Specificities and Efficiencies of Primers Targeting Candidatus Phylum Saccharibacteria in Activated Sludge
Abstract
1. Introduction
2. Results and Discussion
2.1. Assessment of Selected Saccharibacteria-Specific Primers
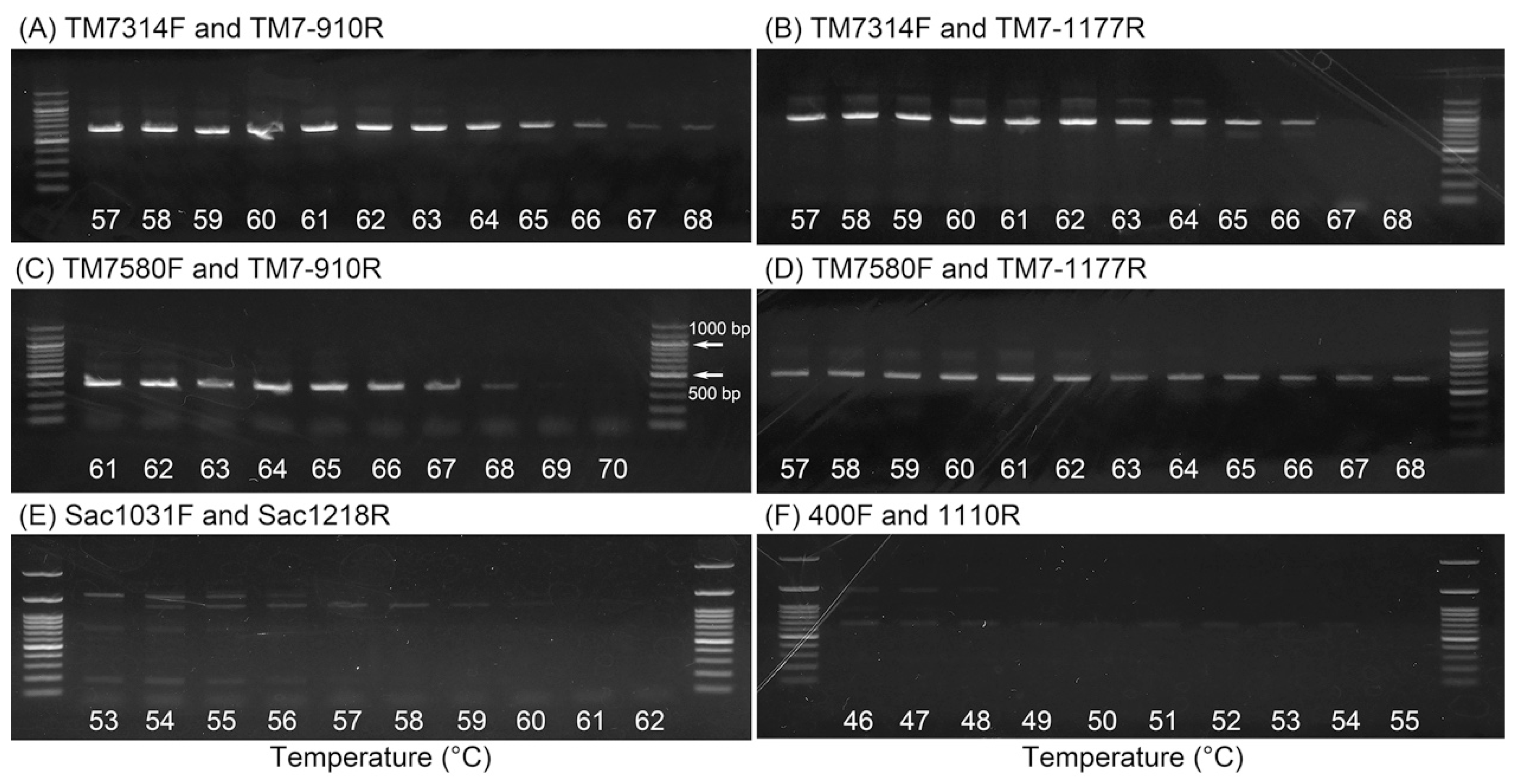
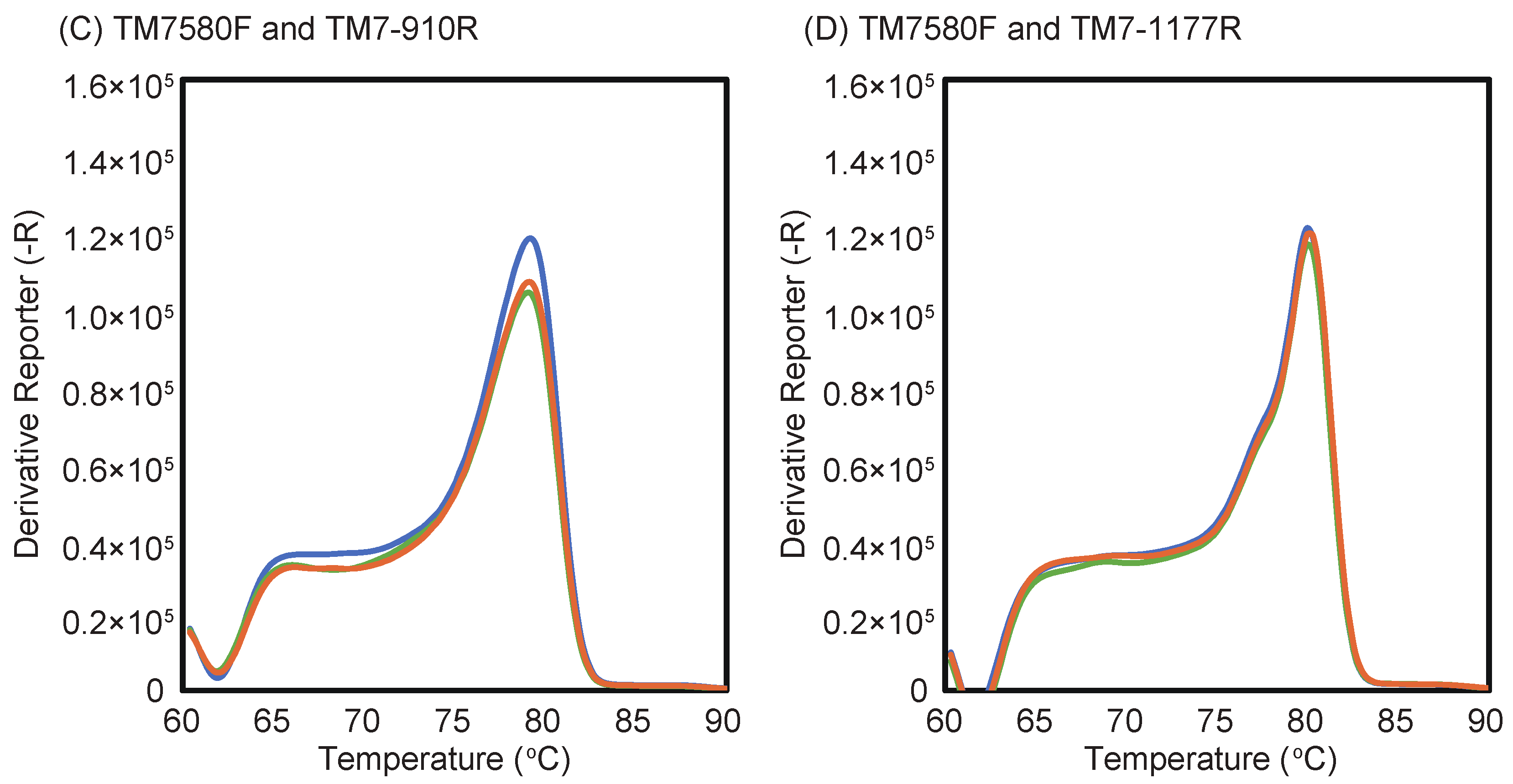
2.2. Determination of Optimal Annealing Temperatures
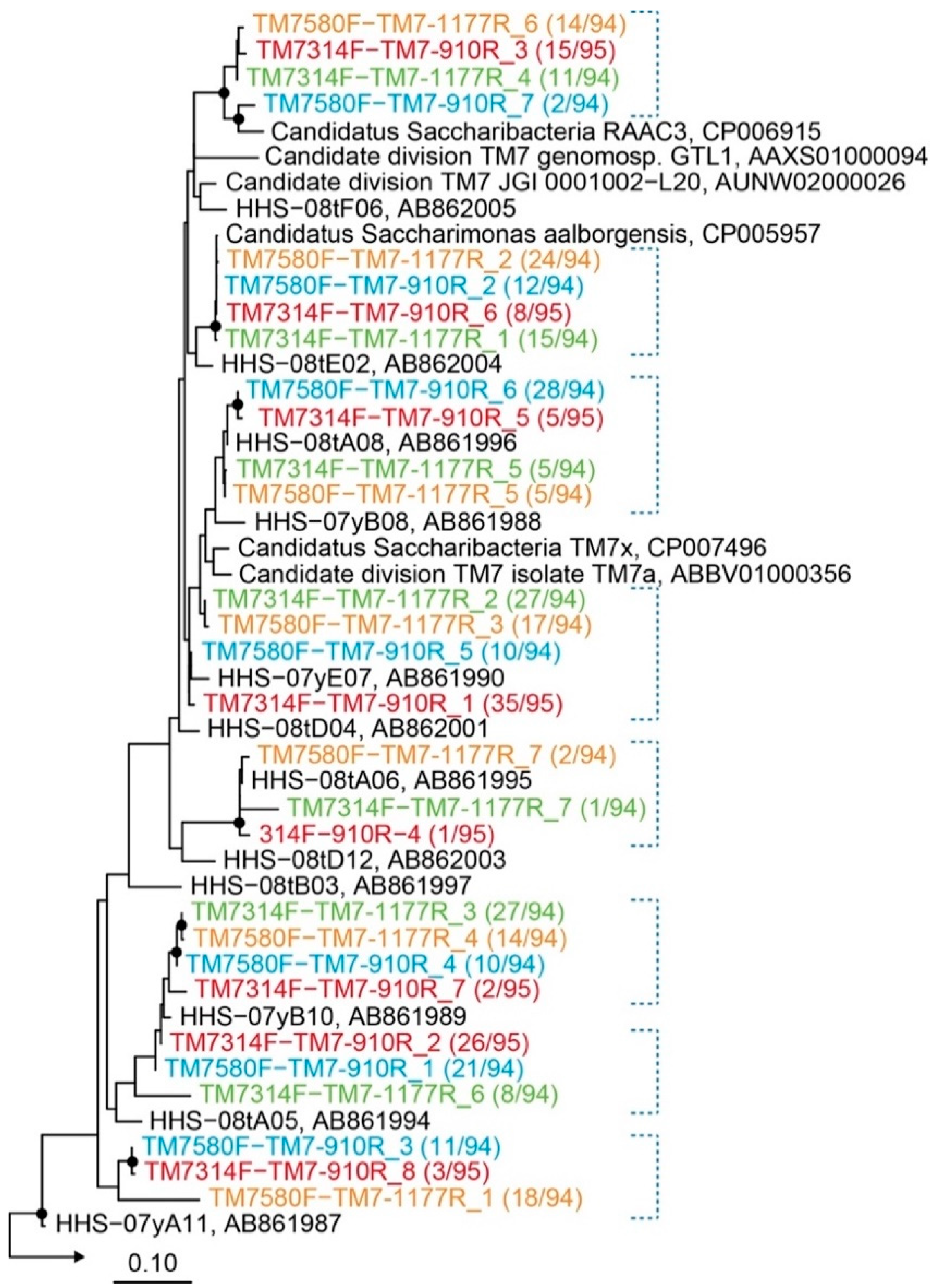
2.3. Evaluation of Primer Specificity by Cloning Analysis
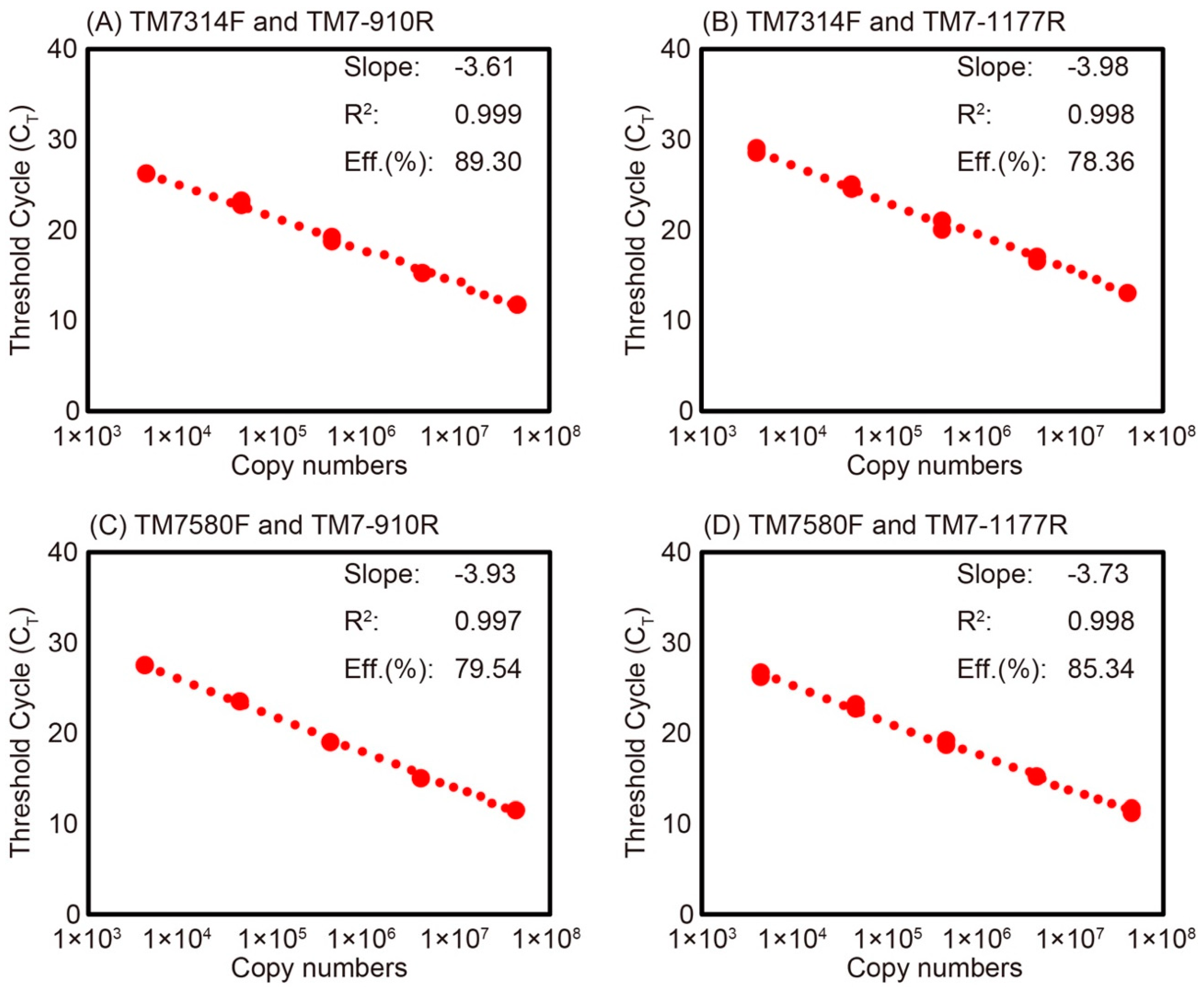
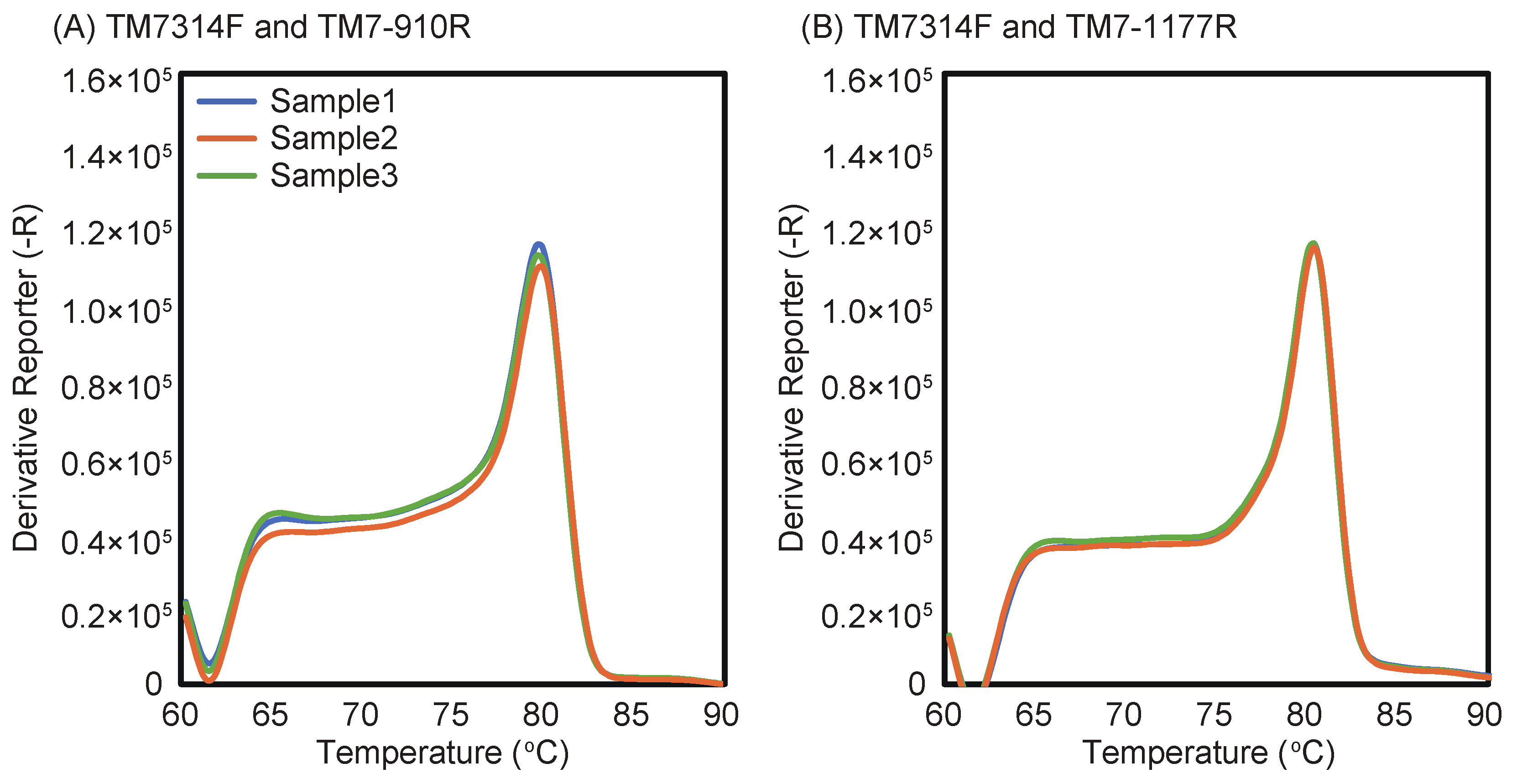
2.4. Evaluation of Primer Specificity by qPCR
3. Materials and Methods
3.1. Primer Selection and Evaluation
3.2. Activated Sludge Samples
3.3. DNA Extraction and PCR Amplification
3.4. Cloning, Sequencing, and Phylogenetic Analysis
3.5. Real-Time Quantitative PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wagner, M.; Amann, R.; Lemmer, H.; Schleifer, K.-H. Probing activated sludge with oligonucleotides specific for proteobacteria: Inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 1993, 59, 1520–1525. [Google Scholar] [PubMed]
- Kindaichi, T.; Kawano, Y.; Ito, T.; Satoh, H.; Okabe, S. Population dynamics and in situ kinetics of nitrifying bacteria in autotrophic nitrifying biofilms as determined by real-time quantitative PCR. Biotechnol. Bioeng. 2006, 94, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Kindaichi, T.; Awata, T.; Suzuki, Y.; Tanabe, K.; Hatamoto, M.; Ozaki, N.; Ohashi, A. Enrichment using an up-flow column reactor and community structure of marine anammox bacteria from coastal sediment. Microbes Environ. 2011, 26, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kindaichi, T.; Nierychlo, M.; Kragelund, C.; Nielsen, J.L.; Nielsen, P.H. High and stable substrate specificities of microorganisms in enhanced biological phosphorus removal plants. Environ. Microbiol. 2013, 15, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Danczak, R.E.; Johnston, M.D.; Kenah, C.; Slattery, M.; Wrighton, K.C.; Wilkins, M.J. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 2017, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, B.; Winsley, T.; Ji, M.; Neilan, B. Insights into the distribution and abundance of the ubiquitous candidatus Saccharibacteria phylum following tag pyrosequencing. Sci. Rep. 2014, 4, 3957. [Google Scholar] [CrossRef] [PubMed]
- Soro, V.; Dutton, L.C.; Sprague, S.V.; Nobbs, A.H.; Ireland, A.J.; Sandy, J.R.; Jepson, M.A.; Micaroni, M.; Splatt, P.R.; Dymock, D.; et al. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl. Environ. Microbiol. 2014, 80, 6480–6489. [Google Scholar] [CrossRef] [PubMed]
- He, X.; McLean, J.S.; Edlund, A.; Yooseph, S.; Hall, A.P.; Liu, S.-Y.; Dorrestein, P.C.; Esquenazi, E.; Hunter, R.C.; Cheng, G.; et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2015, 112, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Tyson, G.W.; Webb, R.I.; Wagner, A.M.; Blackall, L.L. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 2001, 67, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Albertsen, M.; Hugenholtz, P.; Skarshewski, A.; Nielsen, K.L.; Tyson, G.W.; Nielsen, P.H. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 2013, 31, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Kindaichi, T.; Yamaoka, S.; Uehara, R.; Ozaki, N.; Ohashi, A.; Albertsen, M.; Nielsen, P.H.; Nielsen, J.L. Phylogenetic diversity and ecophysiology of Candidate phylum Saccharibacteria in activated sludge. FEMS Microbiol. Ecol. 2016, 92, fiw078. [Google Scholar] [CrossRef] [PubMed]
- Bor, B.; Poweleit, N.; Bois, J.S.; Cen, L.; Bedree, J.K.; Zhou, Z.H.; Gunsalus, R.P.; Lux, R.; McLean, J.S.; He, X.; et al. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont Actinomyces. Microbiol. Ecol. 2016, 71, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Nittami, T.; Speirs, L.B.; Fukuda, J.; Watanabe, M.; Seviour, R.J. Fluorescence in situ hybridization probes targeting members of the phylum Candidatus Saccharibacteria falsely target Eikelboom type 1851 filaments and other Chloroflexi members. Environ. Microbiol. Rep. 2014, 6, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Sizova, M.V.; Doerfert, S.N.; Gavrish, E.; Epstein, S.S. TM7 detection in human microbiome: Are PCR primers and FISH probes specific enough? J. Microbiol. Methods 2015, 114, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Brinig, M.M.; Lepp, P.W.; Ouverney, C.C.; Armitage, G.C.; Relman, D.A. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl. Environ. Microbiol. 2003, 69, 1687–1694. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-W.; Chen, M.-K.; Yang, B.-Y.; Huang, X.-J.; Zhang, X.-R.; He, L.-Q.; Zhang, J.; Hua, Z.-C. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 2015, 81, 6749–6756. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ludwig, W.; Strunk, O.; Westram, R.; Richter, L.; Meier, H.; Yadhukumar; Buchner, A.; Lai, T.; Steppi, S.; Jobb, G.; et al. ARB: A software environment for sequence data. Nucleic Acids Res. 2004, 32, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Tsushima, I.; Kindaichi, T.; Okabe, S. Quantification of anaerobic ammonium-oxidizing bacteria in enrichment cultures by real-time PCR. Water Res. 2007, 41, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Marsh, T.; Cheng, H.; Forney, L.J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1997, 63, 4516–4522. [Google Scholar] [PubMed]
- Nurmiyanto, A.; Kodera, H.; Kindaichi, T.; Ozaki, N.; Aoi, Y.; Ohashi, A. Dominant Candidatus Accumulibacter phosphatis enriched in response to phosphate concentrations in EBPR process. Microbes Environ. 2017, 32, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannokous, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
| Primer | Sequence (5′ to 3′) | Reference |
|---|---|---|
| TM7314F | GAGAGGATGATCAGCCAG | [10] |
| TM7580F | AYTGGGCGTAAAGAGTTGC | [10] |
| Sac1031F | AAGAGAACTGTGCCTTCGG | [17] |
| 400F | TATGAGTGAAGAATATGAC | [13] |
| TM7-910R | GTCCCCGTCAATTCCTTTATG | [16] |
| TM7-1177R | GACCTGACATCATCCCCTCCTTCC | [16] |
| Sac1218R | GCGTAAGGGAAATACTGACC | [17] |
| 1110R | CAGTCCAAGTAGAAAAATAC | [13] |
| Primer Set | Expected Fragment Length (bp) | DNA Band | Annealing Temperature (°C) | Number of Saccharibacteria Sequences a/Number of Total Clones Analyzed (Specificity) |
|---|---|---|---|---|
| TM7314F and TM7-910R | 596 | yes | 64 | 95/95 (100%) |
| TM7314F and TM7-1177R | 863 | yes | 64 | 96/96 (100%) |
| TM7580F and TM7-910R | 330 | yes | 66 | 94/94 (100%) |
| TM7580F and TM7-1177R | 597 | yes | 63 | 94/94 (100%) |
| Sac1031F and Sac1218R | 187 | no | Not determined | Not determined |
| 400F and 1100R | 700 | no | Not determined | Not determined |
| Primer Set | Copy Number of Plasmid (Copies/ng-DNA) a | Measured Copy Number of Plasmid (Copies/ng-DNA) b | Copy Number of Saccharibacteria in Activated Sludge (Copies/g-MLSS) b |
|---|---|---|---|
| TM7314F and TM7-910R | 1.0 × 103 | 1.4 ± 0.1 × 103 | 6.8 ± 0.3 × 106 |
| TM7314F and TM7-1177R | 9.6 × 103 | 1.4 ± 0.1 × 104 | 5.6 ± 1.1 × 106 |
| TM7580F and TM7-910R | 8.9 × 103 | 1.8 ± 0.2 × 104 | 6.7 ± 1.1 × 107 |
| TM7580F and TM7-1177R | 9.9 × 104 | 2.1 ± 0.3 × 105 | 3.5 ± 1.0 × 107 |
| Primer | Sequence of Hydrogenophaga sp. (5′ to 3′) a |
|---|---|
| TM7314F | GAGAGGACGACCAGCCAC |
| TM7580F | ACTGGGCGTAAAGCGTGCG |
| TM7-910R | CTCAAAGGAATTGACGGGGAC |
| TM7-1177R | GGAAGGTGGGGATGACGTCAAGTC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takenaka, R.; Aoi, Y.; Ozaki, N.; Ohashi, A.; Kindaichi, T. Specificities and Efficiencies of Primers Targeting Candidatus Phylum Saccharibacteria in Activated Sludge. Materials 2018, 11, 1129. https://doi.org/10.3390/ma11071129
Takenaka R, Aoi Y, Ozaki N, Ohashi A, Kindaichi T. Specificities and Efficiencies of Primers Targeting Candidatus Phylum Saccharibacteria in Activated Sludge. Materials. 2018; 11(7):1129. https://doi.org/10.3390/ma11071129
Chicago/Turabian StyleTakenaka, Ryota, Yoshiteru Aoi, Noriatsu Ozaki, Akiyoshi Ohashi, and Tomonori Kindaichi. 2018. "Specificities and Efficiencies of Primers Targeting Candidatus Phylum Saccharibacteria in Activated Sludge" Materials 11, no. 7: 1129. https://doi.org/10.3390/ma11071129
APA StyleTakenaka, R., Aoi, Y., Ozaki, N., Ohashi, A., & Kindaichi, T. (2018). Specificities and Efficiencies of Primers Targeting Candidatus Phylum Saccharibacteria in Activated Sludge. Materials, 11(7), 1129. https://doi.org/10.3390/ma11071129





