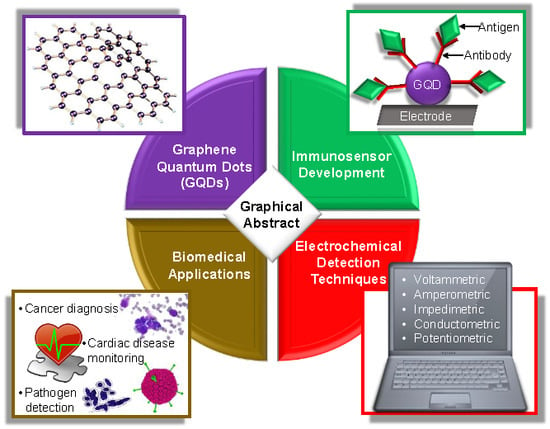
Graphene Quantum Dot-Based Electrochemical Immunosensors for Biomedical Applications
Abstract
1. Introduction
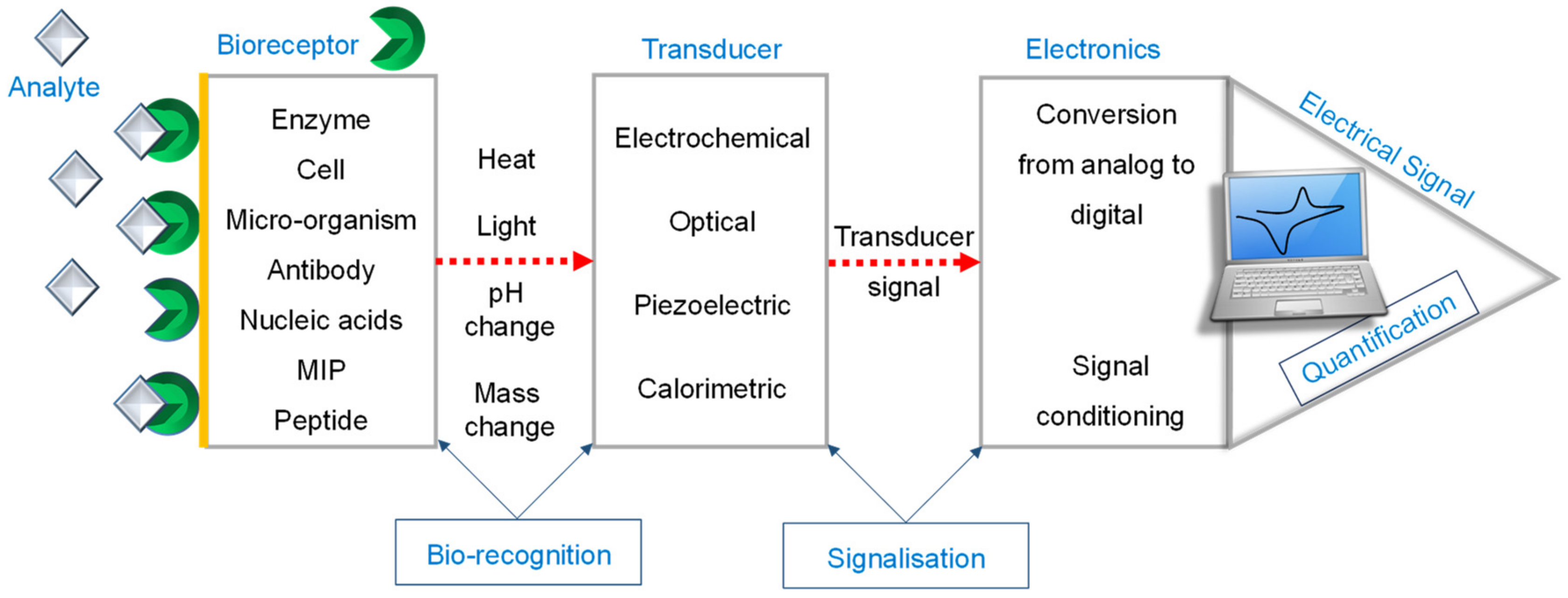
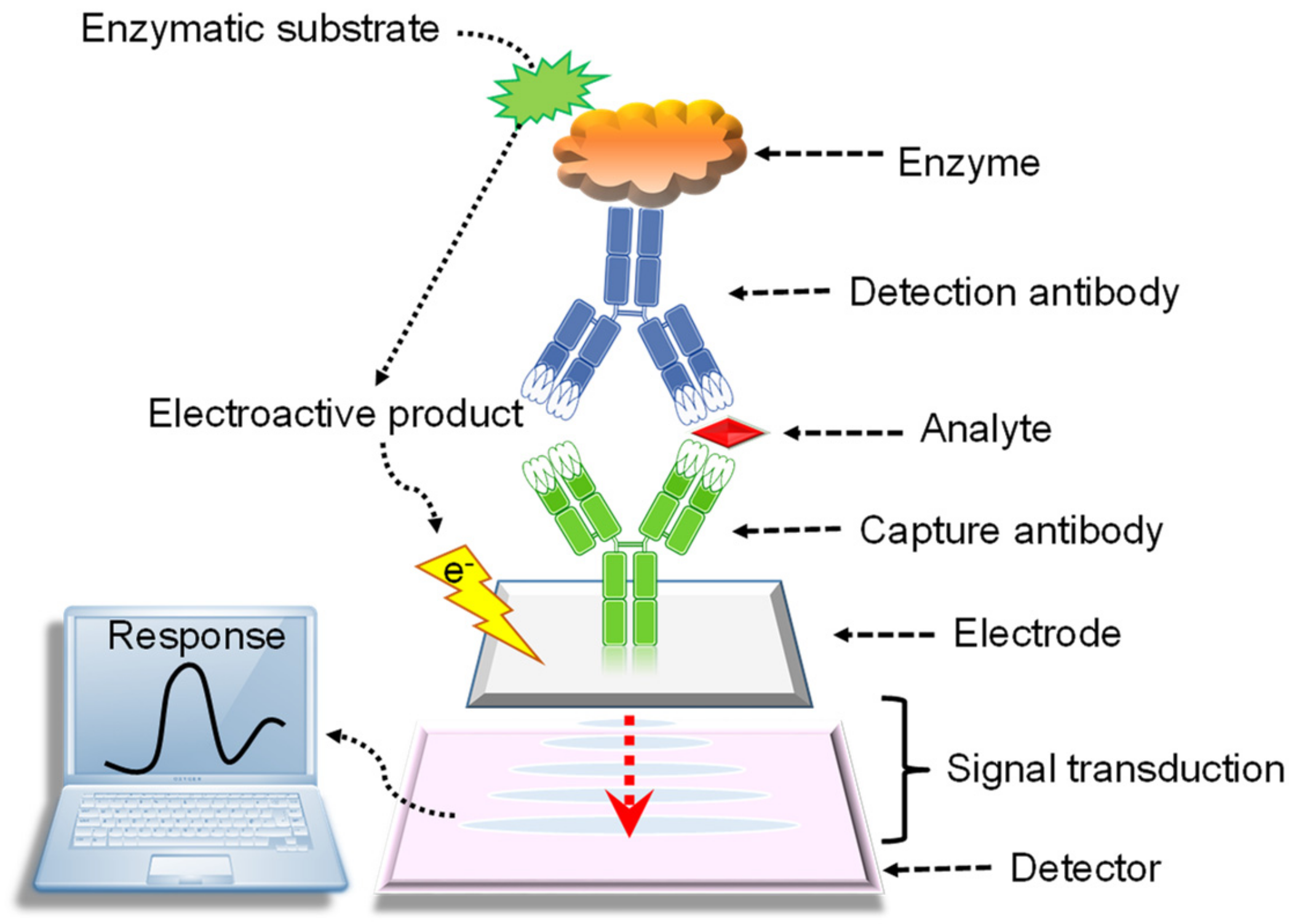
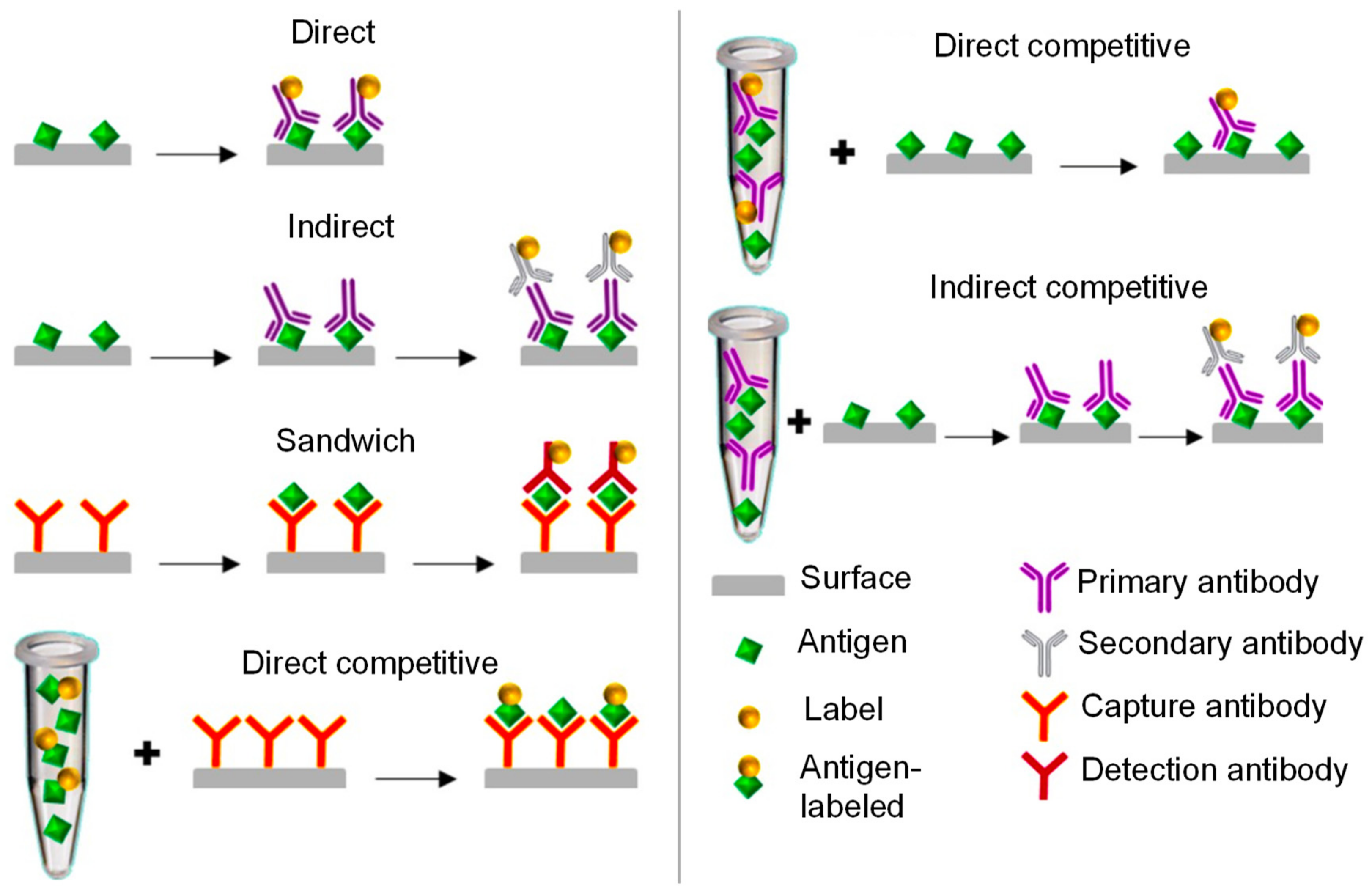
2. Electrochemical Sensors
- (a)
- Capture of the analyte of interest (usually target antigen);
- (b)
- Occlusion of the non-reacted surface; and
- (c)
- Recognition of the analyte.
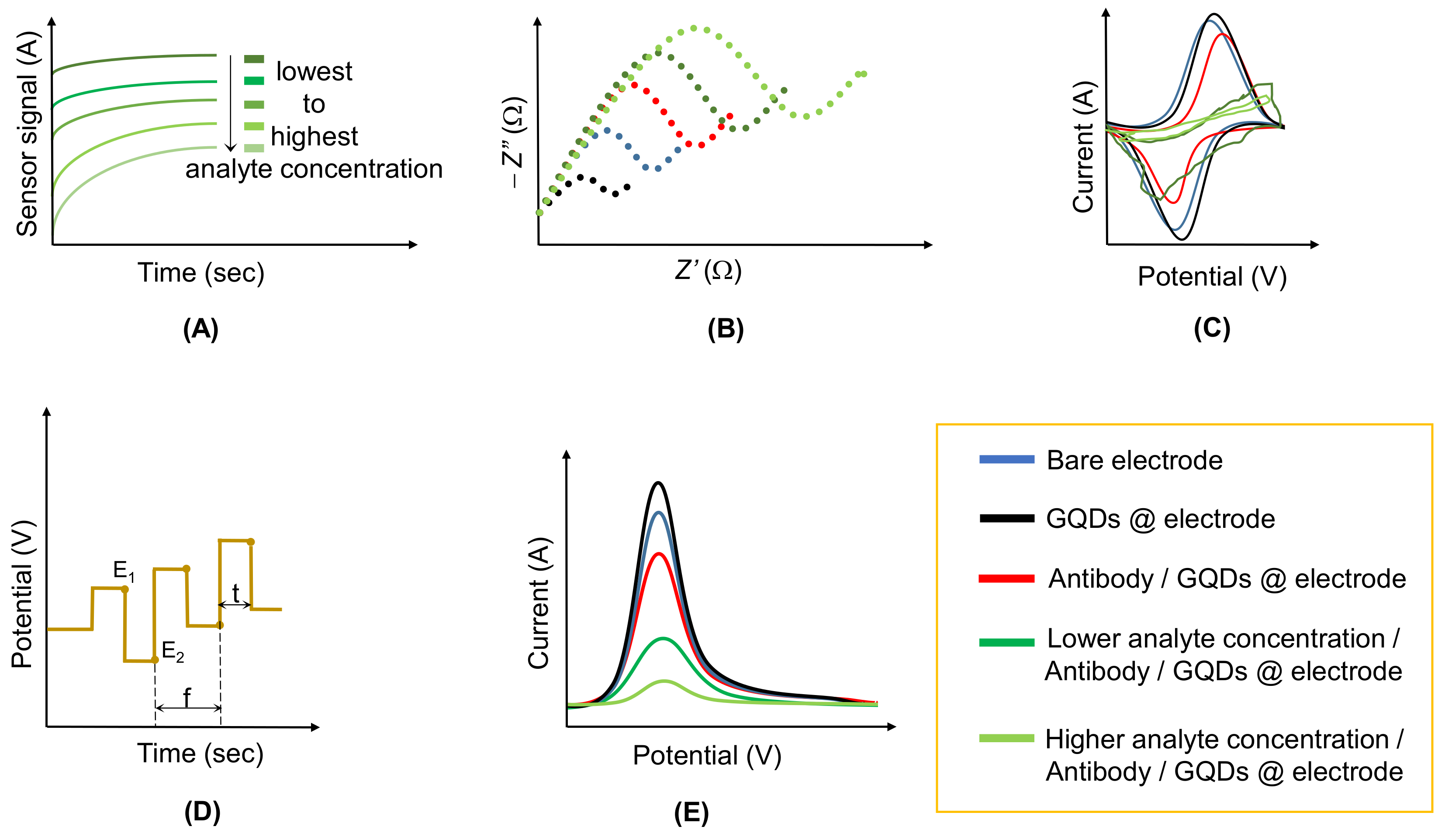
2.1. Amperometric Sensors
2.2. Conductometric Sensors
2.3. Impedimetric Sensors
2.4. Potentiometric Sensors
2.5. Voltammetric Sensors
3. GQD-Based Electrochemical Immunosensors for Cancer Diagnosis
4. GQD-Based Electrochemical Immunosensors for Monitoring Cardiovascular Diseases
5. GQD-Based Electrochemical Immunosensors for the Detection of Infectious Diseases
6. Summary and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Clark, L.C.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Lv, M.; Liu, Y.; Geng, J.; Kou, X.; Xin, Z.; Yang, D. Engineering nanomaterials-based biosensors for food safety detection. Biosens. Bioelectron. 2018, 106, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S. Biosensors for cancer detection applications. Mo. S T’s Peer Peer 2017, 1, 6. [Google Scholar]
- Hernandez-Vargas, G.; Sosa-Hernández, J.E.; Saldarriaga-Hernandez, S.; Villalba-Rodríguez, A.M.; Parra-Saldivar, R.; Iqbal, H.M.N. Electrochemical biosensors: A solution to pollution detection with reference to environmental contaminants. Biosensors 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Current trends in the biosensors for biological warfare agents assay. Materials 2019, 12, 2303. [Google Scholar] [CrossRef]
- Saylan, Y.; Erdem, Ö.; Ünal, S.; Denizli, A. An alternative medical diagnosis method: Biosensors for virus detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef]
- Sharma, T.K.; Ramanathan, R.; Rakwal, R.; Agrawal, G.K.; Bansal, V. Moving forward in plant food safety and security through NanoBioSensors: Adopt or adapt biomedical technologies? Proteomics 2015, 15, 1680–1692. [Google Scholar] [CrossRef]
- Van Dorst, B.; Mehta, J.; Bekaert, K.; Rouah-Martin, E.; De Coen, W.; Dubruel, P.; Blust, R.; Robbens, J. Recent advances in recognition elements of food and environmental biosensors: A review. Biosens. Bioelectron. 2010, 26, 1178–1194. [Google Scholar] [CrossRef]
- Jurado-Sánchez, B. Nanoscale biosensors based on self-propelled objects. Biosensors 2018, 8, 59. [Google Scholar] [CrossRef]
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V.; et al. Enzyme biosensors for biomedical applications: Strategies for safeguarding analytical performances in biological fluids. Sensors 2016, 16, 780. [Google Scholar] [CrossRef]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Bala, C. Peptide-based biosensors: From self-assembled interfaces to molecular probes in electrochemical assays. Bioelectrochemistry 2018, 120, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, I.; Sharma, N.; Vasilescu, A.; Iancu, M.; Badea, G.; Boukherroub, R.; Ogale, S.; Szunerits, S. Electrochemical aptamer-based biosensors for the detection of cardiac biomarkers. ACS Omega 2018, 3, 12010–12018. [Google Scholar] [CrossRef]
- Savas, S.; Ersoy, A.; Gulmez, Y.; Kilic, S.; Levent, B.; Altintas, Z. Nanoparticle enhanced antibody and DNA biosensors for sensitive detection of Salmonella. Materials 2018, 11, 1541. [Google Scholar] [CrossRef] [PubMed]
- Waffo, A.F.T.; Yesildag, C.; Caserta, G.; Katz, S.; Zebger, I.; Lensen, M.C.; Wollenberger, U.; Scheller, F.W.; Altintas, Z. Fully electrochemical MIP sensor for artemisinin. Sens. Actuators B Chem. 2018, 275, 163–173. [Google Scholar] [CrossRef]
- Abdin, M.J.; Altintas, Z.; Tothill, I.E. In silico designed nanoMIP based optical sensor for endotoxins monitoring. Biosens. Bioelectron. 2015, 67, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Mollarasouli, F.; Kurbanoglu, S.; Ozkan, S.A. The role of electrochemical immunosensors in clinical analysis. Biosensors 2019, 9, 86. [Google Scholar] [CrossRef]
- Gharatape, A.; Khosroushahi, A.Y. Optical biomarker-based biosensors for cancer/infectious diseases. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 278–286. [Google Scholar] [CrossRef]
- Pohanka, M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef]
- Sayed, M.; Gul, M.; Shah, N.S.; Khan, J.A.; Khan, Z.U.H.; Rehman, F.; Khan, A.R.; Rauf, S.; Arandiyan, H.; Yang, C.P. In-situ dual applications of ionic liquid coated Co2+ and Fe3+ co-doped TiO2: Superior photocatalytic degradation of ofloxacin at pilot scale level and enhanced peroxidase like activity for calorimetric biosensing. J. Mol. Liq. 2019, 282, 275–285. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Khristunova, Y.; Korotkova, E.; Kratochvil, B.; Barek, J.; Dorozhko, E.; Vyskocil, V.; Plotnikov, E.; Voronova, O.; Sidelnikov, V. Preparation and investigation of silver nanoparticle–antibody bioconjugates for electrochemical immunoassay of tick-borne encephalitis. Sensors 2019, 19, 2103. [Google Scholar] [CrossRef] [PubMed]
- Boverhof, D.R.; Bramante, C.R.; Butala, J.H.; Clancy, S.F.; Lafrancon, M.; West, J.; Gordon, S.C. Comparative assessment of nanomaterial definitions and safety evaluation considerations. Regul. Toxicol. Pharmacol. 2015, 73, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahzadeh, M.; Sajadi, S.M.; Sajjadi, M.; Issaabadi, Z. Applications of nanotechnology in daily life. Interface Sci. Technol. 2019, 28, 113–143. [Google Scholar]
- Lombardo, D.; Kiselev, M.A.; Caccamo, M.T. Smart nanoparticles for drug delivery application: Development of versatile nanocarrier platforms in biotechnology and nanomedicine. J. Nanomater. 2019, 2019, 1–26. [Google Scholar] [CrossRef]
- Gupta, R.; Xie, H. Nanoparticles in daily life: Applications, toxicity and regulations. J. Environ. Pathol. Toxicol. Oncol. 2018, 37, 209–230. [Google Scholar] [CrossRef]
- Baer, D.R.; Engelhard, M.H.; Johnson, G.E.; Laskin, J.; Lai, J.; Mueller, K.; Thevuthasan, S.; Wang, H.; Washton, N.; Elder, A.; et al. Surface characterization of nanomaterials and nanoparticles: Important needs and challenging opportunities. J. Vac. Sci. Technol. A Vac. Surf. Film. 2014, 050820, 1–34. [Google Scholar] [CrossRef]
- Rodríguez-López, J.L. Size effect and shape stability of nanoparticles. Key Eng. Mater. 2010, 444, 47–68. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2017, 12, 908–931. [Google Scholar] [CrossRef]
- Wernsdorfer, W.; Orozco, E.B.; Barbara, B.; Benoit, A.; Mailly, D.; Demoncy, N.; Pascard, H.; Kubo, O.; Nakano, H. Magnetization reversal in individual nanoparticles: Macroscopic quantum tunneling of magnetization. IEEE Trans. Magn. 1998, 34, 973–978. [Google Scholar] [CrossRef]
- Tonelli, D.; Scavetta, E.; Gualandi, I. Electrochemical deposition of nanomaterials for electrochemical sensing. Sensors 2019, 19, 1186. [Google Scholar] [CrossRef] [PubMed]
- Piperno, A.; Scala, A.; Mazzaglia, A.; Neri, G.; Pennisi, R.; Sciortino, M.T.; Grassi, G. Cellular signaling pathways activated by functional graphene nanomaterials. Int. J. Mol. Sci. 2018, 19, 3365. [Google Scholar] [CrossRef] [PubMed]
- Kokkinos, C. Electrochemical DNA biosensors based on labeling with nanoparticles. Nanomaterials 2019, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Paloma, Y.; Pingarr, M. Carbon dots and graphene quantum dots in electrochemical biosensing. Nanomaterials 2019, 9, 634. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh Zeinabad, H.; Ghourchian, H.; Falahati, M.; Fathipour, M.; Azizi, M.; Boutorabi, S.M. Ultrasensitive interdigitated capacitance immunosensor using gold nanoparticles. Nanotechnology 2018, 29, 26. [Google Scholar] [CrossRef]
- Idris, A.O.; Mabuba, N.; Arotiba, O.A. An alpha-fetoprotein electrochemical immunosensor based on a carbon/gold bi-nanoparticle platform. Anal. Methods 2018, 10, 5649–5658. [Google Scholar] [CrossRef]
- Bohli, N.; Belkilani, M.; Mora, L.; Abdelghani, A. Antibody-functionalised gold nanoparticles-based impedimetric immunosensor: Detection methods for better sensitivity. Micro Nano Lett. 2019, 14, 629–633. [Google Scholar] [CrossRef]
- Nikolaev, K.G.; Ermolenko, Y.E.; Offenhäusser, A.; Ermakov, S.S.; Mourzina, Y.G. Multisensor systems by electrochemical nanowire assembly for the analysis of aqueous solutions. Front. Chem. 2018, 6, 1–16. [Google Scholar] [CrossRef]
- Oliveira, T.M.B.F.; Morais, S. New generation of electrochemical sensors based on multi-walled carbon nanotubes. Appl. Sci. 2018, 8, 1925. [Google Scholar] [CrossRef]
- Wang, X.; Gao, D.; Li, M.; Li, H.; Li, C.; Wu, X.; Yang, B. CVD graphene as an electrochemical sensing platform for simultaneous detection of biomolecules. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Raj, A.; John, S.A. Graphene-modified electrochemical sensors. Graphene-Based Electrochem. Sens. Biomol. 2019, 1–41. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. Applications of graphene in electrochemical sensing and biosensing. TrAC Trends Anal. Chem. 2016, 76, 1–14. [Google Scholar] [CrossRef]
- Bettazzi, F.; Natale, A.R.; Torres, E.; Palchetti, I. Glyphosate determination by coupling an immuno-magnetic assay with electrochemical sensors. Sensors 2018, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Helali, S.; Martelet, C.; Abdelghani, A.; Maaref, M.A.; Jaffrezic-Renault, N. A disposable immunomagnetic electrochemical sensor based on functionalised magnetic beads on gold surface for the detection of atrazine. Electrochim. Acta 2006, 51, 5182–5186. [Google Scholar] [CrossRef]
- Wang, Y.; Myers, M.; Staser, J.A. Electrochemical UV sensor using carbon quantum dot/graphene semiconductor. J. Electrochem. Soc. 2018, 165, H3001–H3007. [Google Scholar] [CrossRef]
- Faridbod, F.; Sanati, A.L. Graphene quantum dots in electrochemical sensors/biosensors. Curr. Anal. Chem. 2018, 15, 103–123. [Google Scholar] [CrossRef]
- Sun, H.; Wu, L.; Wei, W.; Qu, X. Recent advances in graphene quantum dots for sensing. Mater. Today 2013, 16, 433–442. [Google Scholar] [CrossRef]
- Pedrero, M.; Campuzano, S.; Pingarrón, J.M. Quantum dots as components of electrochemical sensing platforms for the detection of environmental and food pollutants: A review. J. AOAC Int. 2017, 100, 950–961. [Google Scholar] [CrossRef]
- Mistry, K.K.; Layek, K.; Mahapatra, A.; Chaudhuri, C.R.; Saha, H. A review on amperometric-type immunosensors based on screen-printed electrodes. Analyst 2014, 139, 2289. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, Y.; Liu, Z. Nanowire-based Cu electrode as electrochemical sensor for detection of nitrate in water. Sens. Actuators B Chem. 2016, 232, 336–344. [Google Scholar] [CrossRef]
- Kurniawan, F.; Al Kiswiyah, N.S.; Madurani, K.A.; Tominaga, M. Electrochemical sensor based on single-walled carbon nanotubes-modified gold electrode for uric acid detection. J. Electrochem. Soc. 2018, 165, B515–B522. [Google Scholar] [CrossRef]
- Nanomaterials Definition Matters. Available online: https://www.nature.com/articles/s41565-019-0412-3 (accessed on 23 December 2019).
- Zeng, Z.; Xiao, F.X.; Phan, H.; Chen, S.; Yu, Z.; Wang, R.; Nguyen, T.Q.; Yang, T.T. Unraveling the cooperative synergy of zero-dimensional graphene quantum dots and metal nanocrystals enabled by layer-by-layer assembly. J. Mater. Chem. A 2018, 6, 1700–1713. [Google Scholar] [CrossRef]
- Fan, Z.; Li, S.; Yuan, F.; Fan, L. Fluorescent graphene quantum dots for biosensing and bioimaging. RSC Adv. 2015, 5, 19773–19789. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Shadjou, N. What are the reasons for low use of graphene quantum dots in immunosensing of cancer biomarkers? Mater. Sci. Eng. C 2017, 71, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, S.; Wang, H.; Qu, S.; Zhang, Y.; Zhang, J.; Chen, Q.; Al, W.E.T. Common origin of green luminescence in carbon nanodots and graphene quantum dots. Am. Chem. Soc. 2014, 8, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhu, Y.; Yang, X.; Li, C. Graphene quantum dots: Emergent nanolights for bioimaging, sensors, catalysis and photovoltaic devices. Chem. Commun. 2012, 48, 3686–3699. [Google Scholar] [CrossRef]
- Yan, X.; Cui, X.; Li, L.S. Synthesis of large, stable colloidal graphene quantum dots with tunable size. J. Am. Chem. Soc. 2010, 132, 5944–5945. [Google Scholar] [CrossRef]
- Tachi, S.; Morita, H.; Takahashi, M.; Okabayashi, Y.; Hosokai, T.; Sugai, T.; Kuwahara, S. Quantum yield enhancement in graphene quantum dots via esterification with benzyl alcohol. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Tian, P.; Tang, L.; Teng, K.S.; Lau, S.P. Graphene quantum dots from chemistry to applications. Mater. Today Chem. 2018, 10, 221–258. [Google Scholar] [CrossRef]
- Roushani, M.; Valipour, A. The potentiality of graphene quantum dots functionalized by nitrogen and thiol-doped (GQDs-N-S) to stabilize the antibodies in designing of human chorionic gonadotropin immunosensor. Nano Chem. Res. 2019, 4, 20–26. [Google Scholar]
- Ma, F.; Li, C.C.; Zhang, C.Y. Development of quantum dot-based biosensors: Principles and applications. J. Mater. Chem. B 2018, 6, 6173–6190. [Google Scholar] [CrossRef]
- Savas, S.; Altintas, Z. Graphene quantum dots as nanozymes for electrochemical sensing of Yersinia enterocolitica in milk and human serum. Materials 2019, 12, 2189. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rui, M.; Song, J.; Shen, Z.; Zeng, H. Carbon and graphene quantum dots for optoelectronic and energy devices: A review. Adv. Funct. Mater. 2015, 25, 4929–4947. [Google Scholar] [CrossRef]
- Gupta, S.; Kaushal, A.; Kumar, A.; Kumar, D. Ultrasensitive transglutaminase based nanosensor for early detection of celiac disease in human. Int. J. Biol. Macromol. 2017, 105, 905–911. [Google Scholar] [CrossRef]
- Tuteja, S.K.; Chen, R.; Kukkar, M.; Song, C.K.; Mutreja, R.; Singh, S.; Paul, A.K.; Lee, H.; Kim, K.H.; Deep, A.; et al. A label-free electrochemical immunosensor for the detection of cardiac marker using graphene quantum dots (GQDs). Biosens. Bioelectron. 2016, 86, 548–556. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Q.; Liu, Y.; Cui, J.; Liu, H.; Wang, P.; Li, Y.; Chen, L.; Zhao, Z.; Dong, Y. A novel label-free electrochemical immunosensor based on functionalized nitrogen-doped graphene quantum dots for carcinoembryonic antigen detection. Biosens. Bioelectron. 2017, 90, 31–38. [Google Scholar] [CrossRef]
- Atkinson, A.J., Jr.; Colburn, W.A.; DeGruttola, V.G.; DeMets, D.L.; Downing, G.J.; Spilker, B.A.; Biomarkers Definitions Working Group. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar]
- Stradiotto, N.R.; Yamanaka, H.; Zanoni, M.V.B. Electrochemical sensors: A powerful tool in analytical chemistry. J. Braz. Chem. Soc. 2003, 14, 159–173. [Google Scholar] [CrossRef]
- Lim, S.A.; Ahmed, M.U. Electrochemical immunosensors and their recent nanomaterial-based signal amplification strategies: A review. RCS Adv. 2016, 6, 24995–25014. [Google Scholar] [CrossRef]
- Kokkinos, C.; Economou, A.; Prodromidis, M.I. Electrochemical immunosensors: Critical survey of different architectures and transduction strategies. TrAC Trends Anal. Chem. 2016, 79, 88–105. [Google Scholar] [CrossRef]
- Rama, E.C.; Costa-García, A. Screen-printed electrochemical immunosensors for the detection of cancer and cardiovascular biomarkers. Electroanalysis 2016, 28, 1700–1715. [Google Scholar] [CrossRef]
- Cho, I.H.; Lee, J.; Kim, J.; Kang, M.S.; Paik, J.K.; Ku, S.; Cho, H.M.; Irudayaraj, J.; Kim, D.H. Current technologies of electrochemical immunosensors: Perspective on signal amplification. Sensors 2018, 18, 207. [Google Scholar] [CrossRef] [PubMed]
- Faridbod, F.; Gupta, V.K.; Zamani, H.A. Electrochemical sensors and biosensors. Int. J. Electrochem. 2011, 24, 717. [Google Scholar] [CrossRef]
- Dhahi, T.H.S.; Bin Hashim, U.D.A.; Ahmed, N.M.; Mat Taib, A. A review on the electrochemical sensors and biosensors composed of nanogaps as sensing material. J. Optoelectron. Adv. Mater. 2010, 12, 1857–1862. [Google Scholar]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended defnitions and classification. Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef]
- Altintas, Z.; Fakanya, W.M.; Tothill, I.E. Cardiovascular disease detection using bio-sensing techniques. Talanta 2014, 128, 177–186. [Google Scholar] [CrossRef]
- Pan, D.; Li, G.; Hu, H.; Xue, H.; Zhang, M.; Zhu, M.; Gong, X.; Zhang, Y.; Wan, Y.; Shen, Y. Direct immunoassay for facile and sensitive detection of small molecule aflatoxin B1 based on nanobody. Chem. A Eur. J. 2018, 24, 9869–9876. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Yifeng, E.; Fang, F.; Kuang, G.; Wang, G. Sandwich Immunoassays of multicomponent subtrace pathogenic DNA based on magnetic fluorescent encoded nanoparticles. BioMed Res. Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Dutta, G.; Lillehoj, P.B. Wash-free, label-free immunoassay for rapid electrochemical detection of PfHRP2 in whole blood samples. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Yan, Q.; Yang, Y.; Tan, Z.; Liu, Q.; Liu, H.; Wang, P.; Chen, L.; Zhang, D.; Li, Y.; Dong, Y. A label-free electrochemical immunosensor based on the novel signal amplification system of AuPdCu ternary nanoparticles functionalized polymer nanospheres. Biosens. Bioelectron. 2018, 103, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Mistry, K.K.; Layek, K.; Chell, T.N.; Chaudhuri, C.R.; Saha, H. Design and development of an amperometric immunosensor based on screen-printed electrodes. Anal. Methods 2016, 8, 3096–3101. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Okon, S.L. Nanomaterial-based electrochemical immunosensors for clinically significant biomarkers. Materials 2014, 7, 4669–4709. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.G. Conductometric immunosensors for the detection of staphylococcal enterotoxin B based bio-electrocalytic reaction on micro-comb electrodes. Bioprocess Biosyst. Eng. 2008, 31, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Jaffrezic-Renault, N.; Dzyadevych, S.V. Conductometric microbiosensors for environmental monitoring. Sensors 2008, 8, 2569–2588. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: Routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanalysis 2003, 15, 913–947. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. A review on impedimetric biosensors. Artif. Cells Nanomed. Biotechnol. 2016, 44, 248–262. [Google Scholar] [CrossRef]
- Prodromidis, M. Impedimetric biosensors and immunosensors. Pak. J. Anal. Environ. Chem. 2007, 8, 69–71. [Google Scholar]
- Daniels, J.S.; Pourmand, N. Label-free impedance biosensors: Opportunities and challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef]
- Ravalli, A.; Marrazza, G. Electrochemical-based biosensor technologies in disease detection and diagnostics. In Biosensors and Nanotechnology, 1st ed.; Altintas, Z., Ed.; Wiley: Hoboken, NJ, USA, 2018; pp. 95–124. [Google Scholar]
- Masikini, M.; Mailu, S.N.; Tsegaye, A.; Njomo, N.; Molapo, K.M.; Ikpo, C.O.; Sunday, C.E.; Rassie, C.; Wilson, L.; Baker, P.G.L.; et al. A fumonisins immunosensor based on polyanilino-carbon nanotubes doped with palladium telluride quantum dots. Sensors 2015, 15, 529–546. [Google Scholar] [CrossRef]
- Purvis, D.; Leonardova, O.; Farmakovsky, D.; Cherkasov, V. An ultrasensitive and stable potentiometric immunosensor. Biosens. Bioelectron. 2003, 18, 1385–1390. [Google Scholar] [CrossRef]
- Luppa, P.B.; Sokoll, L.J.; Chan, D.W. Immunosensors—Principles and applications to clinical chemistry. Clin. Chim. Acta 2001, 314, 1–26. [Google Scholar] [CrossRef]
- Farghaly, O.A.; Abdel Hameed, R.S.; Abu-Nawwas, A.A.H. Analytical application using modern electrochemical techniques. Int. J. Electrochem. Sci. 2014, 9, 3287–3318. [Google Scholar]
- Mendoza, S.; Bustos, E.; Manríquez, J.; Godínez, L.A. Voltammetric techniques. In Agric. Food Electroanal., 1st ed.; Escarpa, A., González, M.C., López, M.A., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 21–48. [Google Scholar]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A practical beginner’s guide to cyclic voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
- Mirceski, V.; Gulaboski, R.; Lovric, M.; Bogeski, I.; Kappl, R.; Hoth, M. Square-wave voltammetry: A review on the recent progress. Electroanalysis 2013, 25, 2411–2422. [Google Scholar] [CrossRef]
- Mirceski, V.; Gulaboski, R. Recent advances in square-wave voltammetry: A review. Maced. J. Chem. Chem. Eng. 2014, 33, 1–12. [Google Scholar]
- Bard, A.J.; Faulkner, L.R. Polarography and pulse voltammetry. In Electrochemical Methods: Fundamentals and Applications, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001; pp. 261–304. [Google Scholar]
- Altintas, Z.; Takiden, A.; Utesch, T.; Mroginski, M.A.; Schmid, B.; Scheller, F.W.; Süssmuth, R.D. Integrated approaches toward high-affinity artificial protein binders obtained via computationally simulated epitopes for protein recognition. Adv. Funct. Mater. 2019, 29, 1–11. [Google Scholar] [CrossRef]
- Tchinda, R.; Tutsch, A.; Schmid, B.; Süssmuth, R.D.; Altintas, Z. Recognition of protein biomarkers using epitope-mediated molecularly imprinted films: Histidine or cysteine modified epitopes? Biosens. Bioelectron. 2019, 123, 260–268. [Google Scholar] [CrossRef]
- Li, S.; Zhou, J.; Wu, H.; Lu, Q.; Tai, Y.; Liu, Q.; Wang, C. Oncogenic transformation of normal breast epithelial cells co-cultured with cancer cells. Cell Cycle 2018, 17, 2027–2040. [Google Scholar] [CrossRef]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Dracham, C.B.; Shankar, A.; Madan, R. Radiation induced secondary malignancies: A review article. Radiat. Oncol. J. 2018, 36, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Neagu, M.; Caruntu, C.; Constantin, C.; Boda, D.; Zurac, S.; Spandidos, D.A.; Tsatsakis, A.M. Chemically induced skin carcinogenesis: Updates in experimental models (Review). Oncol. Rep. 2016, 35, 2516–2528. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.H.; Zloza, A. Infection: A cause of and cure for cancer. Curr. Pharmacol. Rep. 2017, 3, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, A.M.; Rudzki, M.; Rudzki, S.; Lewandowski, T.; Laskowska, B. Environmental risk factors for cancer—Review paper. Ann. Agric. Environ. Med. 2019, 26, 1–7. [Google Scholar] [CrossRef]
- Blackadar, C.B. Historical review of the causes of cancer. World J. Clin. Oncol. 2016, 7, 54–86. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical immunosensors for detection of cancer protein biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef]
- Serafín, V.; Valverde, A.; Garranzo-Asensio, M.; Barderas, R.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Simultaneous amperometric immunosensing of the metastasis-related biomarkers IL-13Rα2 and CDH-17 by using grafted screen-printed electrodes and a composite prepared from quantum dots and carbon nanotubes for signal amplification. Microchim. Acta 2019, 18, 411. [Google Scholar] [CrossRef]
- Kmezic, S.; Radenkovic, D.; Pejovic, I.; Antic, A.; Bajec, D. The significance of tumor markers, ca 19-9 and CEA, in the pancreatic cancer staging and evaluation of surgical resectability. HPB 2016, 18, e372. [Google Scholar] [CrossRef][Green Version]
- Grunnet, M.; Sorensen, J.B. Lung cancer carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012, 76, 138–143. [Google Scholar] [CrossRef]
- Cetean, S.; Laszlo, I.; Constantin, A.; Căinap, S. Classic tumor markers in gastric cancer: Current standards and limitations. Clujul Med. 2015, 88, 111–115. [Google Scholar]
- Saito, G.; Sadahiro, S.; Kamata, H.; Miyakita, H.; Okada, K.; Tanaka, A.; Suzuki, T. Monitoring of serum carcinoembryonic antigen levels after curative resection of colon cancer: Cutoff values determined according to preoperative levels enhance the diagnostic. Oncology 2017, 92, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S. The roles of carcinoembryonic antigen in liver metastasis and therapeutic approaches. Gastroenterol. Res. Pract. 2017, 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.K.; Choi, D.H.; Park, H.C.; Park, W.; Yu, J.; Park, Y.S.; Cho, B.; Yun, S.H.; Lee, W.Y. Elevated CEA is associated with worse survival in recurrent rectal cancer. Oncotarget 2017, 8, 105936–105941. [Google Scholar] [CrossRef]
- Press, D. Diagnostic and prognostic value of carcinoembryonic antigen in pancreatic cancer: A systematic review and meta-analysis. OncoTargets Ther. 2017, 10, 4591–4598. [Google Scholar]
- Asad-Ur-Rahman, F.N.U.; Saif, M.W. Elevated level of serum carcinoembryonic antigen (CEA) and search for a malignancy: A case report. Cureus 2016, 8, 8–11. [Google Scholar] [CrossRef]
- Altintas, Z.; Tothill, I. Biomarkers and biosensors for the early diagnosis of lung cancer. Sens. Actuators B Chem. 2013, 188, 988–998. [Google Scholar] [CrossRef]
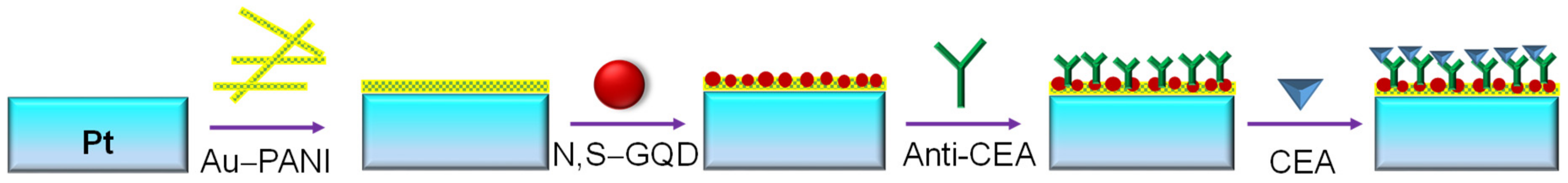
- Ganganboina, A.B.; Doong, R. Graphene quantum dots decorated gold-polyaniline nanowire for impedimetric detection of carcinoembryonic antigen. Nature 2019, 9, 7214. [Google Scholar] [CrossRef]
- Nie, G.; Wang, Y.; Tang, Y.; Zhao, D.; Guo, Q. A graphene quantum dots based electrochemiluminescence immunosensor for carcinoembryonic antigen detection using poly(5-formylindole)/reduced graphene oxide nanocomposite. Biosens. Bioelectron. 2018, 101, 123–128. [Google Scholar] [CrossRef]
- Serafín, V.; Valverde, A.; Martínez-García, G.; Martínez-Periñán, E.; Comba, F.; Garranzo-Asensio, M.; Barderas, R.; Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Graphene quantum dots-functionalized multi-walled carbon nanotubes as nanocarriers in electrochemical immunosensing. Determination of IL-13 receptor A2 in colorectal cells and tumor tissues with different metastatic potential. Sens. Actuators B Chem. 2019, 284, 711–722. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Baghban, H.N.; Shadjou, N.; Mokhtarzadeh, A. Ultrasensitive electrochemical immunosensing of tumor suppressor protein p53 in unprocessed human plasma and cell lysates using a novel nanocomposite based on poly-cysteine/graphene quantum dots/gold nanoparticle. Int. J. Biol. Macromol. 2018, 107, 1348–1363. [Google Scholar] [CrossRef] [PubMed]
- Hasanzadeh, M.; Tagi, S.; Solhi, E.; Mokhtarzadeh, A.; Shadjou, N.; Eftekhari, A.; Mahboob, S. An innovative immunosensor for ultrasensitive detection of breast cancer specific carbohydrate (CA 15-3) in unprocessed human plasma and MCF-7 breast cancer cell lysates using gold nanospear electrochemically assembled onto thiolated graphene quantum dots. Int. J. Biol. Macromol. 2018, 114, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liu, Y.; Wang, Y.; Hu, L.; Ma, H.; Wang, G.; Wei, Q. Label-free electrochemiluminescent immunosensor for detection of prostate specific antigen based on aminated graphene quantum dots and carboxyl graphene quantum dots. Sci. Rep. 2016, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Javadi, A.; Gong, S. Sensitive electrochemical immunosensor for the detection of cancer biomarker using quantum dot functionalized graphene sheets as labels. Sens. Actuators B Chem. 2011, 155, 357–360. [Google Scholar] [CrossRef]
- Yang, H.; Liu, W.; Ma, C.; Zhang, Y.; Wang, X.; Yu, J.; Song, X. Gold-silver nanocomposite-functionalized graphene based electrochemiluminescence immunosensor using graphene quantum dots coated porous PtPd nanochains as labels. Electrochim. Acta 2014, 123, 470–476. [Google Scholar] [CrossRef]
- Okamoto, H.; Yoshimatsu, Y.; Tomizawa, T.; Kunita, A.; Takayama, R.; Morikawa, T.; Komura, D.; Takahashi, K.; Oshima, T.; Sato, M.; et al. Interleukin-13 receptor α2 is a novel marker and potential therapeutic target for human melanoma. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Stewart, J.; Manmathan, G.; Wilkinson, P. Primary prevention of cardiovascular disease: A review of contemporary guidance and literature. JRSM Cardiovasc. Dis. 2017, 6, 2048004016687211. [Google Scholar] [CrossRef]
- Ho, K.J. Cardiovascular diseases. Nutr. Asp. Aging 2018, 2, 75–100. [Google Scholar]
- Ahmad, T.; Fiuzat, M.; Felker, G.M.; O’Connor, C. Novel biomarkers in chronic heart failure. Nat. Rev. Cardiol. 2012, 9, 347–359. [Google Scholar] [CrossRef]
- Braunwald, E. Biomarkers in heart failure. N. Engl. J. Med. 2008, 358, 2148–2159. [Google Scholar] [CrossRef]
- Aydin, S.; Ugur, K.; Aydin, S.; Sahin, İ.; Yardim, M. Biomarkers in acute myocardial infarction: Current perspectives. Vasc. Health Risk Manag. 2019, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mollarasouli, F.; Serafín, V.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M.; Asadpour-Zeynali, K. Ultrasensitive determination of receptor tyrosine kinase with a label-free electrochemical immunosensor using graphene quantum dots-modified screen-printed electrodes. Anal. Chim. Acta 2018, 1011, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, D.; Kaur, I.; Kumar, A. Ultrasensitive cardiac troponin I antibody based nanohybrid sensor for rapid detection of human heart attack. Int. J. Biol. Macromol. 2017, 95, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bing, X.; Wang, G. Label free C-reactive protein detection based on an electrochemical sensor for clinical application. Int. J. Electrochem. Sci. 2017, 12, 6304–6314. [Google Scholar] [CrossRef]
- Batlle, M.; Recarte-Pelz, P.; Roig, E.; Castel, M.A.; Cardona, M.; Farrero, M.; Ortiz, J.T.; Campos, B.; Pulgarín, M.J.; Ramírez, J.; et al. AXL receptor tyrosine kinase is increased in patients with heart failure. Int. J. Cardiol. 2014, 173, 402–409. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, J.; Wen, X.; Chun-Jen, L.C.; Li, J.; Huang, Q.; Yu, Y.; Lin, S.Y.; Li, C. MicroPET/CT imaging of AXL downregulation by HSP90 inhibition in triple-negative breast cancer. Contrast Media Mol. Imaging 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- Mythili, S.; Malathi, N. Diagnostic markers of acute myocardial infarction. Biomed. Rep. 2015, 3, 743–748. [Google Scholar] [CrossRef]
- Nielsen, P.B.; Larsen, T.B.; Gorst-Rasmussen, A.; Skjøth, F.; Lip, G.Y.H. β-Blockers in atrial fibrillation patients with or without heart failure: Association with mortality in a nationwide cohort study. Circ. Heart Fail. 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Galea, R.; Cardillo, M.T.; Caroli, A.; Marini, M.G.; Sonnino, C.; Narducci, M.L.; Biasucci, L.M. Inflammation and C-reactive protein in atrial fibrillation: Cause or effect? Tex. Heart Inst. J. 2014, 41, 461–468. [Google Scholar] [CrossRef]
- Kwon, C.H.; Kang, J.G.; Lee, H.J.; Kim, N.H.; Sung, J.W.; Cheong, E.; Sung, K.C. C-reactive protein and risk of atrial fibrillation in East Asians. Europace 2017, 19, 1643–1649. [Google Scholar] [CrossRef]
- Microbiology by numbers. Nat. Rev. Microbiol. 2011, 9, 628. [CrossRef] [PubMed]
- Christiansen, J. Global Infections by the Numbers. Sci. Am. 2018, 318, 48–49. [Google Scholar]
- Tufa, L.T.; Oh, S.; Tran, V.T.; Kim, J.; Jeong, K.J.; Park, T.J.; Kim, H.J.; Lee, J. Electrochemical immunosensor using nanotriplex of graphene quantum dots, Fe3O4 and Ag nanoparticles for tuberculosis. Electrochim. Acta 2018, 290, 369–377. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Zhang, L.; Gao, J.; Ma, Q. Electrochemiluminescence detection of Escherichia coli O157:H7 based on a novel polydopamine surface imprinted polymer biosensor. ACS Appl. Mater. Interfaces 2017, 9, 5430–5436. [Google Scholar] [CrossRef]
- Ye, W.; Guo, J.; Bao, X.; Chen, T.; Weng, W.; Chen, S.; Yang, M. Rapid and sensitive detection of bacteria response to antibiotics using nanoporous membrane and graphene quantum dot (GQDs)-based electrochemical biosensors. Materials 2017, 10, 603. [Google Scholar] [CrossRef]
- Valipour, A.; Roushani, M. Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: Electrochemical oxidation of riboflavin was used as redox probe. Biosens. Bioelectron. 2017, 89, 946–951. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Takemura, K.; Li, T.-C.; Suzuki, T.; Park, E.Y. Electrical pulse-induced electrochemical biosensor for hepatitis E virus detection. Nat. Commun. 2019, 10, 4–7. [Google Scholar] [CrossRef]
- Ahmed, S.R.; Mogus, J.; Chand, R.; Nagy, E.; Neethirajan, S. Optoelectronic fowl adenovirus detection based on local electric field enhancement on graphene quantum dots and gold nanobundle hybrid. Biosens. Bioelectron. 2018, 103, 45–53. [Google Scholar] [CrossRef]
- Wang, X.; Chen, L.; Su, X.; Ai, S. Electrochemical immunosensor with graphene quantum dots and apoferritin-encapsulated Cu nanoparticles double-assisted signal amplification for detection of avian leukosis virus subgroup J. Biosens. Bioelectron. 2013, 47, 171–177. [Google Scholar] [CrossRef]
- Mehta, J.; Bhardwaj, N.; Bhardwaj, S.K.; Tuteja, S.K.; Vinayak, P.; Paul, A.K.; Kim, K.H.; Deep, A. Graphene quantum dot modified screen printed immunosensor for the determination of parathion. Anal. Biochem. 2017, 523, 1–9. [Google Scholar] [CrossRef]
- Bhardwaj, H.; Singh, C.; Kotnala, R.K.; Sumana, G. Graphene quantum dots-based nano-biointerface platform for food toxin detection. Anal. Bioanal. Chem. 2018, 410, 7313–7323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M. Direct synthesis of graphene quantum dots with different fluorescence properties by oxidation of graphene oxide using nitric acid. Appl. Sci. 2018, 8, 1303. [Google Scholar] [CrossRef]

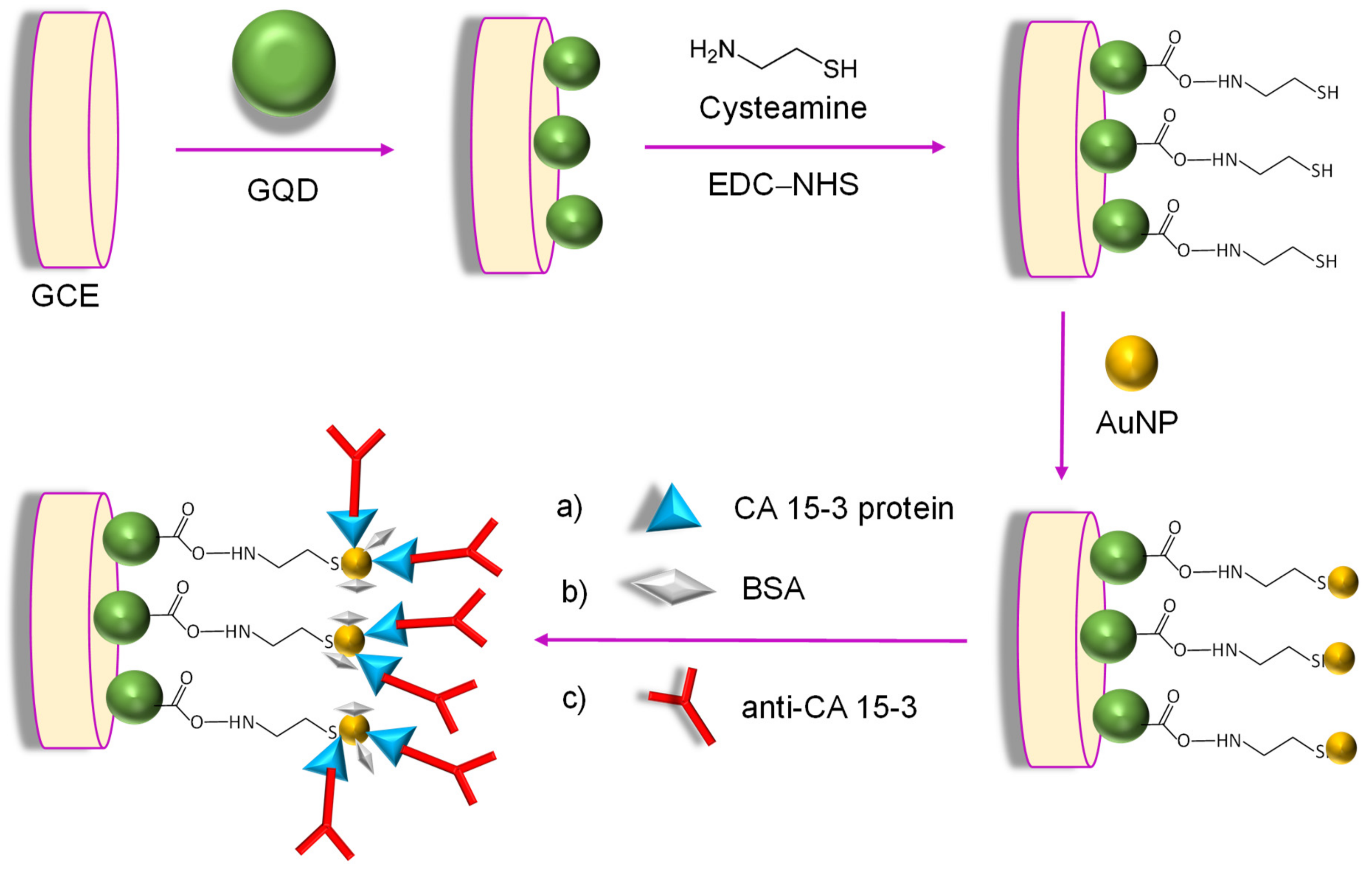

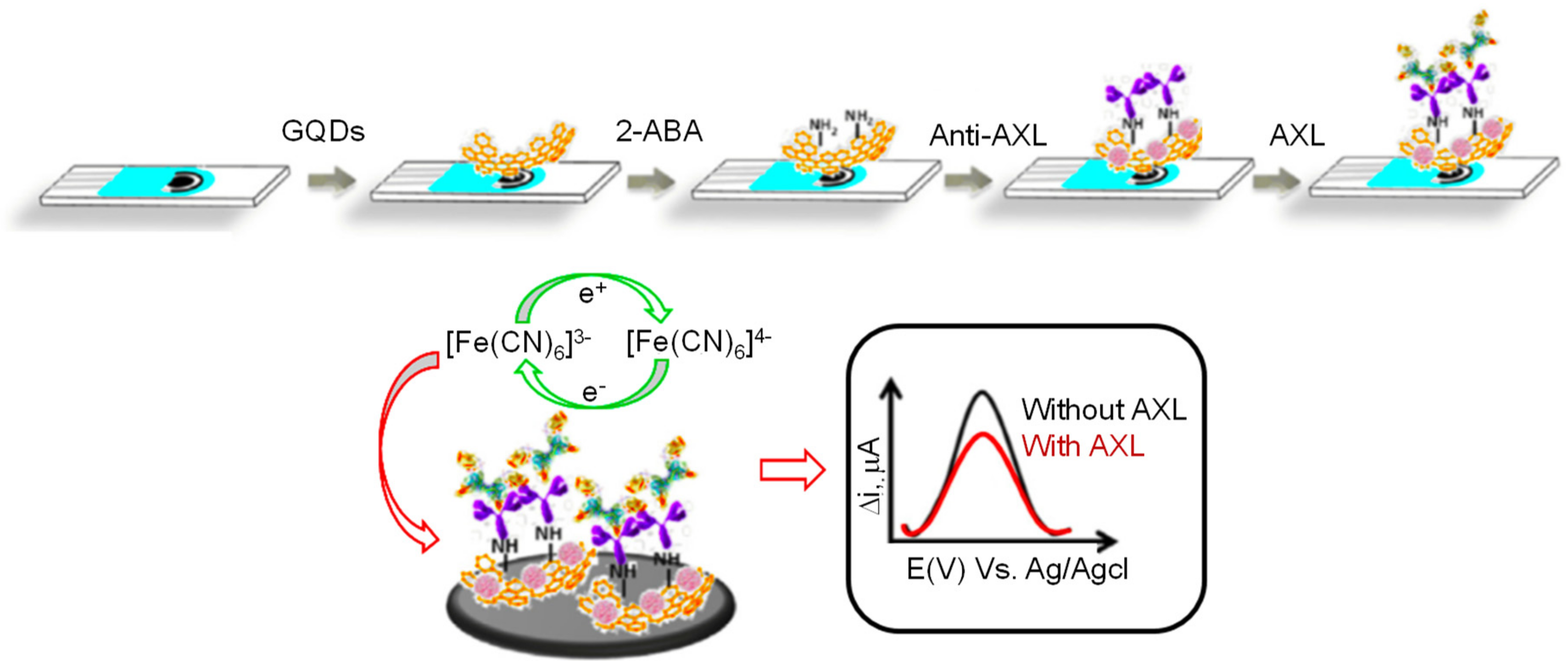
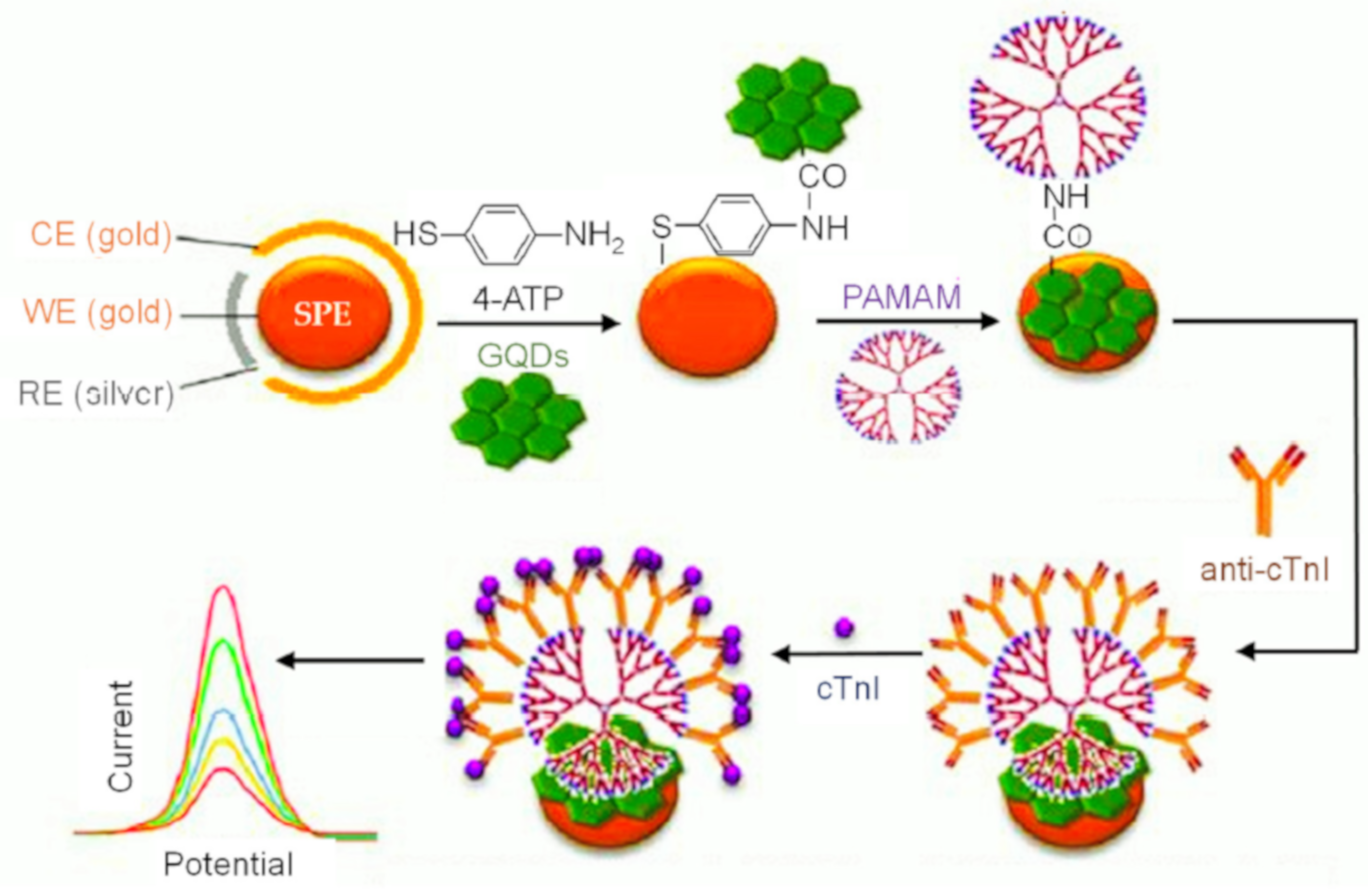
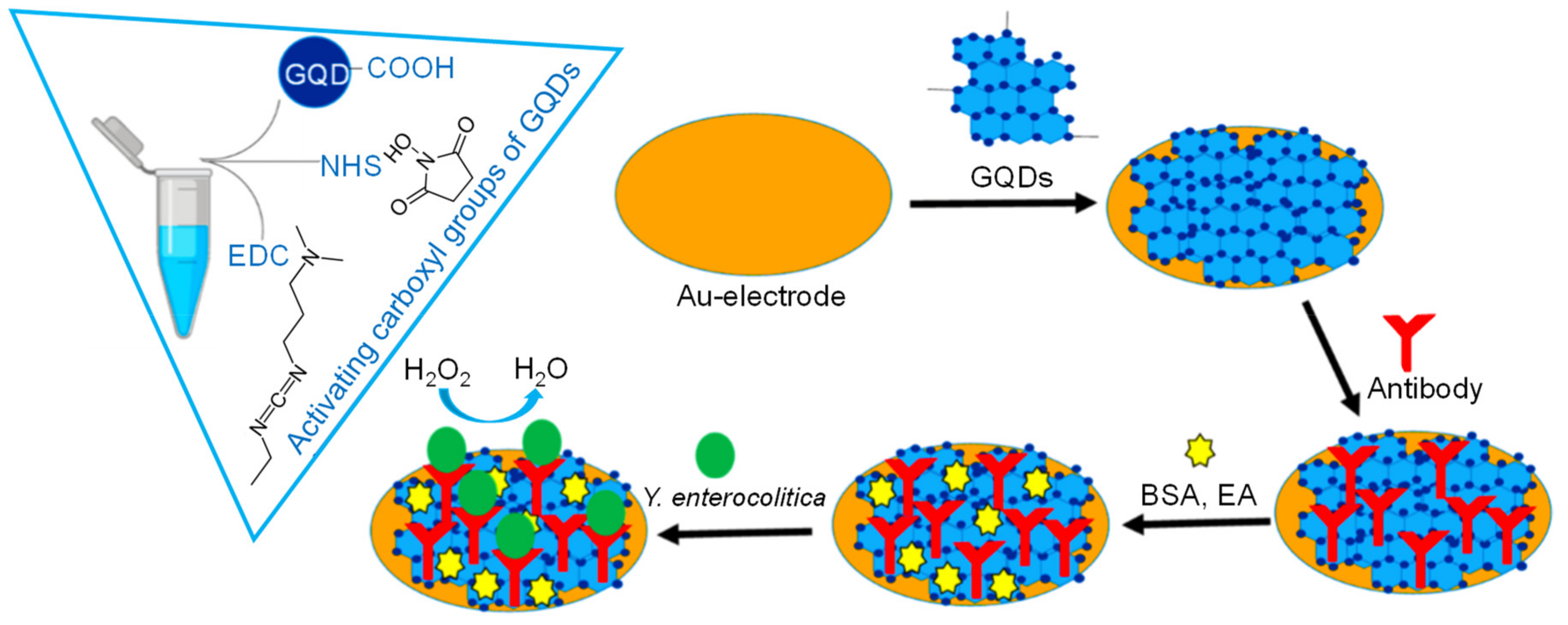
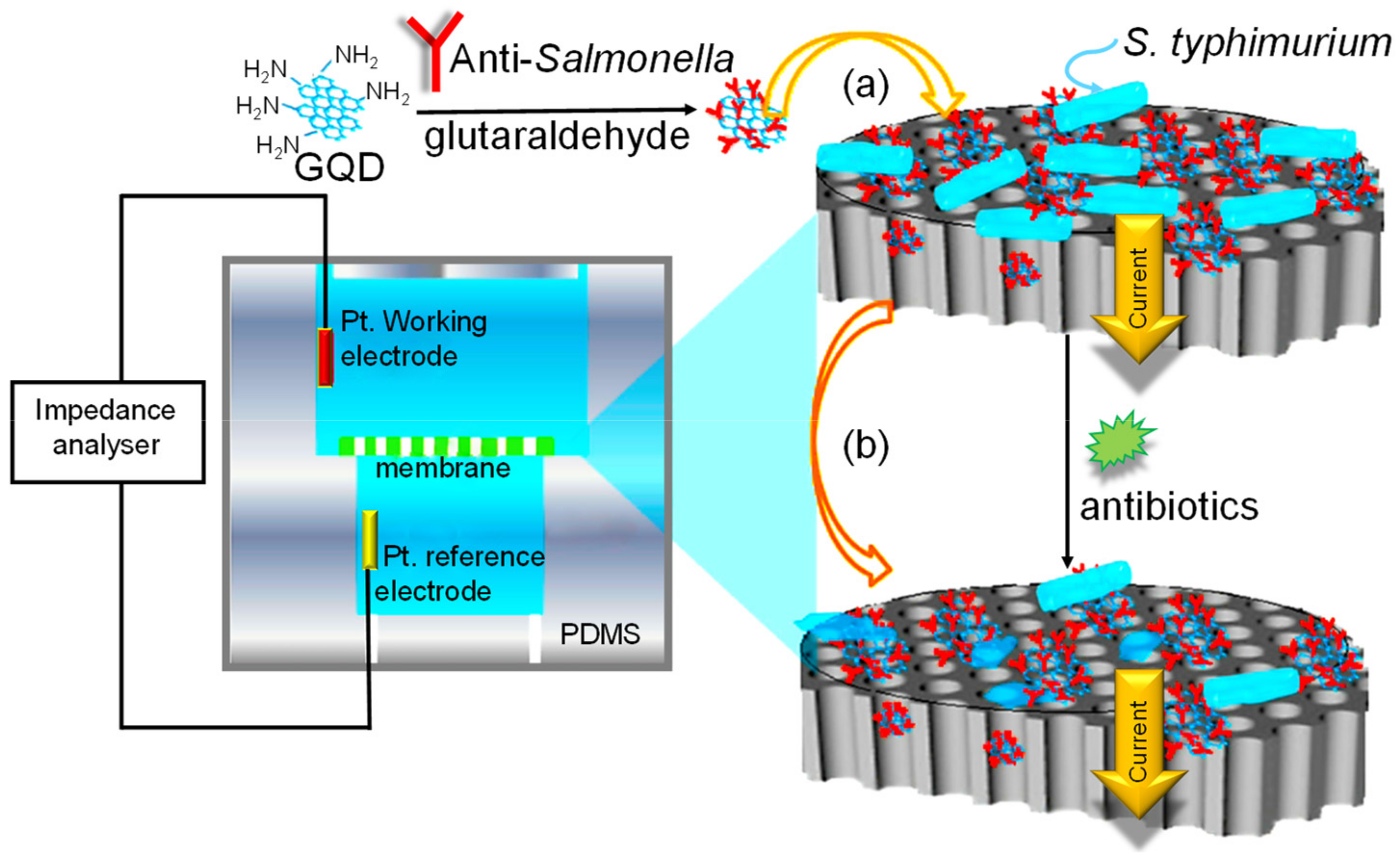
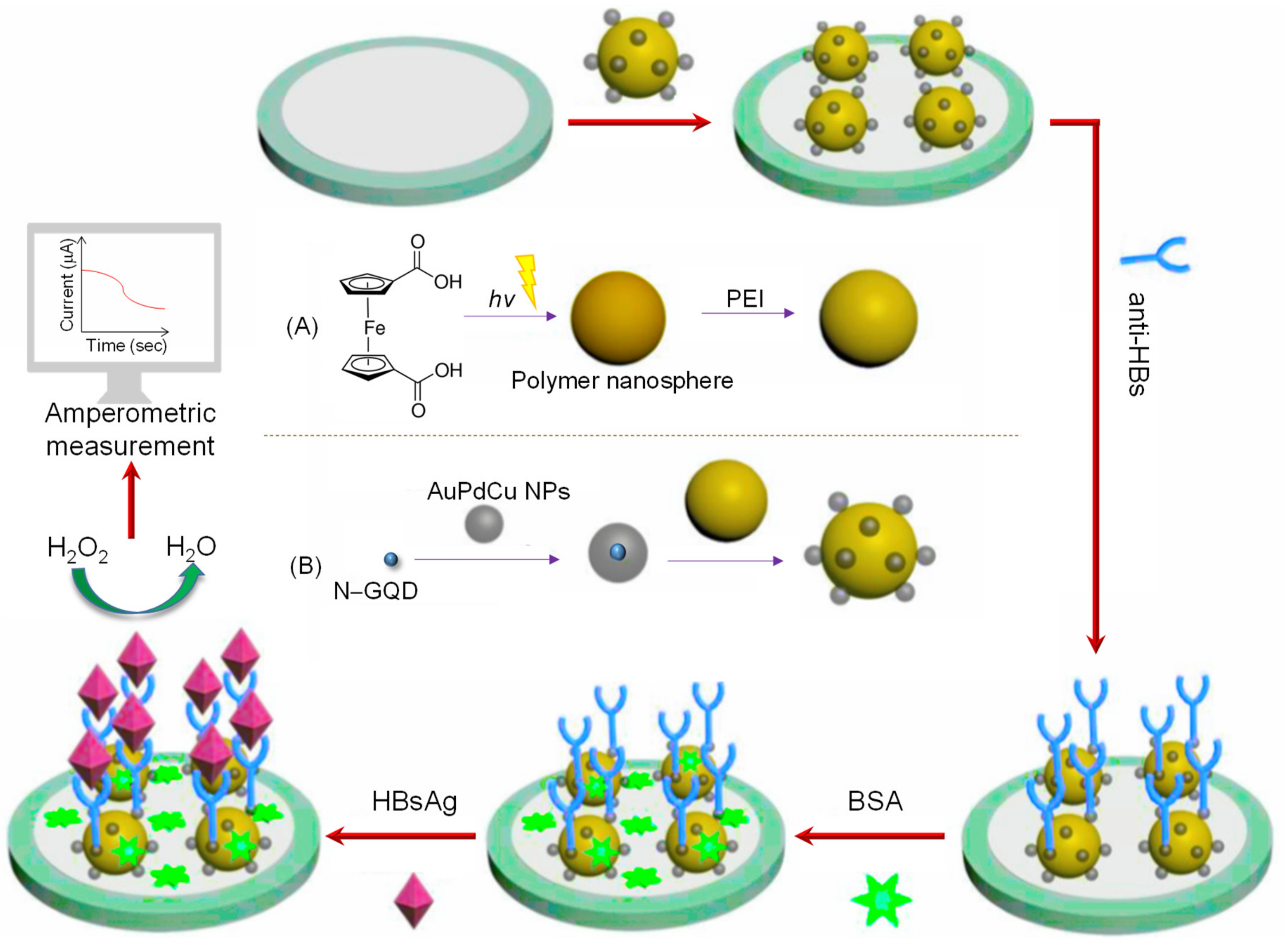
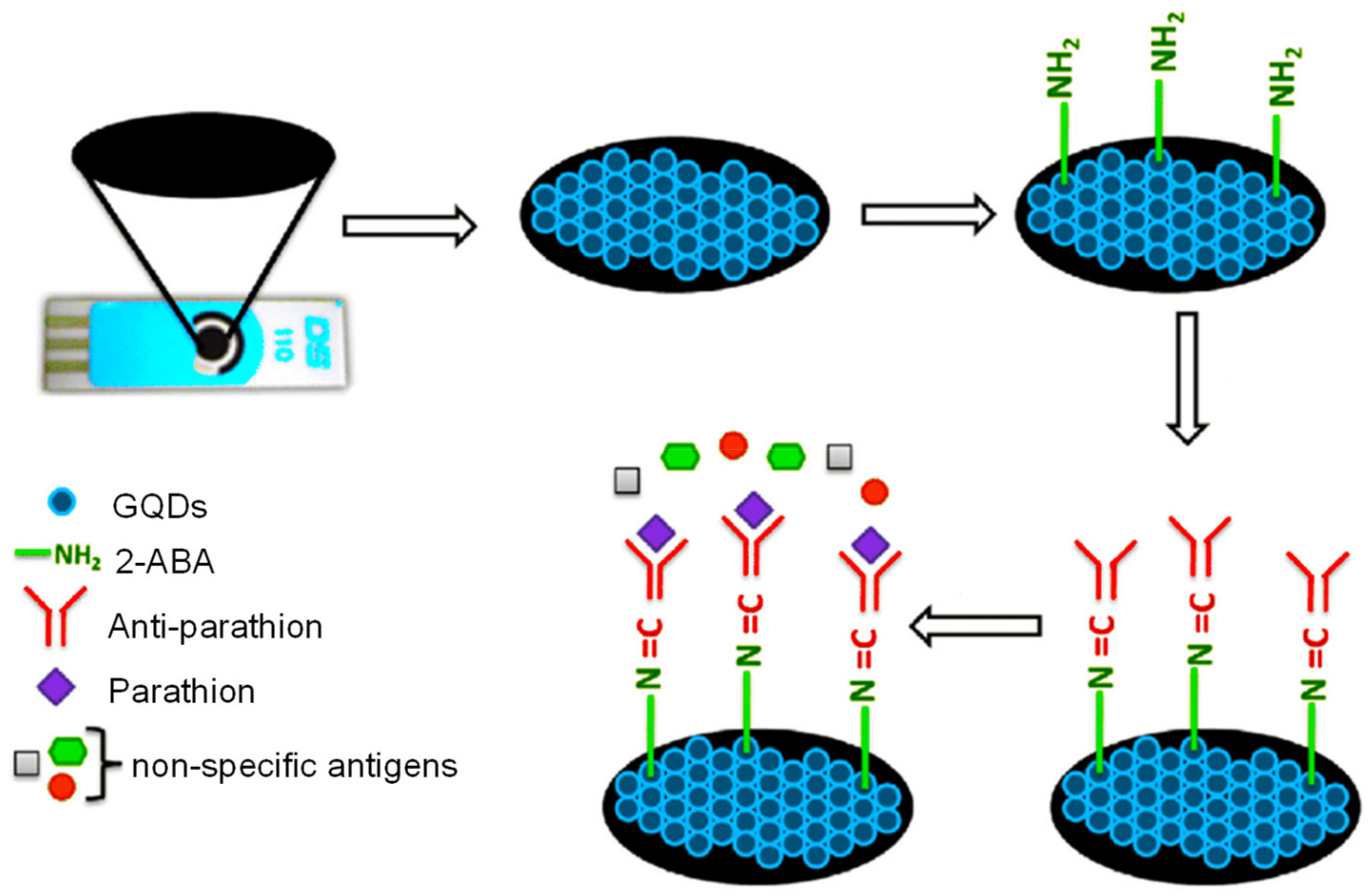

| Electrode | Nanomaterials | Biomarker | Assay Type | Technique(s) | Sample(s) | Linear Range | LOD | Reference |
|---|---|---|---|---|---|---|---|---|
| Pt-electrode | N,S–GQDs/Au–PANI | CEA | Direct | EIS | Human serum | 0.5–1000 ng mL−1 | 0.01 ng mL−1 | [122] |
| GCE | PtPd/N–GQDs/Au | CEA | Direct | Amperometry | Human serum | 5 fg mL−1–50 ng mL−1 | 2 fg mL−1 | [67] |
| GCE | P5FIn/erGO/GQDs/Au | CEA | Sandwich | ECL | Human serum | 0.1–10 ng mL−1 | 3.78 fg mL−1 | [123] |
| SPCE | MWCNTs/GQDs | IL-13Rα2 | Sandwich | Amperometry | Raw cellular lysates from human CRC | 2.7–100 ng mL−1 | 0.8 ng mL−1 | [124] |
| SPdCE | MWCNTs/GQDs | IL-13Rα2, CDH-17 | Sandwich | Amperometry | Raw cellular lysates from human CRC and breast cancer | 4.92–100 ng mL−1 (IL-13sRα2) 0.11–10 ng mL−1 (CDH-17) | 1.44 ng mL−1 (IL-13sRα2) 0.03 ng mL−1 (CDH-17) | [112] |
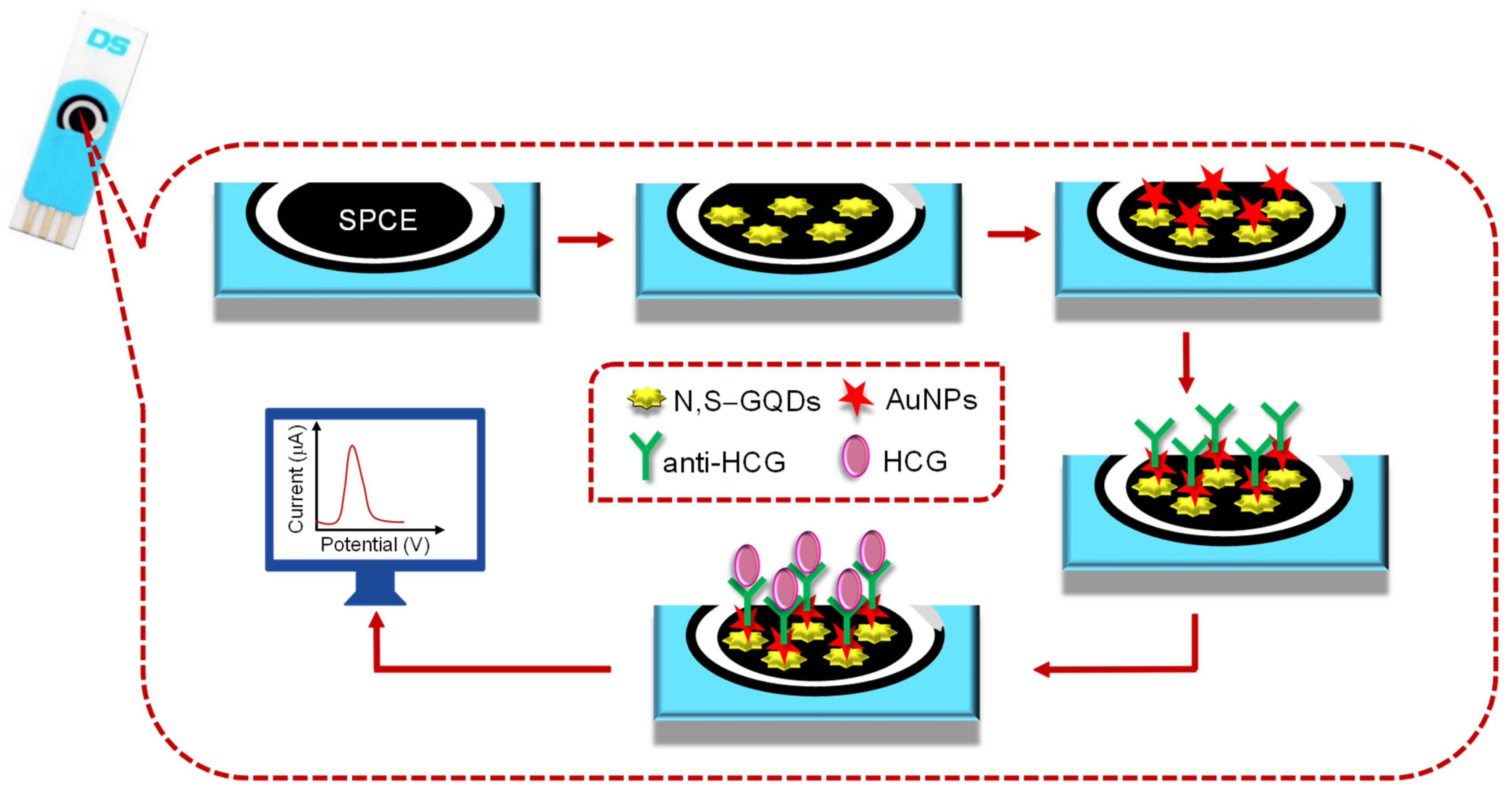
| SPCE | N,S–GQDs/AuNPs | HCG | Direct | CV, SWV, EIS | Human serum | 0.1–125 pg mL−1 | 12.5 fg mL−1 | [61] |
| Au-electrode | P-Cys/GQDs/AuNPs | p53 | Direct | SWV, DPV | Human plasma | 0.0488–12.5 pM | 23.4 fM | [125] |
| GCE | CysA/AuNPs/GQDs | CA 15-3 | Direct | SWV, CV | Human plasma; cellular lysates from human breast cancer | 0.16–125 U mL−1 | 0.11 U mL−1 | [126] |
| GCE | Au/Ag–rGO/GQDs | PSA | Direct | EIS, ECL | Human serum | 1 pg–10 ng mL−1 | 0.29 pg mL−1 | [127] |
| GCE | GQD/GS | PSA | Sandwich | SWV | Human serum | 0.005–10 ng mL−1 | 3 pg mL−1 | [128] |
| GCE | GN–Ag–Au/GQDs | CA 199 | Sandwich | ECL | Human serum | 0.002–70 U mL−1 | 0.96 mU mL−1 | [129] |
| Electrode | Nanomaterials | Biomarker | Technique(s) | Linear Range | LOD | Reference |
|---|---|---|---|---|---|---|
| SPCE | GQDs implanted with 2-ABA | AXL | DPV | 1.7–1000 pg mL−1 | 0.5 pg mL−1 | [136] |
| SPGE | PAMAM/GQDs | cTnI | CV, DPV | 10−6–10 ng mL−1 | 20 fg mL−1 | [137] |
| SPCE | GQDs | cMyo | CV, DPV, EIS | 0.01–100 ng mL−1 | 0.01 ng mL−1 | [66] |
| SPCE | GQDs | CRP | EIS | 0.5–70 nM | 176 pM | [138] |
| Electrode | Nanomaterials | Pathogen | Assay Mode | Technique(s) | Sample(s) | Linear Range | LOD | Reference |
|---|---|---|---|---|---|---|---|---|
| Gold | GQDs | Y. enterocolitica (bacteria) | Direct | Amperometry | Milk and human serum | 1–6.23 × 108 cfu mL−1 (milk); 1–6.23 × 108 cfu mL−1 (serum) | 5 cfu mL−1 (milk); 30 cfu mL−1 (serum) | [63] |
| GCE | Fe3O4@AG/GQDs | CFP-10 (bacteria) | Sandwich | DPV | Human urine | 0.005–500 μg mL−1 | 00.33 ng mL−1 | [147] |
| GCE | PDA/N–GQDs | E. coli (bacteria) | Sandwich | ECL; CV; EIS | Water | 10–107 cfu mL−1 | 8 cfu mL−1 | [148] |
| Platinum | GQDs | S. typhimurium (bacteria) | Direct | EIS | Buffer | 1 pM–100 nM | 1 pM | [149] |
| GCE | AgNPs/thiol–GQDs | HCV (virus) | Direct | DPV | Human serum | 0.05 pg–60 ng mL−1 | 3 fg mL−1 | [150] |
| GCE | N,S–GQDs/AuNPs/PANI | HEV (virus) | Direct | CV; EIS | Buffer, human serum, and feces of HEV-infected monkey | 1–105 fg mL−1 (feces of HEV-infected monkey); 102–107 RNA copies mL−1 (human serum) | 0.8 fg mL−1 (feces of HEV-infected monkey); 96.7 RNA copies mL−1 (human serum) | [151] |
| GCE | AuPd/N–GQDs@PS | HBsAg (virus) | Direct | Amperometry | Human serum | 10 fg mL−1–50 ng mL−1 | 3.3 fg mL−1 | [82] |
| Carbon | GQDs/AuNBs | FAdVs (virus) | Sandwich | CV | Chicken blood | 10–50 pfu mL−1 | 8.75 pfu mL−1 | [152] |
| GCE | Fe3O4/GQDs/Cu-apoferritin | ALVs-J (virus) | Sandwich | DPV | Human serum | 102.08–104.5 TCID50 mL−1 | 115 TCID50 mL−1 | [153] |
| SPCE | GQDs/2-ABA | Parathion (toxin) | Direct | EIS | Food, water, and soil | 0.01–106 ng L−1 | 46 pg L−1 | [154] |
| Glass | GQDs/ITO | AFB1 (toxin) | Direct | CV; EIS | Contaminated maize | 0.1–2.0 ng mL−1 | 0.03 ng mL−1 | [155] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansuriya, B.D.; Altintas, Z. Graphene Quantum Dot-Based Electrochemical Immunosensors for Biomedical Applications. Materials 2020, 13, 96. https://doi.org/10.3390/ma13010096
Mansuriya BD, Altintas Z. Graphene Quantum Dot-Based Electrochemical Immunosensors for Biomedical Applications. Materials. 2020; 13(1):96. https://doi.org/10.3390/ma13010096
Chicago/Turabian StyleMansuriya, Bhargav D., and Zeynep Altintas. 2020. "Graphene Quantum Dot-Based Electrochemical Immunosensors for Biomedical Applications" Materials 13, no. 1: 96. https://doi.org/10.3390/ma13010096
APA StyleMansuriya, B. D., & Altintas, Z. (2020). Graphene Quantum Dot-Based Electrochemical Immunosensors for Biomedical Applications. Materials, 13(1), 96. https://doi.org/10.3390/ma13010096