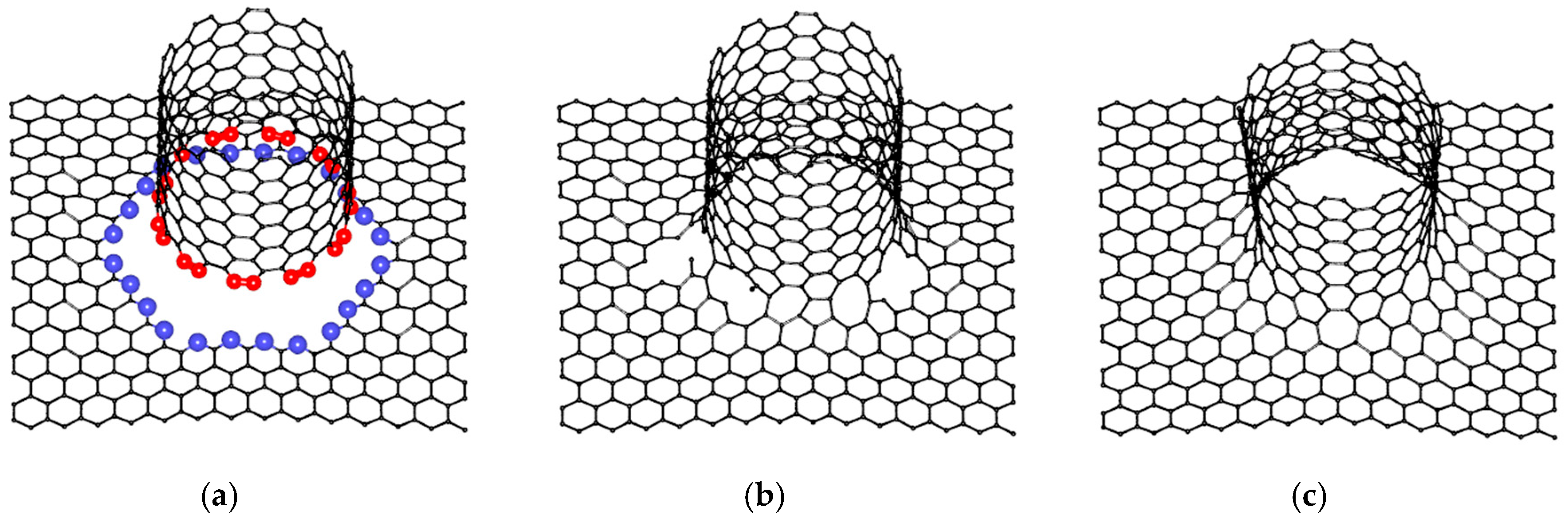
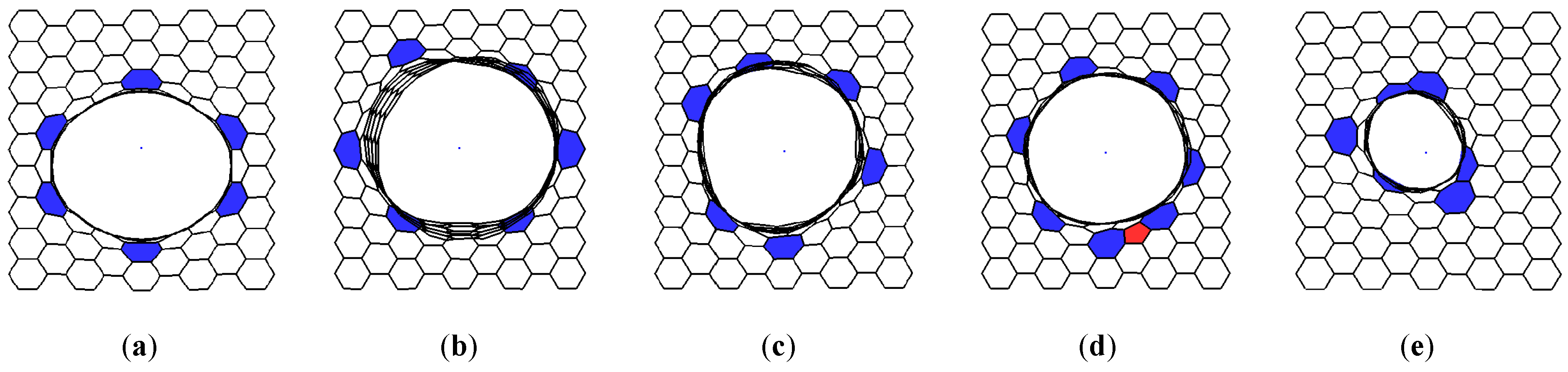
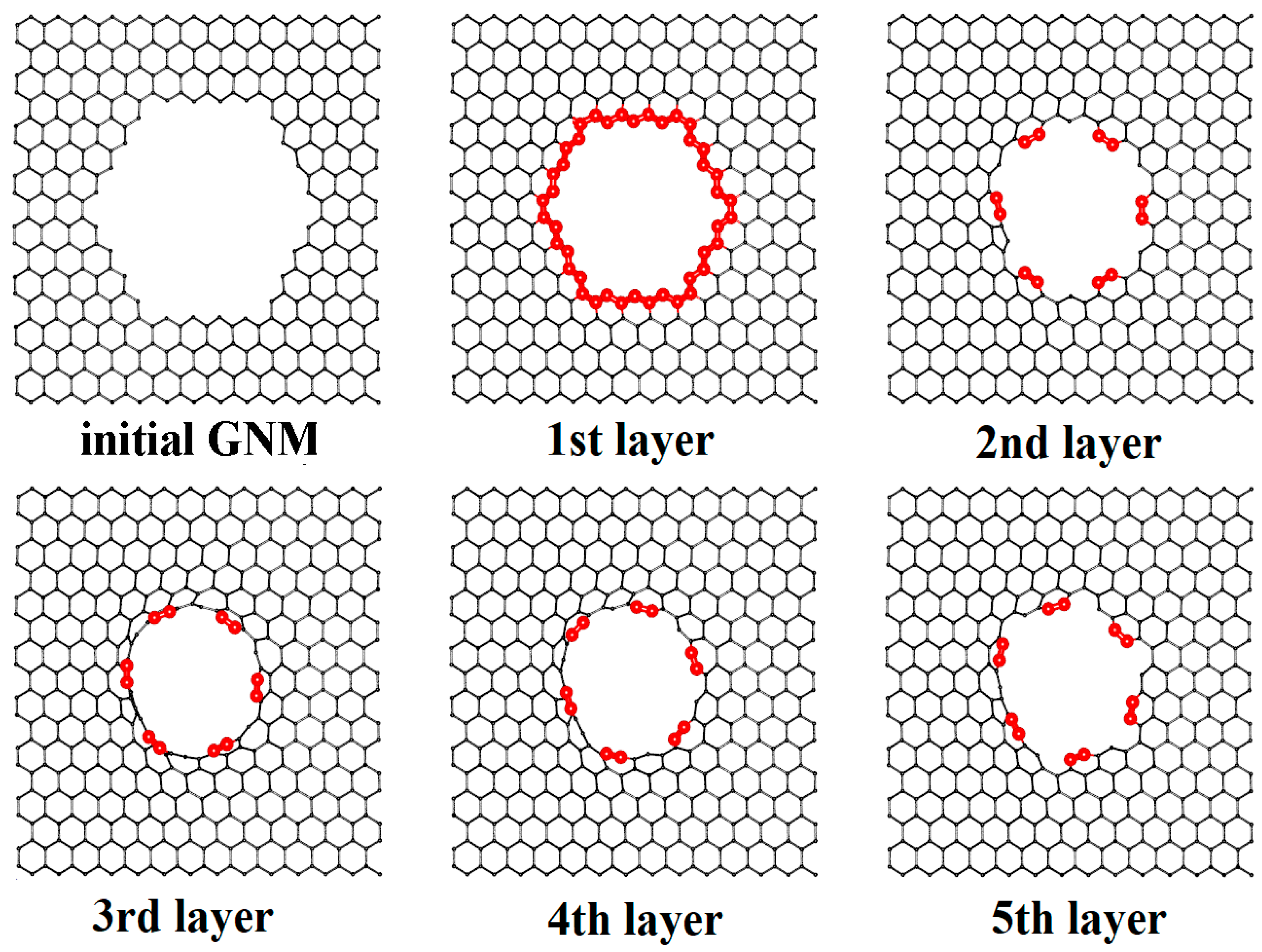
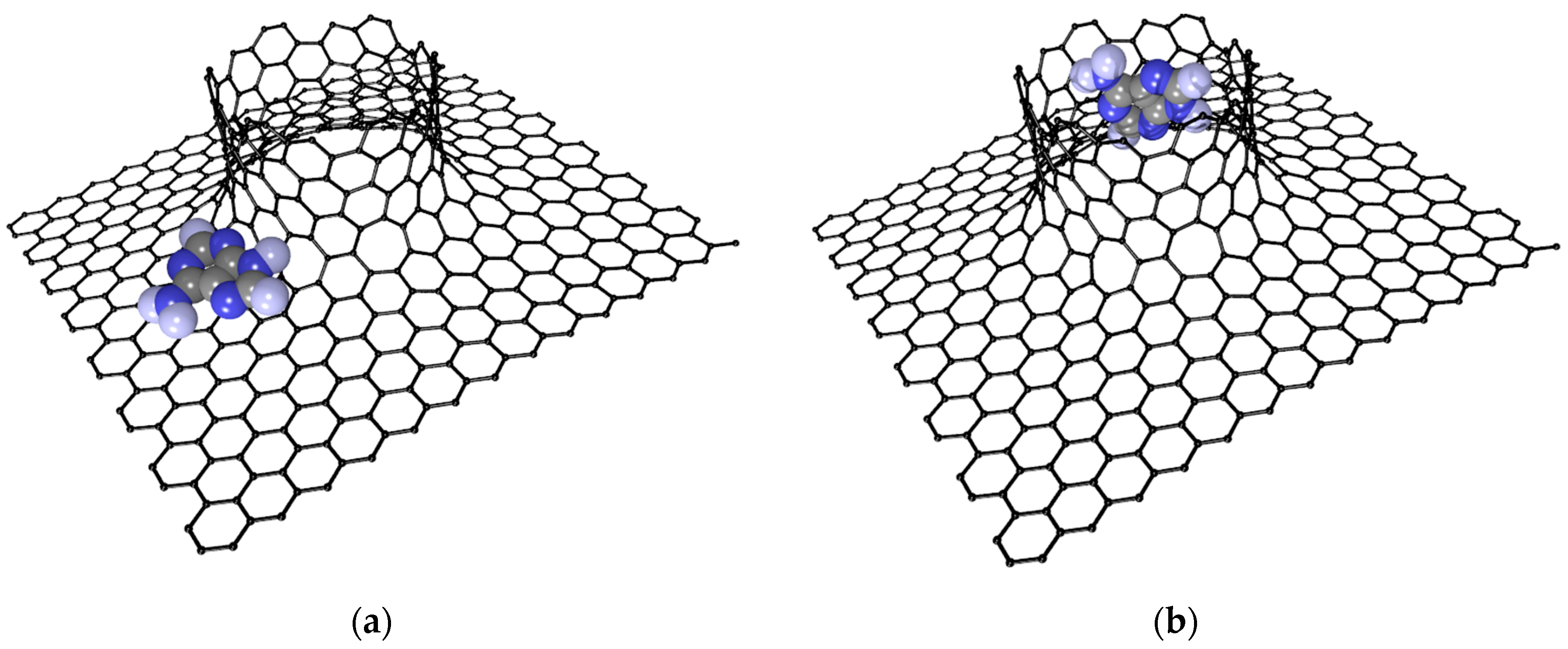
2.1. Construction of VACNT-Graphene Atomic Supercells
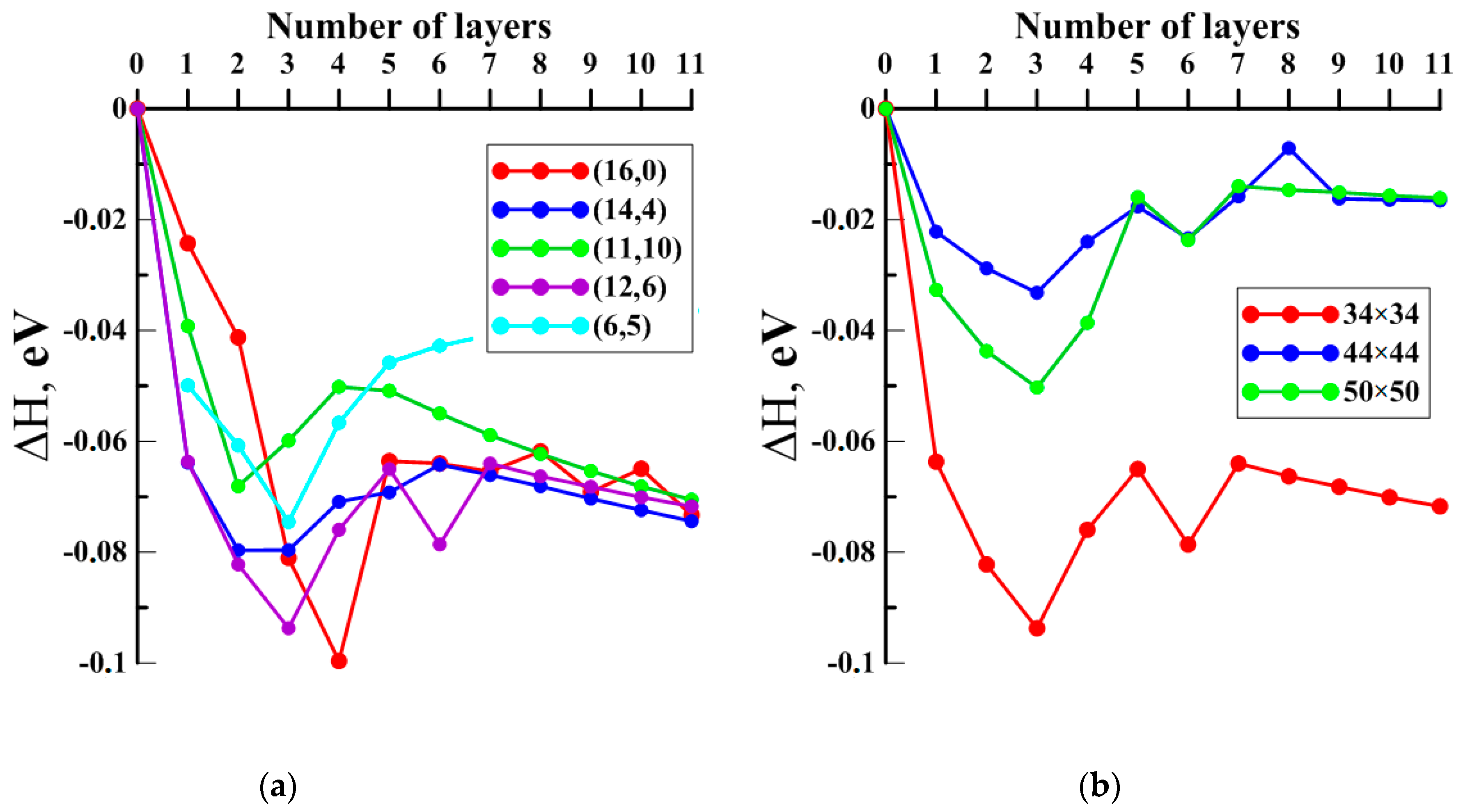
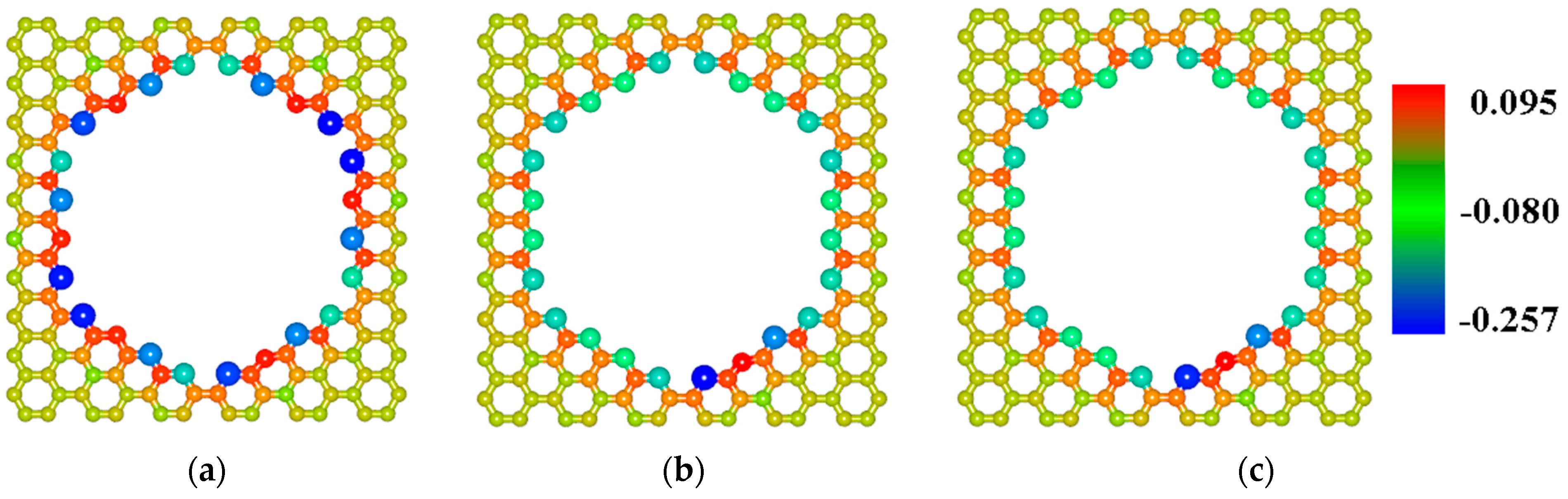
2.2. Finding the Optimal Sizes of the VACNT(12,6)-Graphene Supercell
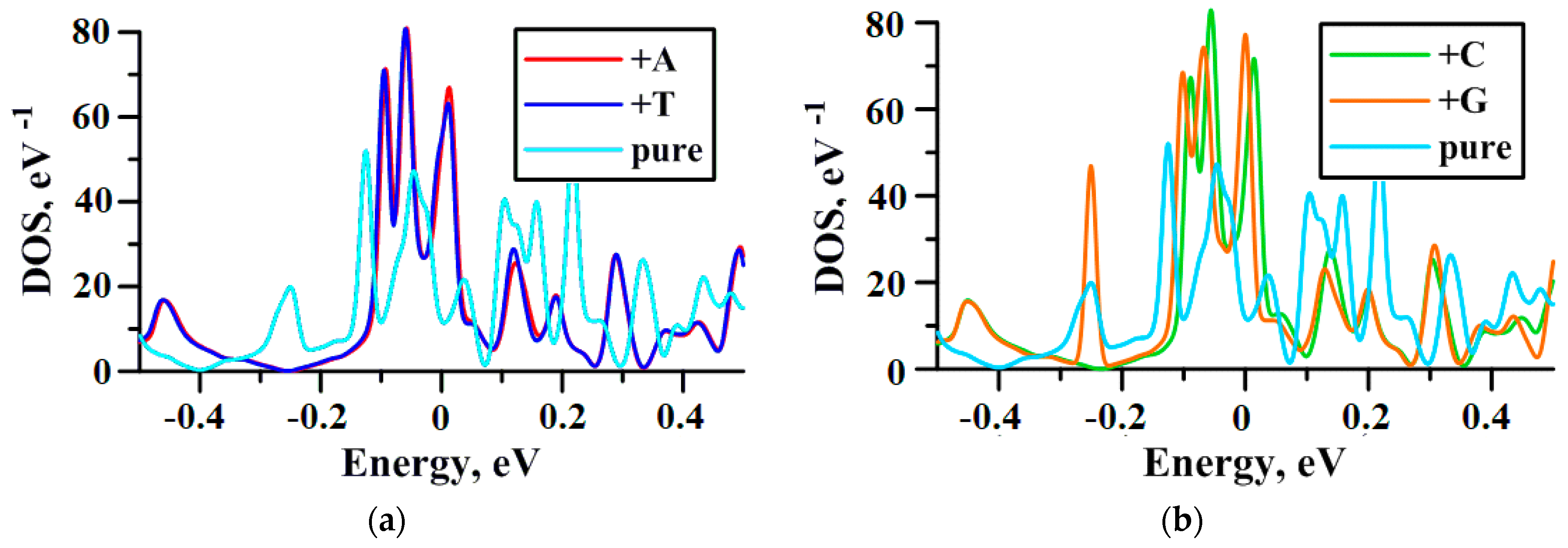
2.3. Influence of DNA Nitrogenous Bases on Electric Properties of the VACNT(12,6)/Graphene
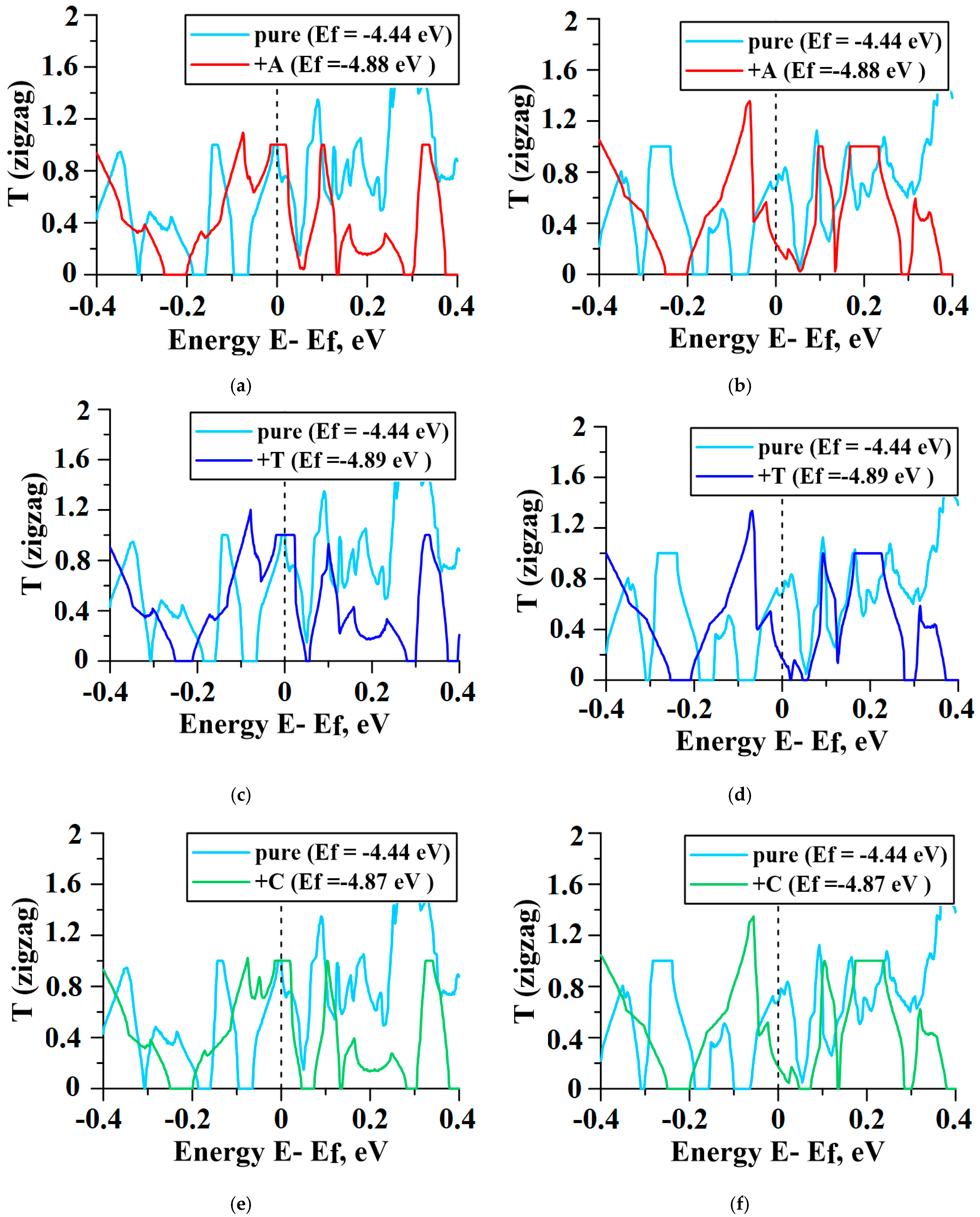
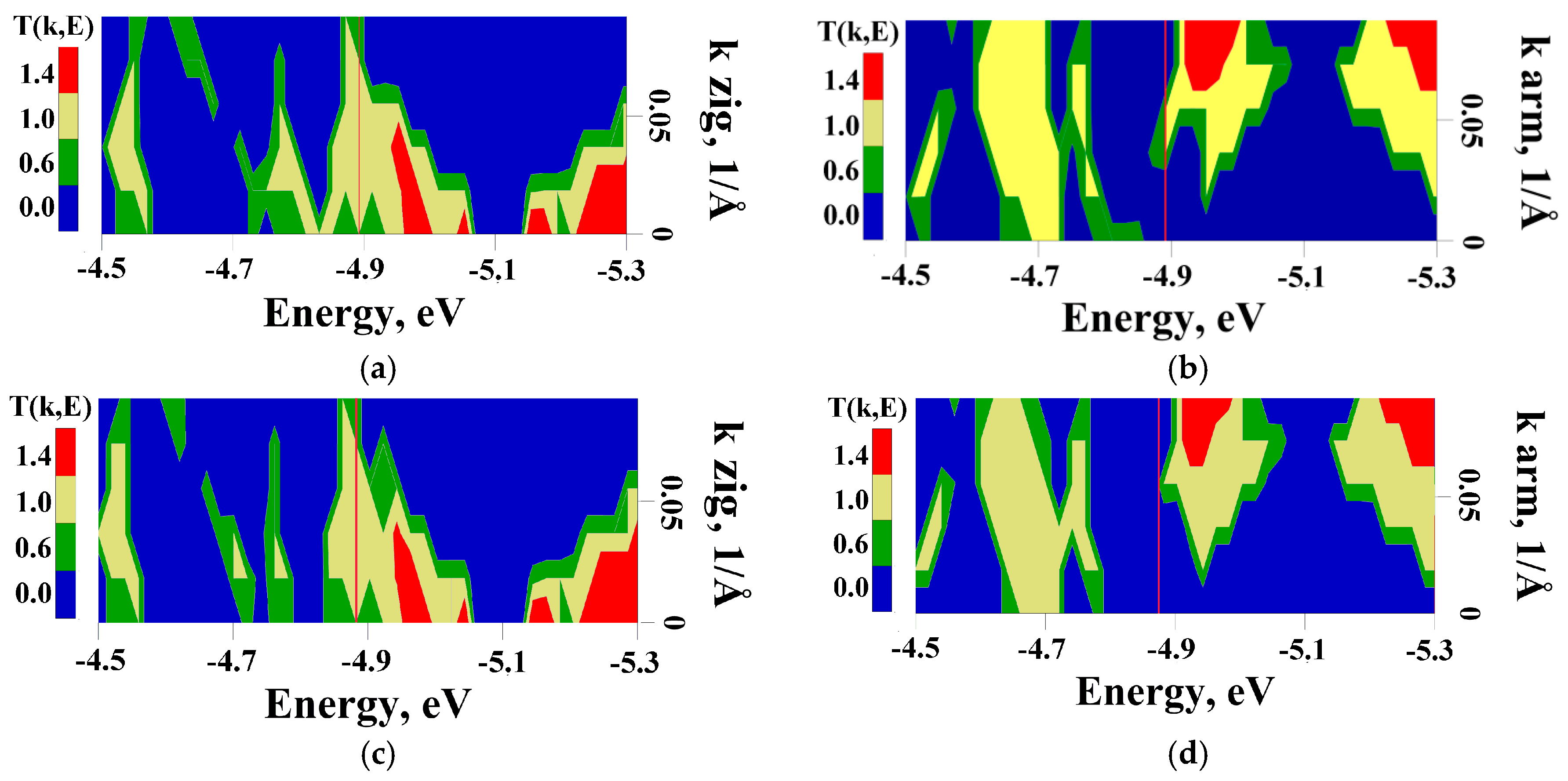
2.4. Influence of DNA Nitrogenous Bases on Conductive Properties of the VACNT(12,6)/Graphene
3. Conclusions and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Tamersit, K.; Djeffal, F. Double-Gate Graphene Nanoribbon Field-Effect Transistor for DNA and Gas Sensing Applications: Simulation Study and Sensitivity Analysis. IEEE Sens. J. 2016, 16, 4180–4191. [Google Scholar] [CrossRef]
- Liu, H.-W.; Chen, L.; Xu, C.; Li, Z.; Zhang, H.; Zhang, X.B.; Tan, W. Recent progresses in small-molecule enzymatic fluorescent probes for cancer imaging. Chem. Soc. Rev. 2018, 47, 7140–7180. [Google Scholar] [PubMed]
- Ye, F.; Zhao, Y.; Ei-Sayed, R.; Muhammed, M.; Hassan, M. Advances in nanotechnology for cancer biomarkers. Nano Today 2018, 18, 103–123. [Google Scholar] [CrossRef]
- Perfezou, M.; Turner, A.; Merkoci, A. Cancer detection using nanoparticle-based sensors. Chem. Soc. Rev. 2012, 41, 2606–2622. [Google Scholar] [CrossRef]
- Zhu, Z. An Overview of Carbon Nanotubes and Graphene for Biosensing Applications. Nano-Micro Lett. 2017, 9, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.; Qu, L.; Tan, Y.; Liu, F.; Wang, Y.; Zhang, W.; Cai, Z.; Mou, L.; Jiang, Y. A fluorescent aptasensor with product-triggered amplification by exonuclease III digestion for highly sensitive ATP detection. Anal. Methods 2017, 9, 4837–4842. [Google Scholar] [CrossRef]
- Li, C.; Shi, G. Carbon nanotube-based fluorescence sensors. J. Photochem. Photobiol. C 2014, 19, 20–34. [Google Scholar] [CrossRef]
- Ou, X.; Zhan, S.; Sun, C.; Cheng, Y.; Wang, X.; Liu, B.; Zhai, T.; Lou, X.; Xia, F. Simultaneous detection of telomerase and miRNA with graphene oxide-based fluorescent aptasensor in living cells and tissue samples. Biosens. Bioelectron. 2019, 124, 199–204. [Google Scholar] [CrossRef]
- Liu, J.; Wang, C.; Jiang, Y.; Hu, Y.; Li, J.; Yang, S.; Li, Y.; Yang, R.; Tan, W.; Huang, C.Z. Graphene Signal Amplification for Sensitive and Real-Time Fluorescence Anisotropy Detection of Small Molecules. Anal. Chem. 2013, 85, 1424–1430. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Y.; Liu, Y.; Liu, H.; Fu, L.; Wen, J.; Li, J.; Wei, P.; Chen, L. A graphene oxide/gold nanoparticle-based amplification method for SERS immunoassay of cardiac troponin. Analyst 2019, 144, 1582–1589. [Google Scholar] [CrossRef]
- Khalil, I.; Yehye, W.A.; Julkapli, N.M.; Rahmati, S.; Sina, A.A.I.; Basirun, W.J.; Johan, M.R. Graphene oxide and gold nanoparticle based dual platform with short DNA probe for the PCR free DNA biosensing using surface-enhanced Raman scattering. Biosens. Bioelectron. 2019, 131, 214–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, L.; Zhang, Q.; Ma, G.; Zhu, L.; Wang, Y.; Chen, Z.; Wang, Y.; Zhao, L. New Dual-Spectroscopic Strategy for the Direct Detection of Aristolochic Acids in Blood and Tissue. Anal. Chem. 2019, 91, 8154–8161. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, L.; Guo, Y.; Gao, X.; Xu, Y.; Wei, Q.; Man, B.; Yang, C. Suspended 3D AgNPs/CNT nanohybrids for the SERS application. Appl. Surf. Sci. 2019, 487, 1077–1083. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Q.; Yang, X.; Wang, K.; Zhang, H.; Nie, W. High sensitivity surface plasmon resonance biosensor for detection of microRNA and small molecule based on graphene oxide-gold nanoparticles composites. Talanta 2017, 174, 521–526. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Pagneux, Q.; Larroulet, I.; Serrano, A.Y.; Pesquera, A.; Zurutuza, A.; Mandler, D.; Boukherroub, R.; Szunerits, S. Label-free femtomolar cancer biomarker detection in human serum using graphene-coated surface plasmon resonance chips. Biosens. Bioelectron. 2017, 89, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Chiu, N.-F.; Lin, T.-L.; Kuo, C.-T. Highly sensitive carboxyl-graphene oxide-based surface plasmon resonance immunosensor for the detection of lung cancer for cytokeratin 19 biomarker in human plasma. Sens. Actuator B Chem. 2018, 265, 264–272. [Google Scholar] [CrossRef]
- Allsop, T.; Arif, R.; Neal, R.; Kalli, K.; Kundrát, V.; Rozhin, A.; Culverhouse, P.; Webb, D.J. Photonic gas sensors exploiting directly the optical properties of hybrid carbon nanotube localized surface plasmon structures. Sens. Light Sci. Appl. 2016, 5, 16036. [Google Scholar] [CrossRef] [Green Version]
- Cheung, W.; Patel, M.; Ma, Y.; Chen, Y.; Xie, Q.; Lockard, J.V.; Gao, Y.; He, H. Plasmon absorption of carbon nanotubes for the selective and sensitive detection of Fe3+ ions. Chem. Sci. 2016, 7, 5192–5199. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Xu, T.; Zhang, X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Cao, X.; Jiang, Y.; He, P.; Fang, Y. Carbon nanotubeenhanced electrochemical DNA biosensor for DNA hybridization detection. Anal. Bioanal. Chem. 2003, 375, 287–293. [Google Scholar] [CrossRef]
- Weber, J.E.; Pillai, S.; Ram, M.K.; Kumar, A.; Singh, S.R. Electrochemical impedance-based DNA sensor using a modified single walled carbon nanotube electrode. Mater. Sci. Eng. C 2011, 31, 821–825. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Rahighi, R. Toward single-DNA electrochemical biosensing by graphene nanowalls. ACS Nano 2012, 6, 2904–2916. [Google Scholar] [CrossRef]
- Salahandish, R.; Ghaffarinejad, A.; Omidinia, E.; Zargartalebi, H.; Majidzadeh-A, K.; Naghib, S.M.; Sanati-Nezhad, A. Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens. Bioelectron. 2018, 120, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Wang, S.; Qiu, J. Preparation and properties of a glucose biosensor electrode based on an ionic liquid-functionalized graphene/carbon nanotube composite. New Carbon Mater. 2020, 35, 12–19. [Google Scholar] [CrossRef]
- Niu, X.; Yang, X.; Mo, Z. Fabrication of an electrochemical chiral sensor via an integrated polysaccharides/3D nitrogen-doped graphene-CNT frame. Bioelectrochemistry 2020, 131, 107396. [Google Scholar] [CrossRef] [PubMed]
- Zarean Mousaabadi, K.; Ensafi, A.A.; Hadadzadeh, H.; Rezaei, B. Reduced graphene oxide and carbon nanotubes composite functionalized by azobenzene, characterization and its potential as a curcumin electrochemical sensor. J. Electroanal Chem. 2020, 873, 114418. [Google Scholar] [CrossRef]
- Cao, M.; Su, J.; Fan, S.; Qiu, H.; Su, D.; Li, L. Wearable piezoresistive pressure sensors based on 3D graphene. Int. J. Chem. Eng. 2021, 406, 126777. [Google Scholar]
- Zhao, P.; Ni, M.; Xu, Y.; Wang, C.; Chen, C.; Zhang, X.; Li, C.; Xie, Y.; Fei, J. A novel ultrasensitive electrochemical quercetin sensor based on MoS2-carbon nanotube@graphene oxide nanoribbons/HS-cyclodextrin/graphene quantum dots composite film. Sens. Actuator B Chem. 2019, 299, 126997. [Google Scholar] [CrossRef]
- Khodadadi, A.; Faghih-Mirzaei, E.; Karimi-Maleh, H. A new epirubicin biosensor based on amplifying DNA interactions with polypyrrole and nitrogen-doped reduced graphene: Experimental and docking theoretical investigations. Sens. Actuator B Chem. 2017, 242, 568–574. [Google Scholar] [CrossRef]
- Pourasl, A.H.; Ahmadi, M.T.; Rahmani, M. Analytical modeling of glucose biosensors based on carbon nanotubes. Nanoscale Res. Lett. 2014, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Akbari, E.; Buntat, Z.; Enzevaee, A.; Mirazimiabarghouei, S.J.; Bahadoran, M.; Shahidi, A.; Nikoukar, A. Correction: An analytical model and ANN simulation for carbon nanotube based ammonium gas sensors. RSC Adv. 2014, 4, 36896–36904. [Google Scholar] [CrossRef]
- Bartneck, M.; Heffels, K.H.; Pan, Y.; Bovi, M.; Zwadlo-Klarwasser, G.; Groll, J. Inducing healing-like human primary macrophage phenotypes by 3D hydrogel coated nanofibres. Biomaterials 2012, 33, 4136–4146. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Li, Z.; Song, R.; Li, Y.; Guo, C.; Sha, Y.; Cui, W.; Xu, S.; Hu, G.; Wang, J. Graphene Biosensor as Affinity Biosensors for Biorecognition between Guanine Riboswitch and Ligand. Appl. Surf. Sci. 2020, 503, 144303. [Google Scholar] [CrossRef]
- Madadi Mahani, N. Dopamine detection by doped single-walled carbon nanotube biosensors: A theoretical study. J. Res. Pharm. 2019, 23, 785–791. [Google Scholar] [CrossRef] [Green Version]
- Mirzaii Babolghani, F.; Mohammadi-Manesh, E. Simulation and experimental study of FET biosensor to detect polycyclic aromatic hydrocarbons. Appl. Surf. Sci. 2019, 488, 662–670. [Google Scholar] [CrossRef]
- Tylianakis, E.; Psofogiannakis, G.M.; Froudakis, G.E. Li-doped pillared graphene oxide: A graphene-based nanostructured material for hydrogen storage. J. Phys. Chem Lett. 2010, 1, 2459–2464. [Google Scholar] [CrossRef]
- Wu, C.-D.; Fang, T.-H.; Lo, J.-Y. Effects of pressure, temperature, and geometric structure of pillared graphene on hydrogen storage capacity. Int. J. Hydrogen Energy. 2012, 37, 14211–14216. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, C.; Yan, Z.; Zhu, Y.; Peng, Z.; Hauge, R.H.; Natelson, D.; Tour, J.M. 3-Dimensional graphene carbon nanotube carpet-based microsupercapacitors with high electrochemical performance. Nano Lett. 2013, 13, 72–78. [Google Scholar] [CrossRef]
- Park, J.; Prakash, V. Thermal transport in 3D pillared SWCNT–graphene nanostructures. J. Mater. Res. 2013, 28, 940–951. [Google Scholar] [CrossRef] [Green Version]
- Duan, K.; Li, L.; Hu, Y.; Wang, X. Pillared graphene as an ultra-high sensitivity mass sensor. Sci. Rep. 2017, 7, 14012. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, X.; Zhao, X.; Yang, F.; Wang, X.; Li, Y. Metallic Catalysts for Structure-Controlled Growth of Single-Walled Carbon Nanotubes. Top. Curr. Chem. 2017, 375, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, S.; Zhu, Z.; Zhang, J.; Wei, F.; Li, Y. Catalysts for single-wall carbon nanotube synthesis—From surface growth to bulk preparation. MRS Bull. 2017, 42, 809–818. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, F.; Chen, J.; Ding, L.; Liu, X.; Yao, F.; Li, M.; Zhang, D.; Zhang, Z.; Liu, X.; et al. Selective Growth of Chirality-Enriched Semiconducting Nanotubes by Using Bimetallic Catalysts from Salt Precursors. Nanoscale 2013, 10, 6922–6927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Glukhova, O.E. New automatic method for generating atomistic models of multi-branched and arbitrary-shaped seamless junctions of carbon nanostructures. Comput. Mater. Sci. 2020, 184, 109943. [Google Scholar] [CrossRef]
- Elstner, M.; Porezag, D.; Jungnickel, G.; Elsner, J.; Haugk, M.; Frauenheim, T.; Suhai, S.; Seifert, G. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 1998, 58, 7260–7268. [Google Scholar] [CrossRef]
- Qian, H.-J.; Eres, G.; Irle, S. Quantum chemical molecular dynamics simulation of carbon nanotube–graphene fusion. Mol. Simul. 2017, 13, 1269–1276. [Google Scholar] [CrossRef]
- Slepchenkov, M.M.; Shmygin, D.S.; Zhang, G.; Glukhova, O.E. Controlling anisotropic electrical conductivity in porous graphene-nanotube thin films. Carbon 2020, 165, 139–149. [Google Scholar] [CrossRef]
- Dimitrakakis, G.K.; Tylianakis, E.; Froudakis, G.E. Pillared Graphene: A New 3-D Network Nanostructure for Enhanced Hydrogen Storage. Nano Lett. 2008, 8, 3166–3170. [Google Scholar] [CrossRef]
- Yang, X.; Yu, D.; Cao, B.; To, A.C. Ultrahigh Thermal Rectification in Pillared Graphene Structure with Carbon Nanotube-graphene Intramolecular Junctions. ACS Appl. Mater. Interfaces 2016, 9, 29–35. [Google Scholar] [CrossRef]
- Bilgili, D.; Kirkayak, L.; Kirca, M. The effects of intertube bridging through graphene nanoribbons on the mechanical properties of pillared graphene. Compos. B Eng. 2017, 120, 1–9. [Google Scholar] [CrossRef]
- Shunaev, V.V.; Savostyanov, G.V.; Slepchenkov, M.M.; Glukhova, O.E. Phenomenon of current occurrence during the motion of a C60 fullerene on substrate-supported graphene. RSC Adv. 2015, 5, 86337–86346. [Google Scholar] [CrossRef]
- Slepchenkov, M.M.; Shunaev, V.V.; Glukhova, O.E. Response to external GHz and THz radiation of K+@C60 endohedral complex in cavity of carbon nanotube containing polymerized fullerenes. J. Appl. Phys. 2019, 125, 244306. [Google Scholar] [CrossRef]
- Gaus, M.; Goez, A.; Elstner, M. Parametrization and Benchmark of DFTB3 for Organic Molecules. J. Chem. Theory Comput. 2013, 9, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Soukup, G.A. Nucleic Acids: General Properties; John Wiley & Sons, Ltd.: New York, NY, USA, 2003. [Google Scholar]
- Mitrofanov, V.V.; Slepchenkov, M.M.; Zhang, G.; Glukhova, O.E. Hybrid carbon nanotube-graphene monolayer films: Regularities of structure, electronic and optical properties. Carbon 2017, 115, 803–810. [Google Scholar] [CrossRef]
- Keldysh, L.V. Diagram technique for nonequilibrium processes. J. Exp. Theor. Phys. Lett. 1965, 20, 1018–1026. [Google Scholar]
- Fratini, A.V.; Kopka, M.L.; Drew, H.R.; Dickerson, R.E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem. 1982, 257, 14686–14707. [Google Scholar]


| G (zig), S | R (zig), kOhm | G (arm), S | R (arm), kOhm | |
|---|---|---|---|---|
| Pure Carcass | 5.3594 × 10−5 | 18.658 | 4.39427 × 10−5 | 22.756 |
| Cytosine | 5.6348 × 10−5 | 17.746 | 2.71896 × 10−5 | 36.778 |
| Thymine | 5.9794 × 10−5 | 16.724 | 2.53869 × 10−5 | 39.390 |
| Guanine | 5.8864 × 10−5 | 16.988 | 2.7131 × 10−5 | 36.858 |
| Adenine | 5.8364 × 10−5 | 17.133 | 2.4219 × 10−5 | 36.739 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shunaev, V.V.; Glukhova, O.E. Pillared Graphene Structures Supported by Vertically Aligned Carbon Nanotubes as the Potential Recognition Element for DNA Biosensors. Materials 2020, 13, 5219. https://doi.org/10.3390/ma13225219
Shunaev VV, Glukhova OE. Pillared Graphene Structures Supported by Vertically Aligned Carbon Nanotubes as the Potential Recognition Element for DNA Biosensors. Materials. 2020; 13(22):5219. https://doi.org/10.3390/ma13225219
Chicago/Turabian StyleShunaev, Vladislav V., and Olga E. Glukhova. 2020. "Pillared Graphene Structures Supported by Vertically Aligned Carbon Nanotubes as the Potential Recognition Element for DNA Biosensors" Materials 13, no. 22: 5219. https://doi.org/10.3390/ma13225219
APA StyleShunaev, V. V., & Glukhova, O. E. (2020). Pillared Graphene Structures Supported by Vertically Aligned Carbon Nanotubes as the Potential Recognition Element for DNA Biosensors. Materials, 13(22), 5219. https://doi.org/10.3390/ma13225219






