Zinc Oxide Nanocomposites—Extracellular Synthesis, Physicochemical Characterization and Antibacterial Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Bacterial Strain
2.2. Extracellular Synthesis of Zinc Oxide Nanocomposites (ZnO NCs)
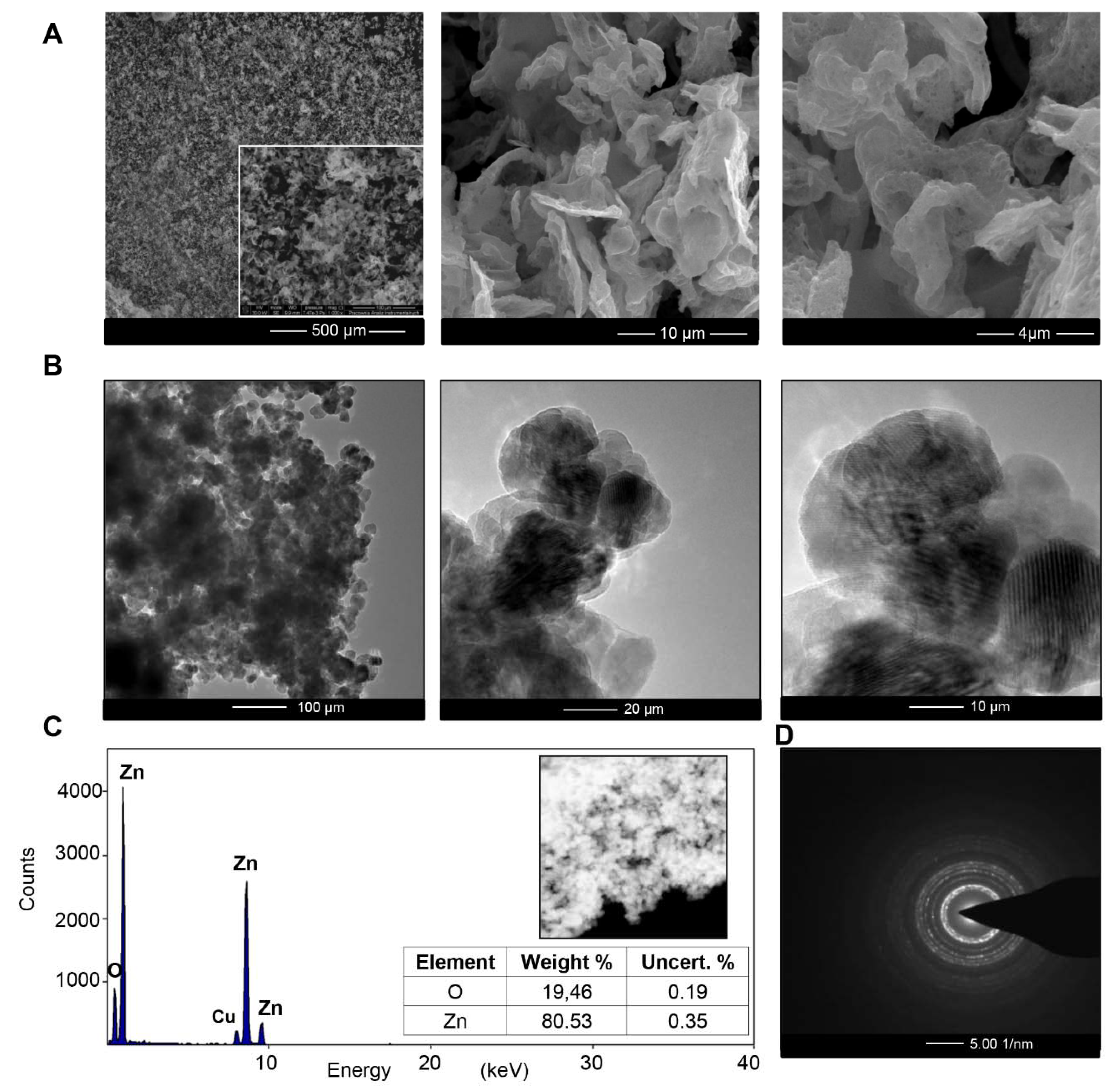
2.3. Physicochemical Characterization of ZnO NCs
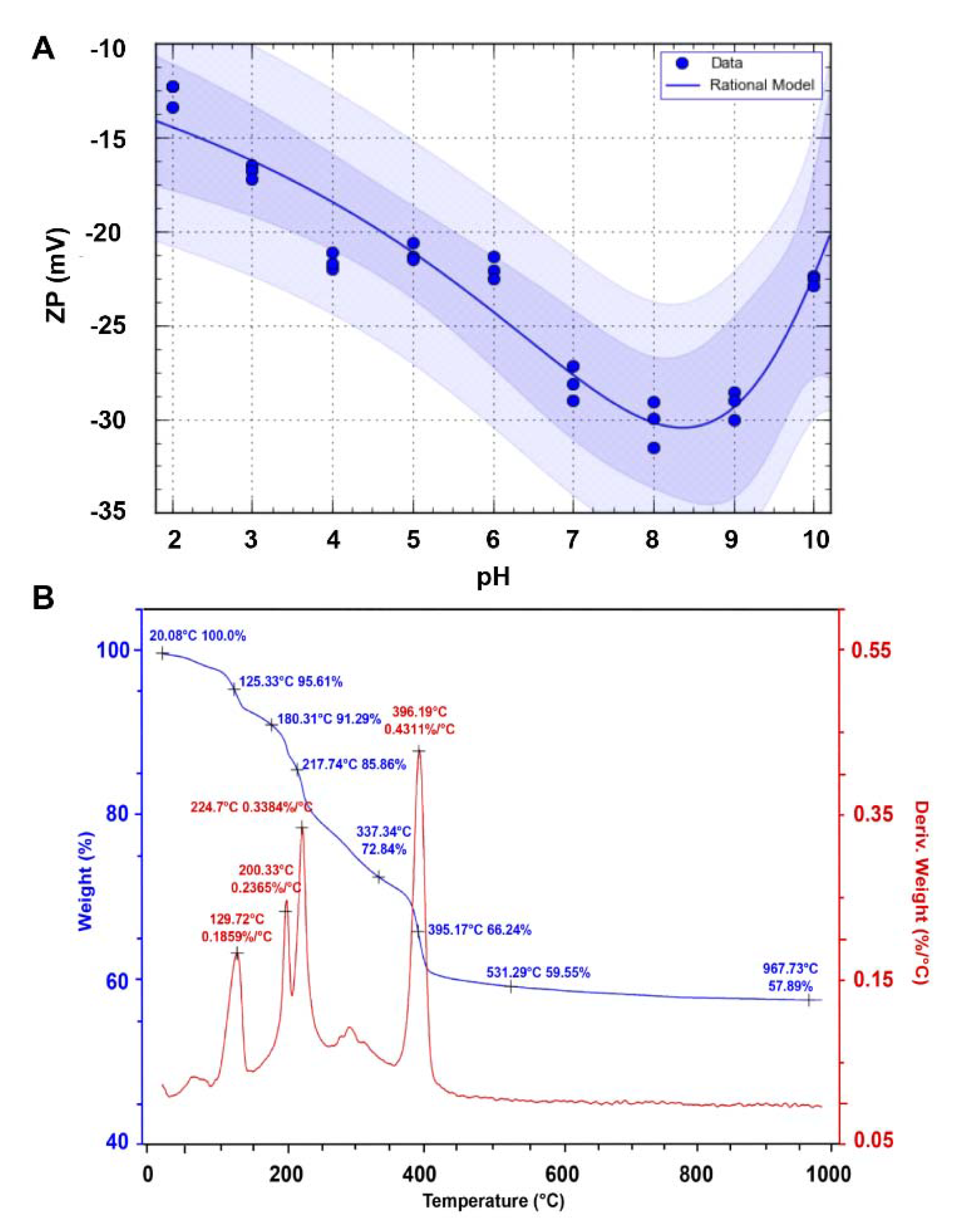
2.3.1. Zeta Potential Measurement
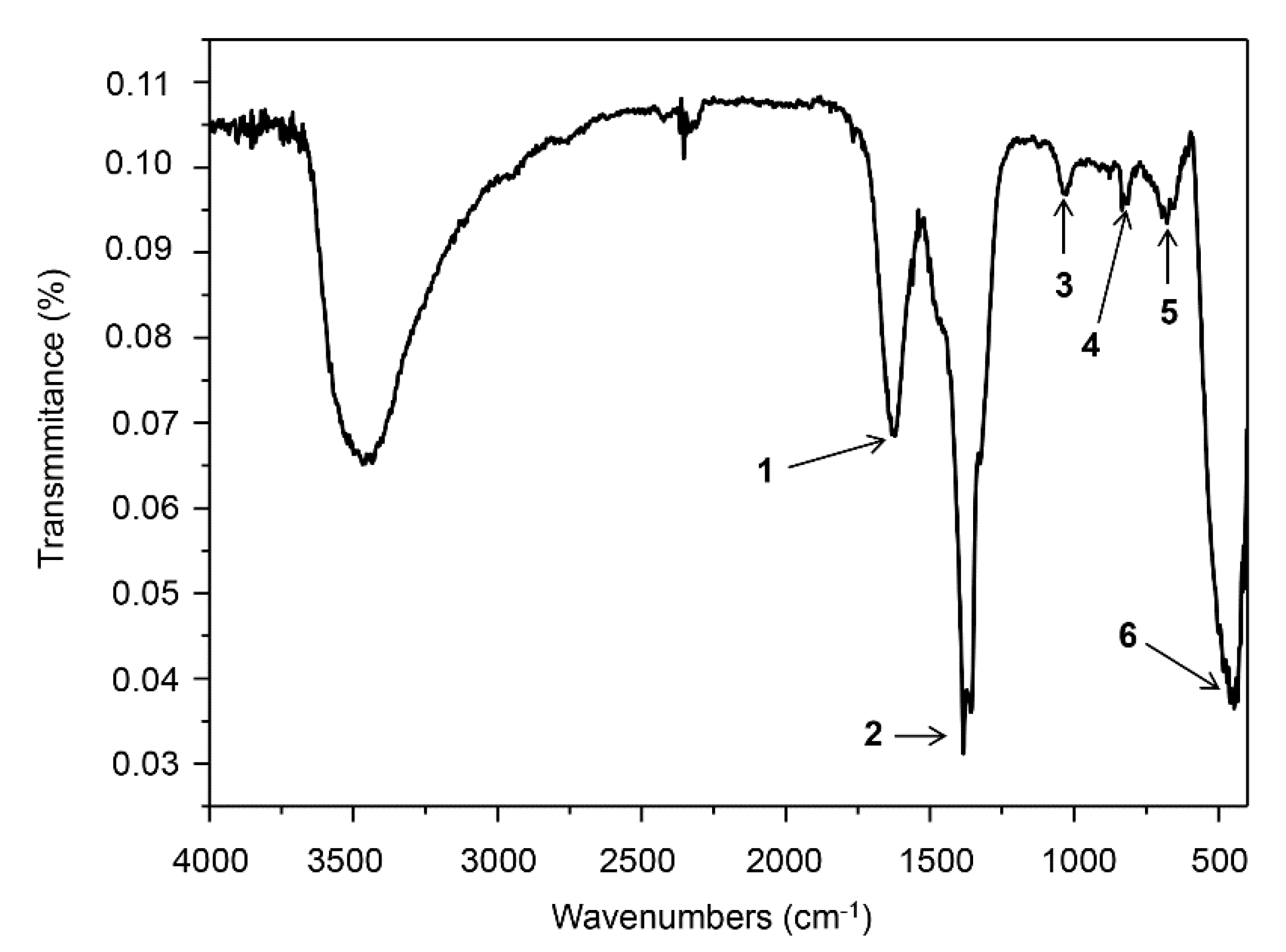
2.3.2. Fourier Transform Infrared Spectroscopy Analysis
2.3.3. Transmission Electron Microscopy (TEM) and Energy Dispersive X-ray (EDX) Analysis
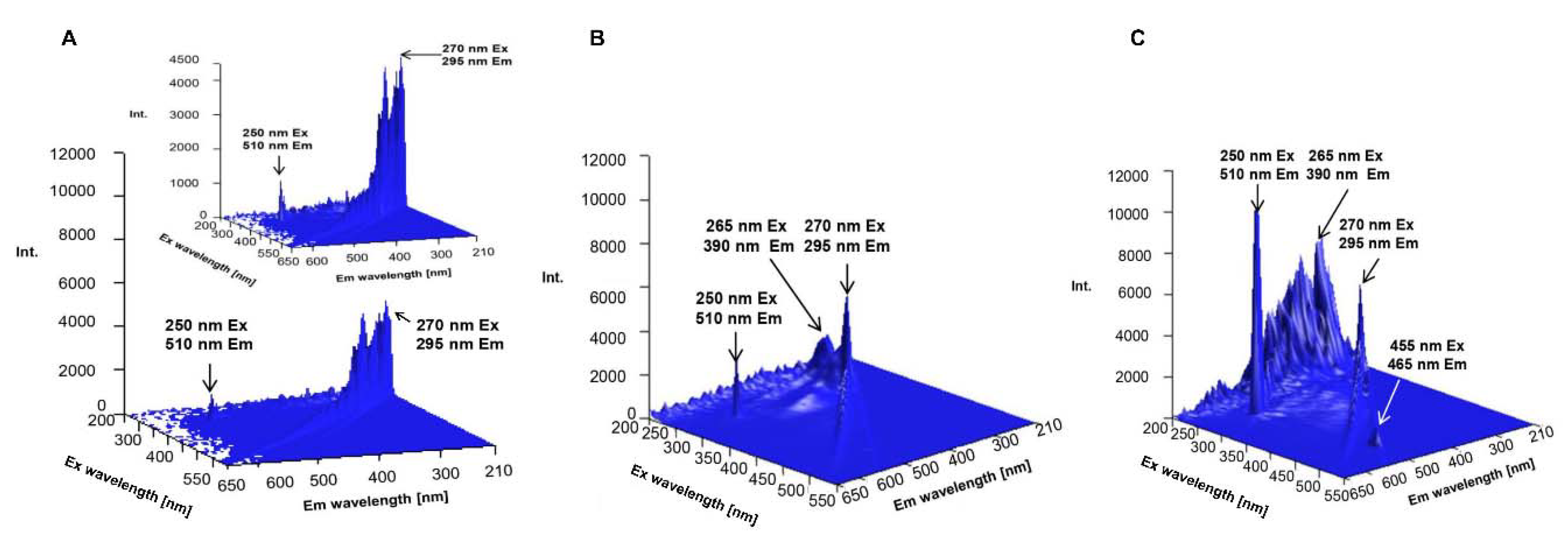
2.3.4. Spectrofluorometric Analysis
2.3.5. X-ray Diffraction Study
2.3.6. Thermogravimetric Analysis (TG-DTA)
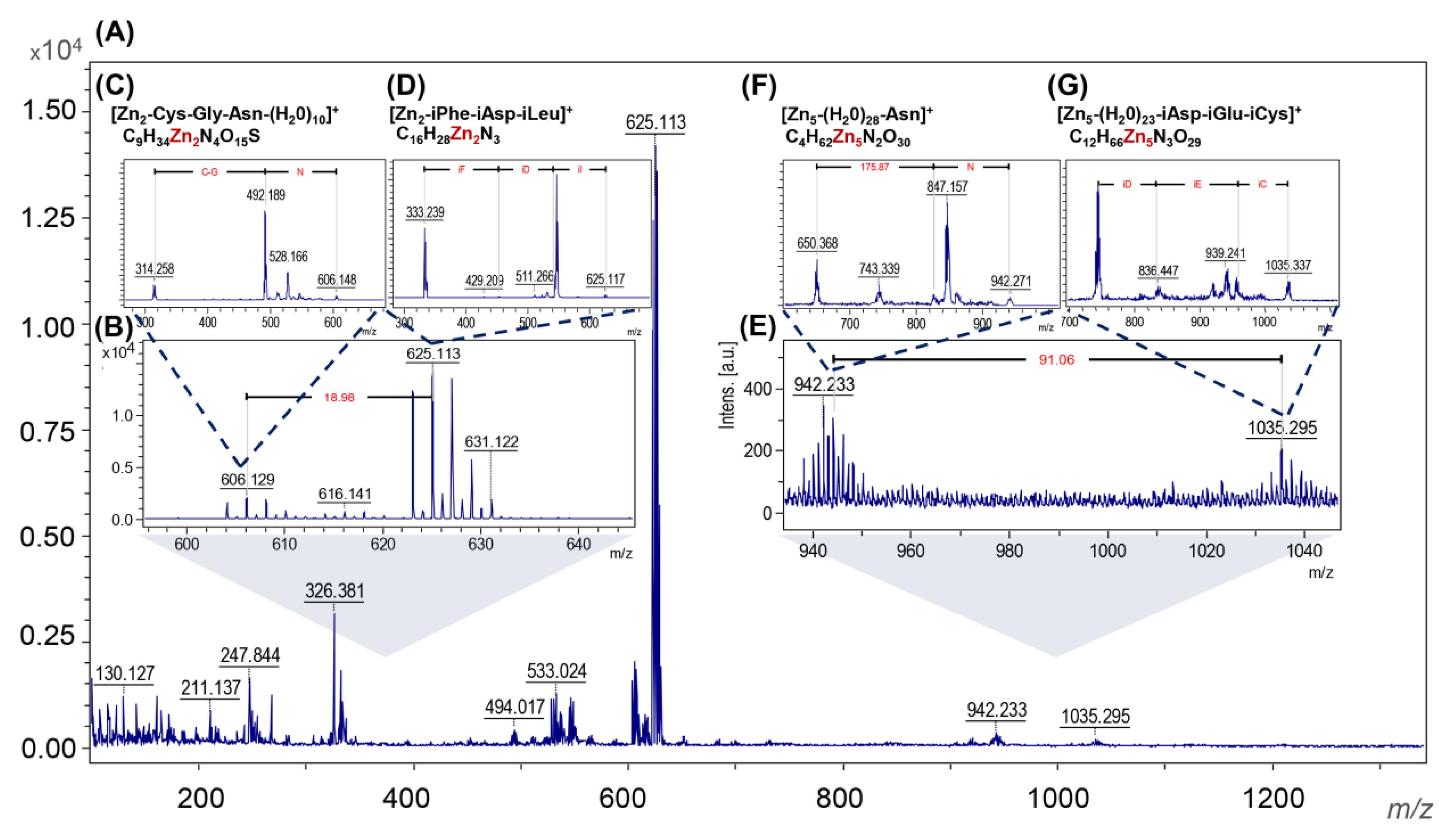
2.3.7. Laser Desorption/Ionization with Mass Spectrometry Analysis (LDI-TOF-MS)
2.4. Antimicrobial Potential of ZnO NCs
Colony Forming Unit (CFU) and Minimal Inhibitory Concentration (MIC)
3. Results and Discussion
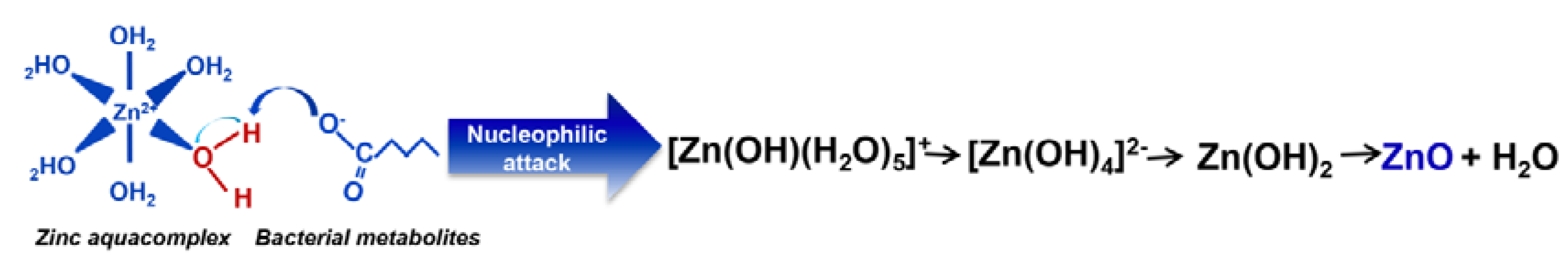
3.1. Physicochemical Characterization of ZnO NCs
3.2. Antimicrobial Activity of ZnO NCs
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Keerthana, S.; Kumar, A. Potential risks and benefits of zinc oxide nanoparticles: A systematic review. Crit. Rev. Toxicol. 2020, 50, 47–71. [Google Scholar] [CrossRef] [PubMed]
- Król, A.; Pomastowski, P.; Rafińska, K.; Railean-Plugaru, V.; Buszewski, B. Zinc oxide nanoparticles: Synthesis, antiseptic activity and toxicity mechanism. Adv. Colloid Interface Sci. 2017, 249, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Ogunyemi, S.O.; Abdallah, Y.; Zhang, M.; Fouad, H.; Hong, X.; Ibrahim, E.; Masum, M.M.I.; Hossain, A.; Mo, J.; Li, B. Green synthesis of zinc oxide nanoparticles using different plant extracts and their antibacterial activity against Xanthomonas oryzae pv. oryzae. Artif. Cells Nanomed. Biotechnol. 2019, 47, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Kalpana, V.N.; Kataru, B.A.S.; Sravani, N.; Vigneshwari, T.; Panneerselvam, A.; Devi Rajeswari, V. Biosynthesis of zinc oxide nanoparticles using culture filtrates of Aspergillus niger: Antimicrobial textiles and dye degradation studies. OpenNano 2018, 3, 48–55. [Google Scholar] [CrossRef]
- Król, A.; Railean-Plugaru, V.; Pomastowski, P.; Buszewski, B. Phytochemical investigation of Medicago sativa L. extract and its potential as a safe source for the synthesis of ZnO nanoparticles: The proposed mechanism of formation and antimicrobial activity. Phytochem. Lett. 2019, 31, 170–180. [Google Scholar] [CrossRef]
- Salvadori, M.R.; Ando, R.A.; Oller Nascimento, C.A.; Corrêa, B. Extra and Intracellular Synthesis of Nickel Oxide Nanoparticles Mediated by Dead Fungal Biomass. PLoS ONE 2015, 10, e0129799. [Google Scholar] [CrossRef]
- Ovais, M.; Khalil, A.; Ayaz, M.; Ahmad, I.; Nethi, S.; Mukherjee, S. Biosynthesis of Metal Nanoparticles via Microbial Enzymes: A Mechanistic Approach. Int. J. Mol. Sci. 2018, 19, 4100. [Google Scholar] [CrossRef]
- Grasso, G.; Zane, D.; Dragone, R. Microbial nanotechnology: Challenges and prospects for green biocatalytic synthesis of nanoscale materials for sensoristic and biomedical applications. Nanomaterials 2020, 10, 11. [Google Scholar] [CrossRef]
- Das, V.L.; Thomas, R.; Varghese, R.T.; Soniya, E.V.; Mathew, J.; Radhakrishnan, E.K. Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area. 3 Biotech 2014, 4, 121–126. [Google Scholar] [CrossRef]
- Railean-Plugaru, V.; Pomastowski, P.P.; Meller, K.; Złoch, M.; Rafińska, K.; Buszewski, B. Lactococcus lactis as a safe and inexpensive source of bioactive silver composites. Appl. Microbiol. Biotechnol. 2017, 101, 7141–7153. [Google Scholar] [CrossRef]
- Kundu, D.; Hazra, C.; Chatterjee, A.; Chaudhari, A.; Mishra, S. Extracellular biosynthesis of zinc oxide nanoparticles using Rhodococcus pyridinivorans NT2: Multifunctional textile finishing, biosafety evaluation and in vitro drug delivery in colon carcinoma. J. Photochem. Photobiol. B Biol. 2014, 140, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Srivastava, S. Antifungal effect of antimicrobial peptides (AMPs LR14) derived from Lactobacillus plantarum strain LR/14 and their applications in prevention of grain spoilage. Food Microbiol. 2014, 42, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Zhu, J.; Gong, S.; Liu, H.; Yu, H. Antimicrobial Characteristics of Lactic Acid Bacteria Isolated from Homemade Fermented Foods. Biomed Res. Int. 2018, 2018, 5416725. [Google Scholar] [CrossRef] [PubMed]
- Selvarajan, E.; Mohanasrinivasan, V. Biosynthesis and characterization of ZnO nanoparticles using Lactobacillus plantarum VITES07. Mater. Lett. 2013, 112, 180–182. [Google Scholar] [CrossRef]
- Al-Zahrani, H.A.A.; El-Waseif, A.A.; El-Ghwas, D.E. Biosynthesis and evaluation of TiO 2 and ZnO nanoparticles from in vitro stimulation of Lactobacillus johnsonii. J. Innov. Pharm. Biol. Sci. 2018, 5, 16–20. [Google Scholar]
- Mohd Yusof, H.; Mohamad, R.; Zaidan, U.H.; Rahman, N.A. Sustainable microbial cell nanofactory for zinc oxide nanoparticles production by zinc-tolerant probiotic Lactobacillus plantarum strain TA4. Microb. Cell Fact. 2020, 19, 10. [Google Scholar] [CrossRef]
- Gaynes, R. The discovery of penicillin—New insights after more than 75 years of clinical use. Emerg. Infect. Dis. 2017, 23, 849–853. [Google Scholar] [CrossRef]
- Li, B.; Webster, T.J. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J. Orthop. Res. 2018, 36, 22–32. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella pneumoniae: An increasing threat to public health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef]
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Wiley: Hoboken, NJ, USA, 2018; Volume 6, pp. 289–316. [Google Scholar]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- De Wit, J.N. Nutritional and Functional Characteristics of Whey Proteins in Food Products. J. Dairy Sci. 1998, 81, 597–608. [Google Scholar] [CrossRef]
- Rodzik, A.; Pomastowski, P.; Sagandykova, G.N.; Buszewski, B. Interactions of whey proteins with metal ions. Int. J. Mol. Sci. 2020, 21, 2156. [Google Scholar] [CrossRef] [PubMed]
- Pomastowski, P.; Złoch, M.; Rodzik, A.; Ligor, M.; Kostrzewa, M.; Buszewski, B. Analysis of bacteria associated with honeys of different geographical and botanical origin using two different identification approaches: MALDI-TOF MS and 16S rDNA PCR technique. PLoS ONE 2019, 14, e0217078. [Google Scholar] [CrossRef]
- Milanowski, M.; Pomastowski, P.; Railean-Plugaru, V.; Rafińska, K.; Ligor, T.; Buszewski, B. Biosorption of silver cations onto Lactococcus lactis and Lactobacillus casei isolated from dairy products. PLoS ONE 2017, 12, e0174521. [Google Scholar] [CrossRef]
- Muniz, F.T.L.; Miranda, M.A.R.; Morilla Dos Santos, C.; Sasaki, J.M. The Scherrer equation and the dynamical theory of X-ray diffraction. Acta Crystallogr. Sect. A Found. Adv. 2016, 72, 385–390. [Google Scholar] [CrossRef]
- Railean-Plugaru, V.; Pomastowski, P.; Kowalkowski, T.; Sprynskyy, M.; Buszewski, B. Physicochemical study of natural fractionated biocolloid by asymmetric flow field-flow fractionation in tandem with various complementary techniques using biologically synthesized silver nanocomposites. Anal. Bioanal. Chem. 2018, 410, 2837–2847. [Google Scholar] [CrossRef]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952–965. [Google Scholar] [CrossRef]
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef]
- Buszewski, B.; Railean-Plugaru, V.; Pomastowski, P.; Rafińska, K.; Szultka-Mlynska, M.; Golinska, P.; Wypij, M.; Laskowski, D.; Dahm, H. Antimicrobial activity of biosilver nanoparticles produced by a novel Streptacidiphilus durhamensis strain. J. Microbiol. Immunol. Infect. 2018, 51, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Nangia, Y.; Wangoo, N.; Goyal, N.; Shekhawat, G.; Suri, C.R. A novel bacterial isolate Stenotrophomonas maltophilia as living factory for synthesis of gold nanoparticles. Microb. Cell Fact. 2009, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Taran, M.; Rad, M.; Alavi, M. Biosynthesis of TiO2 and ZnO nanoparticles by Halomonas elongata IBRC-M 10214 in different conditions of medium. BioImpacts 2018, 8, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Nagashima, K.; Parmiter, D.; de la Cruz, J.; Patri, A.K. SEM X-ray microanalysis of nanoparticles present in tissue or cultured cell thin sections. Methods Mol. Biol. 2011, 697, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Narayana, A.; Bhat, S.A.; Fathima, A.; Lokesh, S.V.; Surya, S.G.; Yelamaggad, C.V. Green and low-cost synthesis of zinc oxide nanoparticles and their application in transistor-based carbon monoxide sensing. RSC Adv. 2020, 10, 13532–13542. [Google Scholar] [CrossRef]
- Gavrilenko, E.A.; Goncharova, D.A.; Lapin, I.N.; Nemoykina, A.L.; Svetlichnyi, V.A.; Aljulaih, A.A.; Mintcheva, N.; Kulinich, S.A. Comparative study of physicochemical and antibacterial properties of zno nanoparticles prepared by laser ablation of zn target in water and air. Materials 2019, 12, 186. [Google Scholar] [CrossRef]
- Yang, Z.X.; Zhong, W.; Au, C.; Wang, J.Y.; Du, Y.W. An environment-benign solvothermal method for the synthesis of flower-like hierarchical nickel and zinc compounds and their transformation to nanoporous NiO and ZnO. CrystEngComm 2011, 13, 1831–1837. [Google Scholar] [CrossRef]
- Wahab, R.; Ansari, S.G.; Kim, Y.S.; Song, M.; Shin, H.S. The role of pH variation on the growth of zinc oxide nanostructures. Appl. Surf. Sci. 2009, 255, 4891–4896. [Google Scholar] [CrossRef]
- Vasile, O.R.; Serdaru, I.; Andronescu, E.; Truşcə, R.; Surdu, V.A.; Oprea, O.; Ilie, A.; Vasile, B.Ş. Influence of the size and the morphology of ZnO nanoparticles on cell viability. Comptes Rendus Chim. 2015, 18, 1335–1343. [Google Scholar] [CrossRef]
- Vasile, O.R.; Andronescu, E.; Ghitulica, C.; Vasile, B.S.; Oprea, O.; Vasile, E.; Trusca, R. Synthesis and characterization of nanostructured zinc oxide particles synthesized by the pyrosol method. J. Nanopart. Res. 2012, 14, 1–13. [Google Scholar] [CrossRef]
- Nicolardi, S.; Palmblad, M.; Dalebout, H.; Bladergroen, M.; Tollenaar, R.A.E.M.; Deelder, A.M.; van der Burgt, Y.E.M. Quality control based on isotopic distributions for high-throughput MALDI-TOF and MALDI-FTICR serum peptide profiling. J. Am. Soc. Mass Spectrom. 2010, 21, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Polfer, N.C.; Oomens, J.; Moore, D.T.; Von Helden, G.; Meijer, G.; Dunbar, R.C. Infrared spectroscopy of phenylalanine Ag(I) and Zn(II) complexes in the gas phase. J. Am. Chem. Soc. 2006, 128, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Hernández, B.; Pflüger, F.; Adenier, A.; Kruglik, S.G.; Ghomi, M. Vibrational analysis of amino acids and short peptides in hydrated media. VIII. Amino acids with aromatic side chains: L-phenylalanine, l-tyrosine, and l-tryptophan. J. Phys. Chem. B 2010, 114, 15319–15330. [Google Scholar] [CrossRef]
- Němec, I.; Mička, Z. FTIR and FT Raman study of L-leucine addition compound with nitric acid. J. Mol. Struct. 1999, 482–483, 23–28. [Google Scholar]
- Kalsi, P.S. Spectroscopy of Organic Compounds; New Age International: New Delhi, India, 2006; ISBN 8122415431. [Google Scholar]
- Nara, M.; Torii, H.; Tasumi, M. Correlation between the vibrational frequencies of the carboxylate group and the types of its coordination to a metal ion: An ab initio molecular orbital study. J. Phys. Chem. 1996, 100, 19812–19817. [Google Scholar] [CrossRef]
- Fischer, G.; Cao, X.; Cox, N.; Francis, M. The FT-IR spectra of glycine and glycylglycine zwitterions isolated in alkali halide matrices. Chem. Phys. 2005, 313, 39–49. [Google Scholar] [CrossRef]
- Parker, S.F. Assignment of the vibrational spectrum of l-cysteine. Chem. Phys. 2013, 424, 75–79. [Google Scholar] [CrossRef]
- Mohamed, M.E.; Mohammed, A.M.A. Experimental and Computational Vibration Study of Amino Acids. Int. Lett. Chem. Phys. Astron. 2013, 15, 1–17. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta—Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef]
- Vodnar, D.C.; Paucean, A.; Dulf, F.V.; Socaciu, C. HPLC characterization of lactic acid formation and FTIR fingerprint of probiotic bacteria during fermentation processes. Not. Bot. Horti Agrobot. Cluj-Napoca 2010, 38, 109–113. [Google Scholar] [CrossRef]
- Keyes, B.M.; Gedvilas, L.M.; Li, X.; Coutts, T.J. Infrared spectroscopy of polycrystalline ZnO and ZnO:N thin films. J. Cryst. Growth 2005, 281, 297–302. [Google Scholar] [CrossRef]
- Król, A.; Railean-Plugaru, V.; Pomastowski, P.; Złoch, M.; Buszewski, B. Mechanism study of intracellular zinc oxide nanocomposites formation. Colloids Surfaces A Physicochem. Eng. Asp. 2018, 553, 349–358. [Google Scholar] [CrossRef]
- Krężel, A.; Maret, W. The biological inorganic chemistry of zinc ions. Arch. Biochem. Biophys. 2016, 611, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Rulíšek, L.; Vondrášek, J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J. Inorg. Biochem. 1998, 71, 115–127. [Google Scholar] [CrossRef]
- Tang, N.; Skibsted, L.H. Zinc bioavailability from whey. Enthalpy-entropy compensation in protein binding. Food Res. Int. 2016, 89, 749–755. [Google Scholar] [CrossRef]
- Pomastowski, P.; Sprynskyy, M.; Buszewski, B. The study of zinc ions binding to casein. Colloids Surfaces B Biointerfaces 2014, 120, 21–27. [Google Scholar] [CrossRef]
- Srikant, V.; Clarke, D.R. On the optical band gap of zinc oxide. J. Appl. Phys. 1998, 83, 5447–5451. [Google Scholar] [CrossRef]
- Szmacinski, H.; Ray, K.; Lakowicz, J.R. Effect of plasmonic nanostructures and nanofilms on fluorescence resonance energy transfer. J. Biophotonics 2009, 2, 243–252. [Google Scholar] [CrossRef]
- Jana, J.; Ganguly, M.; Pal, T. Enlightening surface plasmon resonance effect of metal nanoparticles for practical spectroscopic application. RSC Adv. 2016, 6, 86174–86211. [Google Scholar] [CrossRef]
- Bah, A.; Lim, K.Y.; Wei, F.; Khursheed, A.; Sow, C.H. Fluorescence Invigoration in Carbon-Incorporated Zinc Oxide Nanowires from Passage of Field Emission Electrons. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Das, R.; Phadke, P.; Khichar, N.; Chawla, S. Plasmonic enhancement of blue fluorescence in ZnO nanoparticles. Superlattices Microstruct. 2015, 85, 658–663. [Google Scholar] [CrossRef]
- Kang, M.-J.; Pyun, J.-C.; Lee, J.-C.; Choi, Y.-J.; Park, J.-H.; Park, J.-G.; Lee, J.-G.; Choi, H.-J. Nanowire-assisted laser desorption and ionization mass spectrometry for quantitative analysis of small molecules. Rapid Commun. Mass Spectrom. 2005, 19, 3166–3170. [Google Scholar] [CrossRef]
- Pomastowski, P.; Buszewski, B. Complementarity of matrix-and nanostructure-assisted laser desorption/ionization approaches. Nanomaterials 2019, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kawasaki, H.; Yonezawa, T.; Arakawa, R. Surface-assisted laser desorption/ionization mass spectrometry (SALDI-MS) of low molecular weight organic compounds and synthetic polymers using zinc oxide (ZnO) nanoparticles. J. Mass Spectrom. 2008, 43, 1063–1071. [Google Scholar] [CrossRef]
- Degen, A.; Kosec, M. Effect of pH and impurities on the surface charge of zinc oxide in aqueous solution. J. Eur. Ceram. Soc. 2000, 20, 667–673. [Google Scholar] [CrossRef]
- Fatehah, M.O.; Aziz, H.A.; Stoll, S. Stability of ZnO Nanoparticles in Solution. Influence of pH, Dissolution, Aggregation and Disaggregation Effects. J. Colloid Sci. Biotechnol. 2014, 3, 75–84. [Google Scholar] [CrossRef]
- Reichle, R.A.; McCurdy, K.G.; Hepler, L.G. Zinc Hydroxide: Solubility Product and Hydroxy-complex Stability Constants from 12.5–75 °C. Can. J. Chem. 1975, 53, 3841–3845. [Google Scholar] [CrossRef]
- Prochowicz, D.; Sokołowski, K.; Lewiński, J. Zinc hydroxides and oxides supported by organic ligands: Synthesis and structural diversity. Coord. Chem. Rev. 2014, 270–271, 112–126. [Google Scholar] [CrossRef]
- Siddiqui, K.A.; Bharati, A.K.; Lama, P. Zinc-orotate coordination polymer: Synthesis, thermogravimetric analysis and luminescence properties. SN Appl. Sci. 2020, 2, 1–11. [Google Scholar] [CrossRef]
- Sujamol, M.S.; Athira, C.J.; Sindhu, Y.; Mohanan, K. Synthesis, spectroscopic characterization, electrochemical behaviour and thermal decomposition studies of some transition metal complexes with an azo derivative. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2010, 75, 106–112. [Google Scholar] [CrossRef]
- Basha, S.K.; Lakshmi, K.V.; Kumari, V.S. Ammonia sensor and antibacterial activities of green zinc oxide nanoparticles. Sens. Bio-Sens. Res. 2016, 10, 34–40. [Google Scholar] [CrossRef]
- Ebadi, M.; Zolfaghari, M.R.; Aghaei, S.S.; Zargar, M.; Shafiei, M.; Zahiri, H.S.; Noghabi, K.A. A bio-inspired strategy for the synthesis of zinc oxide nanoparticles (ZnO NPs) using the cell extract of cyanobacterium: Nostoc sp. EA03: From biological function to toxicity evaluation. RSC Adv. 2019, 9, 23508–23525. [Google Scholar] [CrossRef]
- Jayaseelan, C.; Rahuman, A.A.; Kirthi, A.V.; Marimuthu, S.; Santhoshkumar, T.; Bagavan, A.; Gaurav, K.; Karthik, L.; Rao, K.V.B. Novel microbial route to synthesize ZnO nanoparticles using Aeromonas hydrophila and their activity against pathogenic bacteria and fungi. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2012, 90, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Peton, V.; Le Loir, Y. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 2014, 21, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Kittana, H.; Gomes-Neto, J.C.; Heck, K.; Geis, A.L.; Segura Muñoz, R.R.; Cody, L.A.; Schmaltz, R.J.; Bindels, L.B.; Sinha, R.; Hostetter, J.M.; et al. Commensal Escherichia coliStrains can promote intestinal inflammation via differential interleukin-6 production. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Lade, H.; Park, J.H.; Chung, S.H.; Kim, I.H.; Kim, J.-M.; Joo, H.-S.; Kim, J.-S. Biofilm Formation by Staphylococcus aureus Clinical Isolates is Differentially Affected by Glucose and Sodium Chloride Supplemented Culture Media. J. Clin. Med. 2019, 8, 1853. [Google Scholar] [CrossRef]
- Allaker, R.P.; Yuan, Z. Nanoparticles and the control of oral biofilms. In Nanobiomaterials in Clinical Dentistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 243–275. ISBN 9780128158869. [Google Scholar]
- Qayyum, S.; Khan, A.U. Nanoparticles: Vs. biofilms: A battle against another paradigm of antibiotic resistance. Medchemcomm 2016, 7, 1479–1498. [Google Scholar] [CrossRef]
- Lellouche, J.; Friedman, A.; Lahmi, R.; Gedanken, A.; Banin, E. Antibiofilm surface functionalization of catheters by magnesium fluoride nanoparticles. Int. J. Nanomed. 2012, 7, 1175–1188. [Google Scholar] [CrossRef]
- Iannitelli, A.; Grande, R.; di Stefano, A.; di Giulio, M.; Sozio, P.; Bessa, L.J.; Laserra, S.; Paolini, C.; Protasi, F.; Cellini, L. Potential antibacterial activity of carvacrol-loaded poly(DL-lactide-co-glycolide) (PLGA) nanoparticles against microbial biofilm. Int. J. Mol. Sci. 2011, 12, 5039–5051. [Google Scholar] [CrossRef]
- Eshed, M.; Lellouche, J.; Matalon, S.; Gedanken, A.; Banin, E. Sonochemical coatings of ZnO and CuO nanoparticles inhibit streptococcus mutans biofilm formation on teeth model. Langmuir 2012, 28, 12288–12295. [Google Scholar] [CrossRef] [PubMed]
- Jasim, N.A.; Al-Gasha’a, F.A.; Al-Marjani, M.F.; Al-Rahal, A.H.; Abid, H.A.; Al-Kadhmi, N.A.; Jakaria, M.; Rheima, A.M. ZnO nanoparticles inhibit growth and biofilm formation of vancomycin-resistant S. aureus (VRSA). Biocatal. Agric. Biotechnol. 2020, 29, 101745. [Google Scholar] [CrossRef]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomedicine 2017, 12, 1227–1249. [Google Scholar] [CrossRef]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-strategies to fight multidrug resistant bacteria-“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.S.H.; Karthikeyan, C.; Ahamed, A.P.; Thajuddin, N.; Alharbi, N.S.; Alharbi, S.A.; Ravi, G. In vitro antibacterial activity of ZnO and Nd doped ZnO nanoparticles against ESBL producing Escherichia coli and Klebsiella pneumoniae. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Ansari, M.A.; Khan, H.M.; Khan, A.A.; Sultan, A.; Azam, A. Synthesis and characterization of the antibacterial potential of ZnO nanoparticles against extended-spectrum β-lactamases-producing escherichia coli and klebsiella pneumoniae isolated from a tertiary care hospital of North India. Appl. Microbiol. Biotechnol. 2012, 94, 467–477. [Google Scholar] [CrossRef]


| Tested Material | Klebsiella Pneumoniae | Escherichia Coli | Pseudomonas Aeruginosa | Staphylococcus Aureus |
|---|---|---|---|---|
| Access no. | ATCC BAA-1144 | ATCC 25922 | ATCC 15441 | ATCC 11632 |
| ZnO NCs | 172.5 μg/mL | 172.5 μg/mL | - | 86.25 μg/mL |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pomastowski, P.; Król-Górniak, A.; Railean-Plugaru, V.; Buszewski, B. Zinc Oxide Nanocomposites—Extracellular Synthesis, Physicochemical Characterization and Antibacterial Potential. Materials 2020, 13, 4347. https://doi.org/10.3390/ma13194347
Pomastowski P, Król-Górniak A, Railean-Plugaru V, Buszewski B. Zinc Oxide Nanocomposites—Extracellular Synthesis, Physicochemical Characterization and Antibacterial Potential. Materials. 2020; 13(19):4347. https://doi.org/10.3390/ma13194347
Chicago/Turabian StylePomastowski, Paweł, Anna Król-Górniak, Viorica Railean-Plugaru, and Bogusław Buszewski. 2020. "Zinc Oxide Nanocomposites—Extracellular Synthesis, Physicochemical Characterization and Antibacterial Potential" Materials 13, no. 19: 4347. https://doi.org/10.3390/ma13194347
APA StylePomastowski, P., Król-Górniak, A., Railean-Plugaru, V., & Buszewski, B. (2020). Zinc Oxide Nanocomposites—Extracellular Synthesis, Physicochemical Characterization and Antibacterial Potential. Materials, 13(19), 4347. https://doi.org/10.3390/ma13194347









