Electrochemical Bacterial Enrichment from Natural Seawater and Its Implications in Biocorrosion of Stainless-Steel Electrodes
Abstract
1. Introduction
2. Materials and Methods
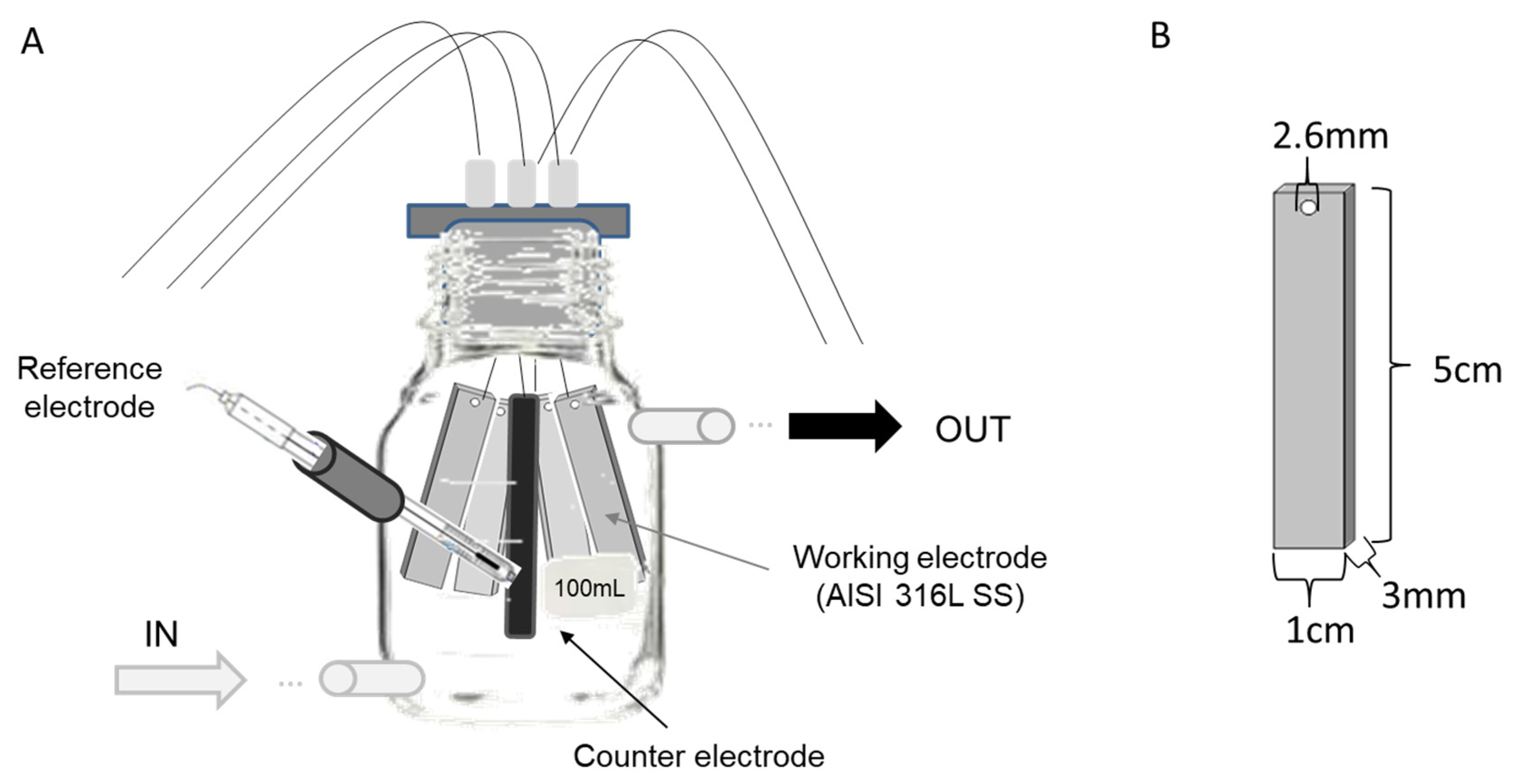
2.1. Reactor Configuration
2.2. Surface Analysis for Corrosion Evaluation
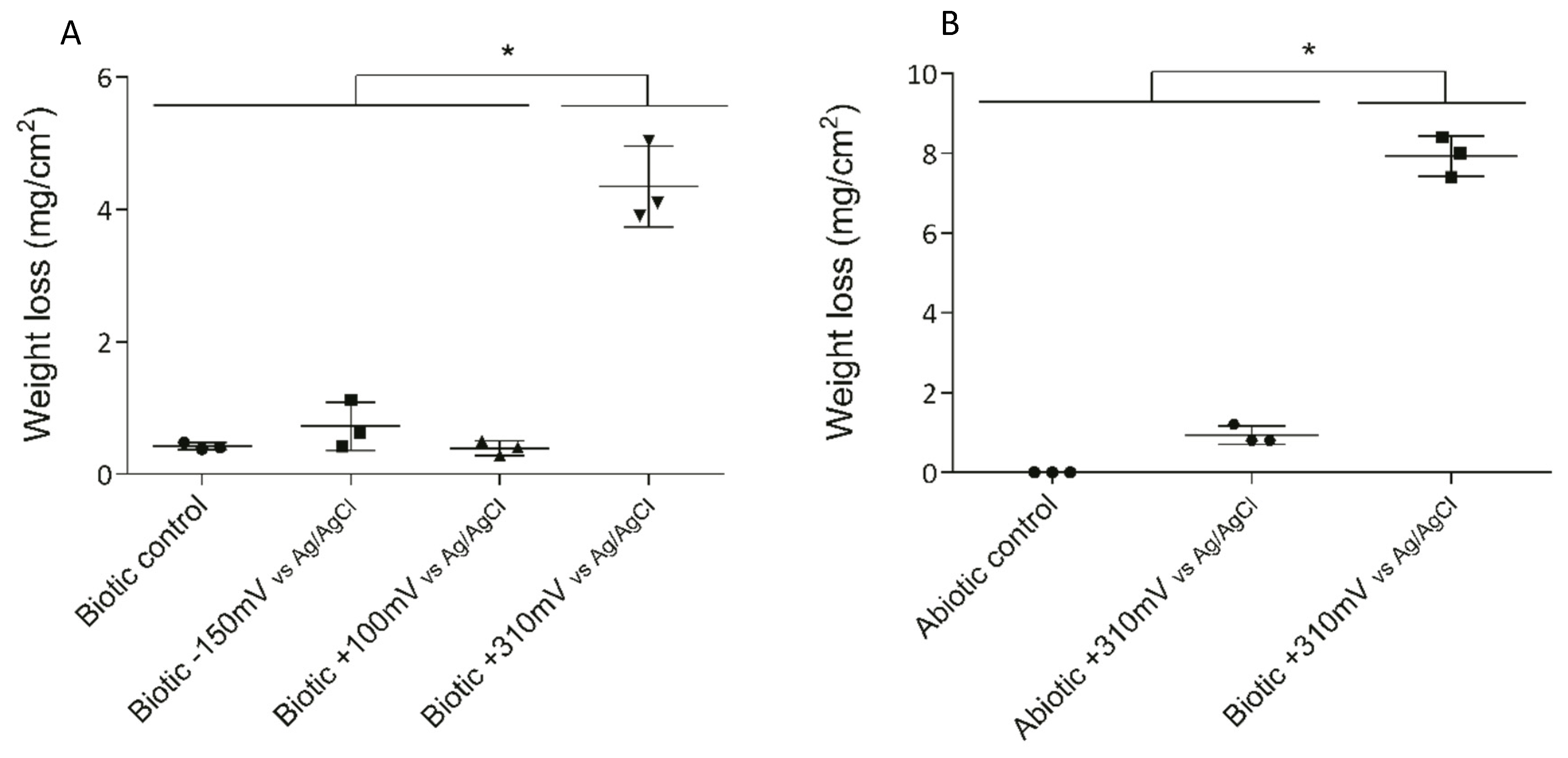
2.2.1. Weight Loss
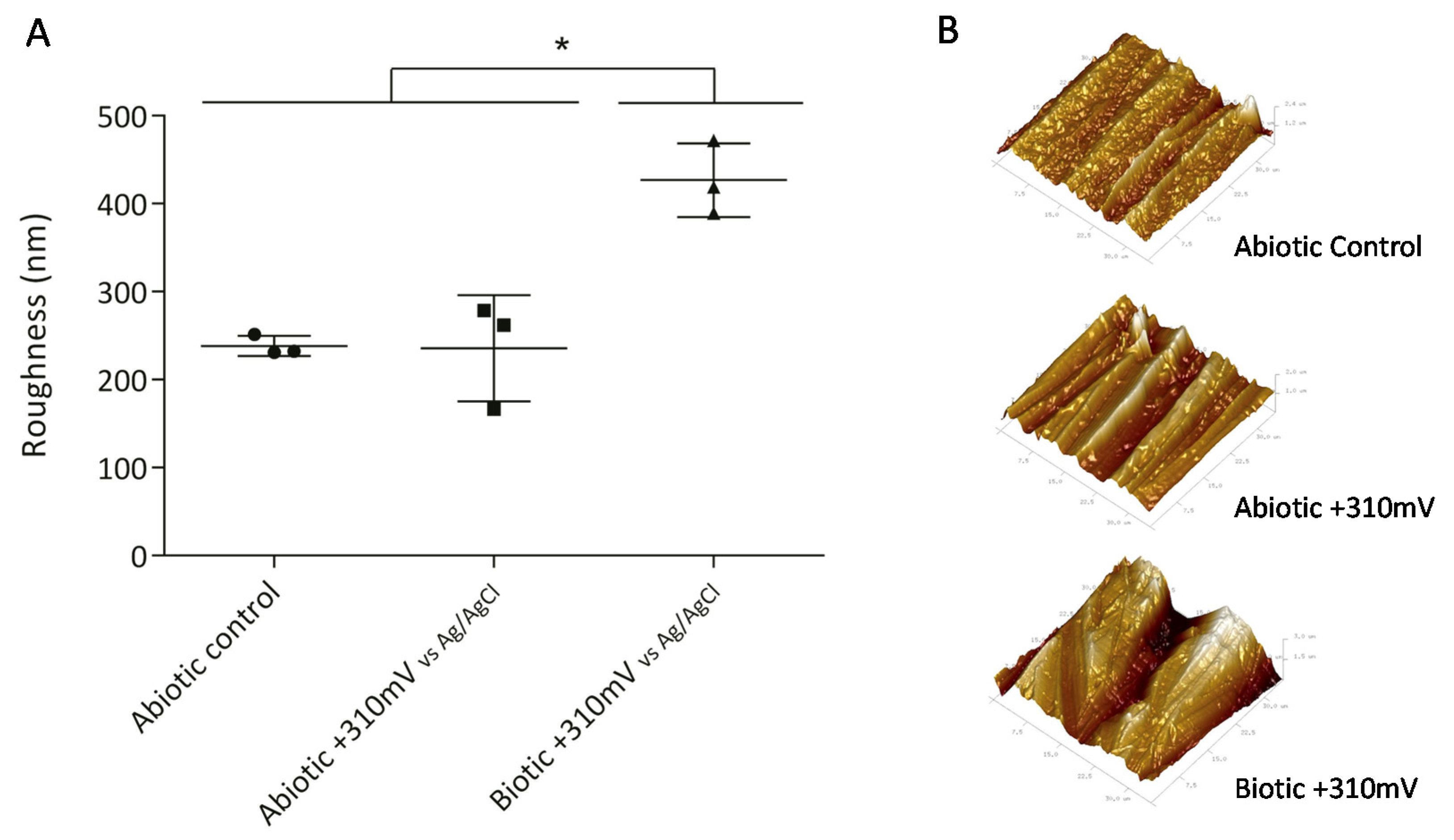
2.2.2. Atomic Force Microscopy (AFM)
2.3. Microbial Community Analysis
2.3.1. DNA Extraction
2.3.2. Fragment Analysis (FA) of 16S rRNA Genes
2.3.3. 16S rRNA Gene Clone Library Construction
2.3.4. Analysis of Clone Sequences
2.3.5. Amplicon Analysis of 16 rRNA Gene Sequences
2.3.6. Statistical Analysis of Fragment and Amplicon Analysis Profiles
2.3.7. Similarity Analysis between Cloning and Amplicon Sequences
2.3.8. Microscopy Analysis
3. Results and Discussion
3.1. Electrode Deterioration
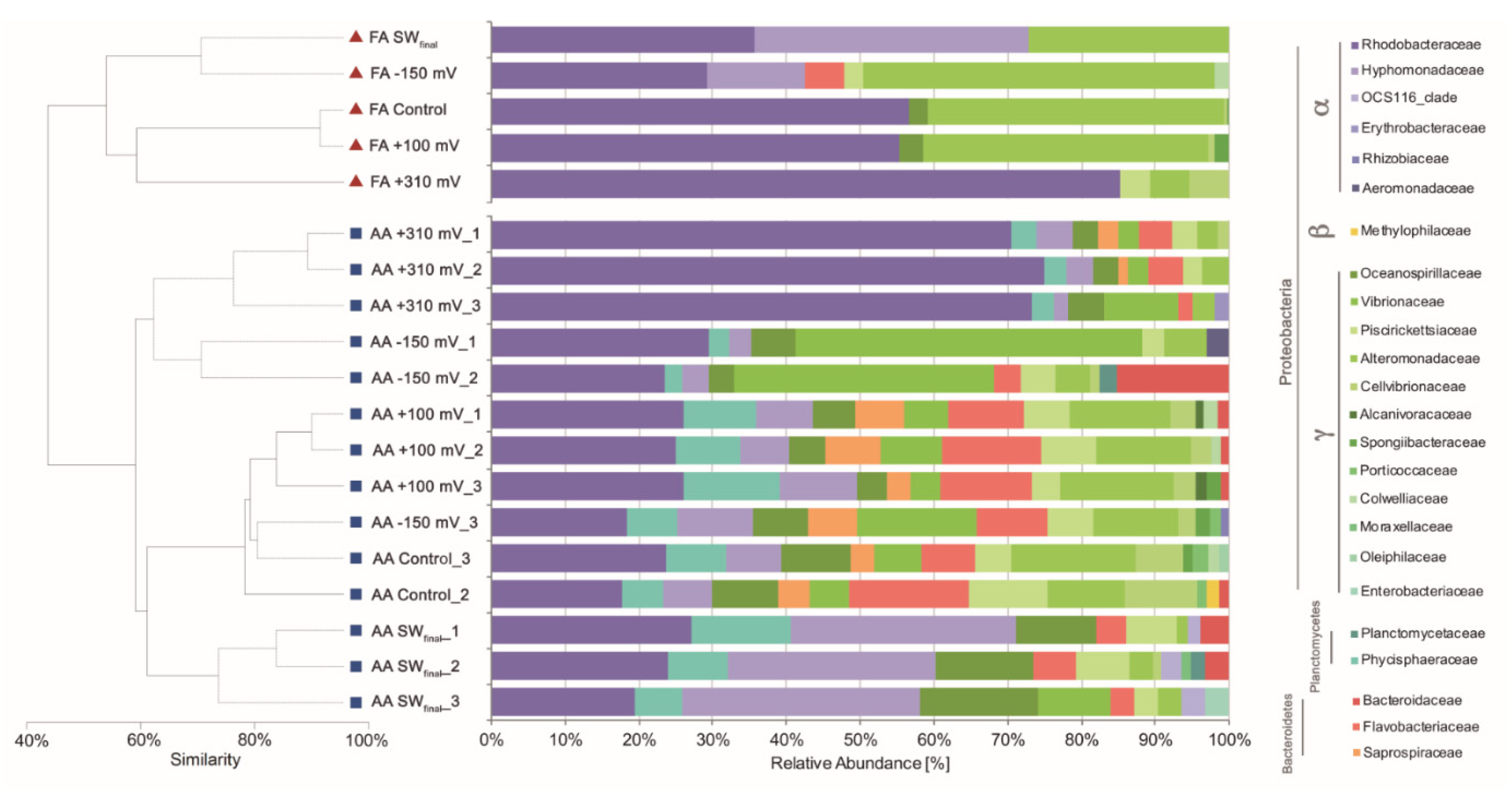
3.2. Bacterial Enrichment Using Overpotentials
3.3. Metabolic Inference of Electrochemically Enriched Bacterial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rao, J.R.; Richter, G.J.; Von Sturm, F.; Weidlich, E. The Performance of Glucose Electrodes and the Characteristics of Different. Biofuel Cell Constructions. Bioelectrochemistry Bioenerg. 1976, 3, 139–150. [Google Scholar] [CrossRef]
- Roller, S.D.; Bennetto, H.P.; Delaney, G.M.; Mason, J.R.; Stirling, J.L.; Thurston, C.F. Electron-transfer coupling in microbial fuel cells: 1. Comparison of redox-mediator reduction rates and respiratory rates of bacteria. J. Chem. Technol. Biotechnol. 1984, 34, 3–12. [Google Scholar] [CrossRef]
- He, Z.; Kan, J.; Wang, Y.; Huang, Y.; Mansfeld, F.; Nealson, K.H. Electricity production coupled to ammonium in a microbial fuel cell. Environ. Sci. Technol. 2009, 43, 3391–3397. [Google Scholar] [CrossRef] [PubMed]
- Berks, B.C.; Ferguson, S.J.; Moir, J.W.B.; Richardson, D.J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1995, 2728, 97–173. [Google Scholar] [CrossRef]
- Li, W.-W.; Yu, H.-Q. Stimulating sediment bioremediation with benthic microbial fuel cells. Biotechnol. Adv. 2015, 33, 1–12. [Google Scholar] [CrossRef]
- Jiang, C.; Yang, Q.; Wang, D.; Zhong, Y.; Chen, F.; Li, X.; Zeng, G.; Li, X. Simultaneous perchlorate and nitrate removal coupled with electricity generation in autotrophic denitrifying biocathode microbial fuel cell. Chem. Eng. J. 2017, 308, 783–790. [Google Scholar] [CrossRef]
- Virdis, B.; Rabaey, K.; Rozendal, R.A.; Yuan, Z.; Keller, J. Simultaneous nitrification, denitrification and carbon removal in microbial fuel cells. Water Res. 2010, 44, 2970–2980. [Google Scholar] [CrossRef]
- Wang, H.; Ren, Z.J. A comprehensive review of microbial electrochemical systems as a platform technology. Biotechnol. Adv. 2013, 31, 1796–1807. [Google Scholar] [CrossRef]
- Rowe, A.R.; Chellamuthu, P.; Lam, B.; Okamoto, A.; Nealson, K.H. Marine sediments microbes capable of electrode oxidation as a surrogate for lithotrophic insoluble substrate metabolism. Front. Microbiol. 2015, 6, 1–15. [Google Scholar] [CrossRef]
- Logan, B.E. Scaling up microbial fuel cells and other bioelectrochemical systems. Appl. Microbiol. Biotechnol. 2010, 85, 1665–1671. [Google Scholar] [CrossRef]
- Kim, J.R.; Jung, S.H.; Regan, J.M.; Logan, B.E. Electricity generation and microbial community analysis of alcohol powered microbial fuel cells. Bioresour. Technol. 2007, 98, 2568–2577. [Google Scholar] [CrossRef] [PubMed]
- Baudler, A.; Schmidt, I.; Langner, M.; Greiner, A.; Schröder, U. Does it have to be carbon? Metal anodes in microbial fuel cells and related bioelectrochemical systems. Energy Environ. Sci. 2015, 8, 2048–2055. [Google Scholar] [CrossRef]
- Selembo, P.A.; Merrill, M.D.; Logan, B.E. The use of stainless steel and nickel alloys as low-cost cathodes in microbial electrolysis cells. J. Power Sources 2009, 190, 271–278. [Google Scholar] [CrossRef]
- Dumas, C.; Mollica, A.; Féron, D.; Basséguy, R.; Etcheverry, L.; Bergel, A. Marine microbial fuel cell: Use of stainless steel electrodes as anode and cathode materials. Electrochim. Acta 2007, 53, 468–473. [Google Scholar] [CrossRef]
- Call, D.F.; Merrill, M.D.; Logan, B.E. High Surface Area Stainless Steel Brushes as Cathodes in Microbial Electrolysis Cells. Environ. Sci. Technol. 2009, 43, 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Pocaznoi, D.; Calmet, A.; Etcheverry, L.; Erable, B.; Bergel, A. Stainless steel is a promising electrode material for anodes of microbial fuel cells. Energy Environ. Sci. 2012, 5, 9645–9652. [Google Scholar] [CrossRef]
- Tender, L.M.; Reimers, C.E.; Iii, H.A.S.; Holmes, D.E.; Bond, D.R.; Lowy, D.A.; Pilobello, K.; Fertig, S.J.; Lovley, D.R. Harnessing microbially generated power on the seafloor. Nat. Biotechnol. 2002, 20, 821–825. [Google Scholar] [CrossRef]
- Miceli, J.F.; Parameswaran, P.; Kang, D.; Krajmalnik-brown, R. Enrichment and analysis of anode-respiring bacteria from diverse anaerobic inocula. Environ. Sci. Technol. 2012, 46, 10349–10355. [Google Scholar] [CrossRef]
- Zhan, G.; Zhang, L.; Tao, Y.; Wang, Y.; Zhu, X.; Li, D. Anodic ammonia oxidation to nitrogen gas catalyzed by mixed biofilms in bioelectrochemical systems. Electrochim. Acta 2014, 135, 345–350. [Google Scholar] [CrossRef]
- Wang, Z.; Leary, D.H.; Malanoski, A.H.P.; Li, R.W.; Judson Hervey, W.; Eddie, B.J.; Tender, G.S.; Yanosky, S.G.; Vora, G.J.; Tender, L.M.; et al. A previously uncharacterized, nonphotosynthetic member of the chromatiaceae is the primary CO2-fixing constituent in a self-regenerating biocathode. Appl. Environ. Microbiol. 2015, 81, 699–712. [Google Scholar] [CrossRef]
- Torres, C.I.; Krajmalnik-Brown, R.; Parameswaran, P.; Marcus, A.K.; Wanger, G.; Gorby, Y.A.; Rittmann, B.E. Selecting anode-respiring bacteria based on anode potential: Phylogenetic, electrochemical, and microscopic characterization. Environ. Sci. Technol. 2009, 43, 9519–9524. [Google Scholar] [CrossRef] [PubMed]
- Juretschko, S.; Loy, A.; Lehner, A.; Wagner, M. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 2002, 25, 84–99. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.A.; Daille, L.; Aguirre, J.; Galarce, C.; Armijo, F.; la Iglesia, R.D.; Pizarro, G.; Vargas, I.; Walczak, M. Corrosion of stainless steel in simulated tide of fresh natural seawater of south east pacific. Int. J. Electrochem. Sci. 2016, 11, 6873–6885. [Google Scholar] [CrossRef]
- Ter Heijne, A.; Strik, D.P.B.T.B.; Hamelers, H.V.; Buisman, C.J. Cathode potential and mass transfer determine performance of oxygen reducing biocathodes in microbial fuel cells. Environ. Sci. Technol. 2010, 44, 7151–7156. [Google Scholar] [CrossRef]
- Boivin, M.E.Y.; Greve, G.D.; Kools, S.A.E.; van der Wurff, A.W.G.; Leeflang, P.; Smit, E.; Breure, A.M.; Rutgers, M.; van Straalen, N.M. Discriminating between effects of metals and natural variables in terrestrial bacterial communities. Appl. Soil Ecol. 2006, 34, 103–113. [Google Scholar] [CrossRef]
- Henríquez-castillo, C.; Rodríguez-marconi, S.; Rubio, F.; Trefault, N.; Andrade, S.; Iglesia, R. De Eukaryotic picophytoplankton community response to copper enrichment in a metal-perturbed coastal environment. Phycol. Res. 2015, 63, 189–196. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Osborne, C.A.; Rees, G.N.; Bernstein, Y.; Janssen, P.H. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Appl. Environ. Microbiol. 2006, 72, 1270–1278. [Google Scholar] [CrossRef][Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.E.W.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Beals, E.W. Bray-curtis ordination: An effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res. 1984, 14, 1–55. [Google Scholar] [CrossRef]
- Warwick, R.M.; Clarke, K.R. Change in marine communities: An approach to statistical analysis and interpretation. Am. J. Clin. Nutr. 2007, 85, 1236–1243. [Google Scholar]
- Olesen, B.H.; Avci, R.; Lewandowski, Z. Manganese dioxide as a potential cathodic reactant in corrosion of stainless steels. Corros. Sci. 2000, 42, 211–227. [Google Scholar] [CrossRef]
- Trigodet, F.; Larché, N.; Morrison, H.G.; Jebbar, M.; Thierry, D.; Maignien, L. Electroactive bacteria associated with stainless steel ennoblement in seawater. Front. Microbiol. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Chamritski, I.G.; Burns, G.R.; Webster, B.J.; Laycock, N.J. Effect of iron-oxidizing bacteria on pitting of stainless steel. Corrosion 2004, 60, 658–669. [Google Scholar] [CrossRef]
- Eddie, B.J.; Wang, Z.; Malanoski, A.P.; Hall, R.J.; Oh, S.D.; Heiner, C.; Lin, B.; Strycharz-glaven, S.M.; Eddie, B.J. ‘Candidatus Tenderia electrophaga’, an uncultivated electroautotroph from a biocathode enrichment. Int. J. Syst. Evol. Microbiol. 2016, 66, 2178–2185. [Google Scholar] [CrossRef]
- Eddie, B.J.; Wang, Z.; Iv, W.J.H.; Leary, D.H.; Malanoski, A.P.; Tender, L.M.; Lin, B.; Strycharz-glaven, S.M. Metatranscriptomics Supports the Mechanism for Biocathode Electroautotrophy by “Candidatus Tenderia electrophaga”. Mol. Biol. Physiol. 2017, 2, 1–13. [Google Scholar] [CrossRef]
- Compère, C.; Jaffré, P.; Festy, D. Aging of type 316L stainless steel in seawater: Relationship between open-circuit potential, exposure time, and pitting potential. Corros. Sci. 1996, 52, 496–501. [Google Scholar] [CrossRef]
- Erable, B.; Byrne, N.; Etcheverry, L.; Achouak, W.; Bergel, A. Single medium microbial fuel cell: Stainless steel and graphite electrode materials select bacterial communities resulting in opposite electrocatalytic activities. Int. J. Hydrogen Energy 2017, 42, 26059–26067. [Google Scholar] [CrossRef]
- van Dorst, J.; Bissett, A.; Palmer, A.S.; Brown, M.; Snape, I.; Stark, J.S.; Raymond, B.; Mckinlay, J.; Ji, M.; Winsley, T.; et al. Community fingerprinting in a sequencing world. Fems Microbiol. Ecol. 2014, 89, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Gobet, A.; Boetius, A.; Ramette, A. Ecological coherence of diversity patterns derived from classical fingerprinting and next generation sequencing techniques. Environ. Microbiol. 2014, 16, 2672–2681. [Google Scholar] [CrossRef]
- Vandecandelaere, I.; Segaert, E.; Mollica, A.; Faimali, M.; Vandamme, P. Phaeobacter caeruleus sp. nov., a blue-coloured, colony-forming bacterium isolated from a marine electroactive biofilm. Int. J. Syst. Evol. Microbiol. 2009, 59, 1209–1214. [Google Scholar] [CrossRef]
- Parot, S.; Vandecandelaere, I.; Cournet, A.; Délia, M.; Vandamme, P.; Bergé, M.; Roques, C.; Bergel, A. Catalysis of the electrochemical reduction of oxygen by bacteria isolated from electro-active biofilms formed in seawater. Bioresour. Technol. 2011, 102, 304–311. [Google Scholar] [CrossRef]
- Allgaier, M.; Uphoff, H.; Felske, A.; Wagner-do, I. aerobic anoxygenic photosynthesis in roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 2003, 69, 5051–5059. [Google Scholar] [CrossRef]
- Li, H.; Zhou, E.; Zhang, D.; Xu, D.; Xia, J.; Yang, C.; Feng, H.; Jiang, Z.; Li, X.; Gu, T.; et al. Microbiologically influenced corrosion of 2707 hyper-duplex stainless steel by marine pseudomonas aeruginosa biofilm. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Xia, J.; Yang, C.; Xu, D.; Sun, D.; Nan, L.; Sun, Z.; Li, Q.; Gu, T.; Yang, K. Laboratory investigation of the microbiologically influenced corrosion (MIC) resistance of a novel Cu-bearing 2205 duplex stainless steel in the presence of an aerobic marine Pseudomonas aeruginosa biofilm. Biofouling 2015, 31, 481–492. [Google Scholar] [CrossRef]
- Rajasekar, A.; Ting, Y.P. Role of inorganic and organic medium in the corrosion behavior of bacillus megaterium and pseudomonas sp. in stainless steel SS 304. Ind. Eng. Chem. Res. 2011, 50, 12534–12541. [Google Scholar] [CrossRef]
- Vandecandelaere, I.; Nercessian, O.; Faimali, M.; Segaert, E.; Mollica, A.; Achouak, W.; De Vos, P.; Vandamme, P. Bacterial diversity of the cultivable fraction of a marine electroactive biofilm. Bioelectrochemistry 2010, 78, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Erable, B.; Roncato, M.-A.; Achouak, W.; Bergel, A. Sampling natural biofilms: A new route to build efficient microbial anodes. Environ. Sci. Technol. 2009, 43, 3194–3199. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.; Peixoto, L.; Ribeiro, D.C.; Parpot, P.; Brito, A.G.; Nogueira, R. Bioelectrochemistry towards implementation of a benthic microbial fuel cell in lake Furnas (Azores): Phylogenetic af fi liation and electrochemical activity of sediment bacteria. Bioelectrochemistry 2010, 78, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Ringelberg, D.B.; Foley, K.L.; Reynolds, C.M. Electrogenic capacity and community composition of anodic biofilms in soil-based bioelectrochemical systems. Appl. Microbiol. Biotechnol. 2011, 90, 1805–1815. [Google Scholar] [CrossRef]
- Malanoski, A.P.; Lin, B.; Brian, J.; Wang, Z.; Iv, W.J.H.; Glaven, S.M. Relative abundance of ‘Candidatus Tenderia electrophaga’ is linked to cathodic current in an aerobic biocathode community. Microb. Biotechnol. 2018, 11, 98–111. [Google Scholar] [CrossRef]
- Barco, R.A.; Emerson, D.; Sylvan, J.B.; Orcutt, B.N.; Meyers, E.J.; Ramírez, G.A.; Zhong, J.D.; Edwards, J. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl. Environ. Microbiol. 2015, 81, 5927–5937. [Google Scholar] [CrossRef]
- Oldham, A.L.; Steinberg, M.K.; Duncan, K.E.; Makama, Z.; Duncan, K.E. Molecular methods resolve the bacterial composition of natural marine biofilms on galvanically coupled stainless steel cathodes. J. Ind. Microbiol. Biotechnol. 2017, 44, 167–180. [Google Scholar] [CrossRef]
- Holland, R.L.; Cooper, B.H.; Helgeson, N.G.; Mccracken, A.W. Automated detection of microbial growth in blood automated detection of microbial growth in blood cultures by using stainless-steel electrodes. J. Clin. Microbiol. 1980, 12, 180–184. [Google Scholar] [CrossRef]
- Strycharz-glaven, S.M.; Glaven, R.H.; Wang, Z.; Zhou, J.; Vora, G.J.; Tender, L.M. Electrochemical investigation of a microbial solar cell reveals a nonphotosynthetic biocathode catalyst. Appl. Environ. Microbiol. 2013, 79, 3933–3942. [Google Scholar] [CrossRef]
- Leary, D.H.; Hervey, J.W.; Malanoski, A.P.; Wang, Z.; Eddie, B.J.; Tender, G.S.; Vora, G.J.; Tender, L.M.; Lin, B.; Strycharz-Glaven, S.M. Metaproteomic evidence of changes in protein expression following a change in electrode potential in a robust biocathode microbiome. Proteomics 2015, 15, 3486–3496. [Google Scholar] [CrossRef]
- Malik, S.; Drott, E.; Grisdela, P.; Lee, J.; Lee, C.; Lowy, D.A.; Tender, L.M. A self-assembling self-repairing microbial photoelectrochemical solar cell. Energy Environ. Sci. 2009, 2, 292–298. [Google Scholar] [CrossRef]
- Rimboud, M.; Bergel, A.; Erable, B. Multiple electron transfer systems in oxygen reducing biocathodes revealed by different conditions of aeration/agitation. Bioelectrochemistry 2016, 110, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Moran, M.A. Evolutionary ecology of the marine roseobacter clade. Microbiol. Mol. Biol. Rev. 2014, 78, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2000, 66, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Li, T.; Chen, M.; Huang, G. Cross-ocean distribution of rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 2008, 74, 52–60. [Google Scholar] [CrossRef]
- Chen, G.W.; Cha, J.H.; Choi, S.J.; Lee, T.H.; Kim, C.W. Characterization of an open biocathode microbial fuel cell for electricity generation and effluent polish. Korean J. Chem. Eng. 2010, 27, 828–835. [Google Scholar] [CrossRef]
- Abraham, W.; Strompl, C.; Meyer, H.; Lindholst, S.; Moore, E.R.B.; Christ, R.; Vancanneyt, M.; Tindali, B.J.; Bennasar, A.; Smit, J.; et al. Phylogeny and polyphasic taxonomy of caulobacter species. Proposal of maricaulis gen. nov. with maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera brevundirnonas and caulobacter. Int. J. Syst. Bacteriol. 1999, 49, 1053–1073. [Google Scholar] [CrossRef]
- Moura, V.; Ribeiro, I.; Moriggi, P.; Capão, A.; Salles, C.; Bitati, S.; Procópio, L. The influence of surface microbial diversity and succession on microbiologically influenced corrosion of steel in a simulated marine environment. Arch. Microbiol. 2018, 200, 1447–1456. [Google Scholar] [CrossRef]
- Yuan, L.N.; Ren, L.F.; Li, Y.T.; Han, W.J.; Yu, Y.; Chu, Y.N.; Liu, G.M.; Yu, D.; Teng, M.J.; Wang, L.; et al. A complete genome assembly of glaciecola mesophila sp. nov. sequenced by using BIGIS-4 sequencer system. Sci. China Life Sci. 2011, 54, 835–840. [Google Scholar] [CrossRef][Green Version]
- Darus, L.; Ledezma, P.; Keller, J.; Freguia, S. Marine phototrophic consortia transfer electrons to electrodes in response to reductive stress. Photosynth Res. 2016, 127, 347–354. [Google Scholar] [CrossRef]
- Schwermer, C.U.; Lavik, G.; Abed, R.M.M.; Dunsmore, B.; Ferdelman, T.G.; Stoodley, P.; Gieseke, A.; De Beer, D. Impact of nitrate on the structure and function of bacterial biofilm communities in pipelines used for injection of seawater into oil fields. Appl. Environ. Microbiol. 2008, 74, 2841–2851. [Google Scholar] [CrossRef] [PubMed]
- Janvier, M.; Grimont, P.A.D. The genus Methylophaga, a new line of descent within phylogenetic branch γ of proteobacteria. Res. Microbiol. 1995, 146, 543–550. [Google Scholar] [CrossRef]
- Lo, N.; Kim, K.H.; Baek, K.; Jia, B.; Jeon, C.O. Aestuariicella hydrocarbonica gen. nov., sp. Nov., an aliphatic hydrocarbon-degrading bacterium isolated from a sea tidal flat. Int. J. Syst. Evol. Microbiol. 2015, 65, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.D. Lewinella agarilytica sp. nov., a novel marine bacterium of the phylum bacteroidetes, isolated from beach sediment. Int. J. Syst. Evol. Microbiol. 2007, 57, 2814–2818. [Google Scholar] [CrossRef]
- Graeber, I.; Kaesler, I.; Borchert, M.S.; Dieckmann, R.; Pape, T.; Lurz, R.; Nielsen, P.; von Döhren, H.; Michaelis, W.; Szewzyk, U. Spongiibacter marinus gen. nov., sp. nov., a halophilic marine bacterium isolated from the boreal sponge Haliclona sp. 1. Int. J. Syst. Evol. Microbiol. 2008, 58, 585–590. [Google Scholar] [CrossRef]
| Family | Genus | Relative Abundance (%) | |||
|---|---|---|---|---|---|
| AA | |||||
| +100 mV | −150 mV | +310 mV | Control | ||
| Rhodobacteraceae | Roseobacter | 4.3 | 3.9 | 34.0 | 6.4 |
| Phaeobacter | 6.0 | 4.8 | 18.8 | 5.9 | |
| Sulfitobacter | 7.2 | 7.9 | 15.2 | 3.5 | |
| Ruegeria | 1.3 | 1.0 | 0.5 | 1.2 | |
| Labrenzia | 1.1 | 0.0 | 0.1 | 0.1 | |
| Vibrionaceae | Vibrio | 4.0 | 3.3 | 3.8 | 5.0 |
| Photobacterium | 0.1 | 0.0 | 0.0 | 0.0 | |
| Aliivibrio | 0.0 | 0.0 | 0.0 | 0.0 | |
| Hyphomonadaceae | Hyphomonas | 4.3 | 3.4 | 3.6 | 5.1 |
| Maricaulis | 1.0 | 0.3 | 0.7 | 0.8 | |
| Flavobacteriaceae | Muricauda | 5.3 | 2.1 | 1.7 | 3.7 |
| Maribacter | 0.0 | 0.0 | 0.0 | 0.0 | |
| Cellulophaga | 0.0 | 0.0 | 0.0 | 0.0 | |
| Alteromonadaceae | Alteromonas | 0.6 | 0.1 | 2.5 | 11.5 |
| Glaciecola | 7.3 | 5.9 | 0.0 | 0.6 | |
| Marinobacter | 2.9 | 0.2 | 0.0 | 0.2 | |
| Phycisphaeraceae | Plantomycete | 10.3 | 4.2 | 2.2 | 6.6 |
| Oceanospirillaceae | Neptuniibacter | 3.7 | 4.3 | 3.3 | 6.5 |
| Amphritea | 0.1 | 0.0 | 0.0 | 0.0 | |
| Oleibacter | 0.7 | 0.1 | 0.0 | 1.1 | |
| Piscirickettsiaceae | Methylophaga | 5.1 | 3.3 | 2.1 | 6.9 |
| Cellvibrionaceae | Aestuariicella | 2.3 | 0.8 | 0.5 | 4.5 |
| Saprospiraceae | Lewinella | 5.5 | 1.7 | 1.3 | 1.7 |
| Bacteroidaceae | Bacteroides | 1.1 | 4.6 | 0.0 | 0.5 |
| Colwelliaceae | Colwellia | 0.9 | 0.0 | 0.0 | 0.7 |
| Spongiibacteraceae | Spongiibacter | 0.5 | 0.5 | 0.0 | 0.6 |
| Family | Most Abundant Genus Identified by AA | % Similarity | Most Abundant Genus Identified by FA | Metabolism and Environments Wherein They Have Been Reported |
|---|---|---|---|---|
| Rhodobacteraceae | Roseobacter | 90 | Roseobacter | 1. Aerobic anoxygenic photosynthesis [47]. |
| 2. Identified as primary colonizers on surfaces exposed to seawater [64]. | ||||
| 3. Reported as EAB with high efficiency in the catalysis of the oxygen reduction reaction [46]. | ||||
| Pheobacter | 93 | Pheobacter | 1. Reported in polarized stainless steel cathode [45]. | |
| 2. Association with Roseobacter during primary colonization [45]. | ||||
| 3. Aerobic anoxygenic photosynthesis [45]. | ||||
| Sulfitobacter | 80 | Sulfitobacter | 1. Reported on the bacterial communities associated with the early stages of marine corrosion of carbon steel [65]. | |
| 2. Reported as EAB with high efficiency in the catalysis of the oxygen reduction reaction [46]. | ||||
| 3. Has been found in both anodic and cathodic biofilms [42]. | ||||
| Vibrionaceae | Vibrio | - | - | 1. Reported in graphite bioanodes present in bioelectrochemical systems [54]. |
| Hyphomonadaceae | Hyphomonas | 99 | Hyphomonas | 1. Identified in graphite biocathodes present in an MFC [66]. |
| Maricaulis | 90 | Maricaulis | 1. Identified as a typical bacterioplankton in marine ecosystems [67]. | |
| 2. Reported as the primary colonizer in biofilm developed on stainless steel [68]. | ||||
| Flavobacteriaceae | Muricauda | 87 | Muricauda | 1. Reported in a biocathode microbial community [52]. |
| Alteromonadaceae | Alteromonas | 85 | Alteromonas | 1. Reported on electrochemically active biofilms [51]. |
| 2. Chemo-heterotrophic halophytes [51]. | ||||
| Glaciecola | 87 | Glaciecola | 1. Aerobic chemo-heterotrophic bacteria [69]. | |
| 2. Reported as predominant bacteria in electroactive biofilms on stainless steel electrodes [42]. | ||||
| Phycisphaeraceae | Planctomycetes | - | - | 1. Reported in marine phototrophic consortia that can transfer electrons to electrodes in response to reductive stress [70]. |
| Oceanospirillaceae | Neptuniibacter | - | - | 1. Reported in microbial community associated with stainless steel coupons [56]. |
| Piscirickettsiaceae | Methylophaga | - | - | 1. Reported in stainless steel and carbon steel cathodes [57]. |
| 2. Has been reported to reduce nitrate to nitrite [71]. | ||||
| 3. Halophilic methylotrophic metabolism [72]. | ||||
| Cellvibrionaceae | Aestuariicella | - | - | 1. Aliphatic hydrocarbon-degrading bacterium [73]. |
| 2. Not yet reported in biofilms associated with stainless steel [73]. | ||||
| Saprospiraceae | Lewinella | - | - | 1. Isolated from marine sediment [74]. |
| 2. Not yet reported in biofilms associated with stainless steel [74]. | ||||
| Bacteroidaceae | Bacteroides | - | - | 1. Reported on stainless steel electrodes [58]. |
| Colwelliaceae | Colwellia | 89 | Colwellia | 1. Reported on carbon steel cathodes [71]. |
| 2. Identified in a marine biofilm exposed to high concentration of nitrate [71]. | ||||
| Spongiibacteraceae | Spongiibacter | 92 | Spongiibacter | 1. Halophilic marine bacterium [75]. |
| 2. Not yet reported in biofilms associated with stainless steel [75]. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De La Fuente, M.J.; Daille, L.K.; De la Iglesia, R.; Walczak, M.; Armijo, F.; Pizarro, G.E.; Vargas, I.T. Electrochemical Bacterial Enrichment from Natural Seawater and Its Implications in Biocorrosion of Stainless-Steel Electrodes. Materials 2020, 13, 2327. https://doi.org/10.3390/ma13102327
De La Fuente MJ, Daille LK, De la Iglesia R, Walczak M, Armijo F, Pizarro GE, Vargas IT. Electrochemical Bacterial Enrichment from Natural Seawater and Its Implications in Biocorrosion of Stainless-Steel Electrodes. Materials. 2020; 13(10):2327. https://doi.org/10.3390/ma13102327
Chicago/Turabian StyleDe La Fuente, María José, Leslie K. Daille, Rodrigo De la Iglesia, Magdalena Walczak, Francisco Armijo, Gonzalo E. Pizarro, and Ignacio T. Vargas. 2020. "Electrochemical Bacterial Enrichment from Natural Seawater and Its Implications in Biocorrosion of Stainless-Steel Electrodes" Materials 13, no. 10: 2327. https://doi.org/10.3390/ma13102327
APA StyleDe La Fuente, M. J., Daille, L. K., De la Iglesia, R., Walczak, M., Armijo, F., Pizarro, G. E., & Vargas, I. T. (2020). Electrochemical Bacterial Enrichment from Natural Seawater and Its Implications in Biocorrosion of Stainless-Steel Electrodes. Materials, 13(10), 2327. https://doi.org/10.3390/ma13102327






