A Single-Step Surface Modification of Electrospun Silica Nanofibers Using a Silica Binding Protein Fused with an RGD Motif for Enhanced PC12 Cell Growth and Differentiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Characterization of SNF Substrates
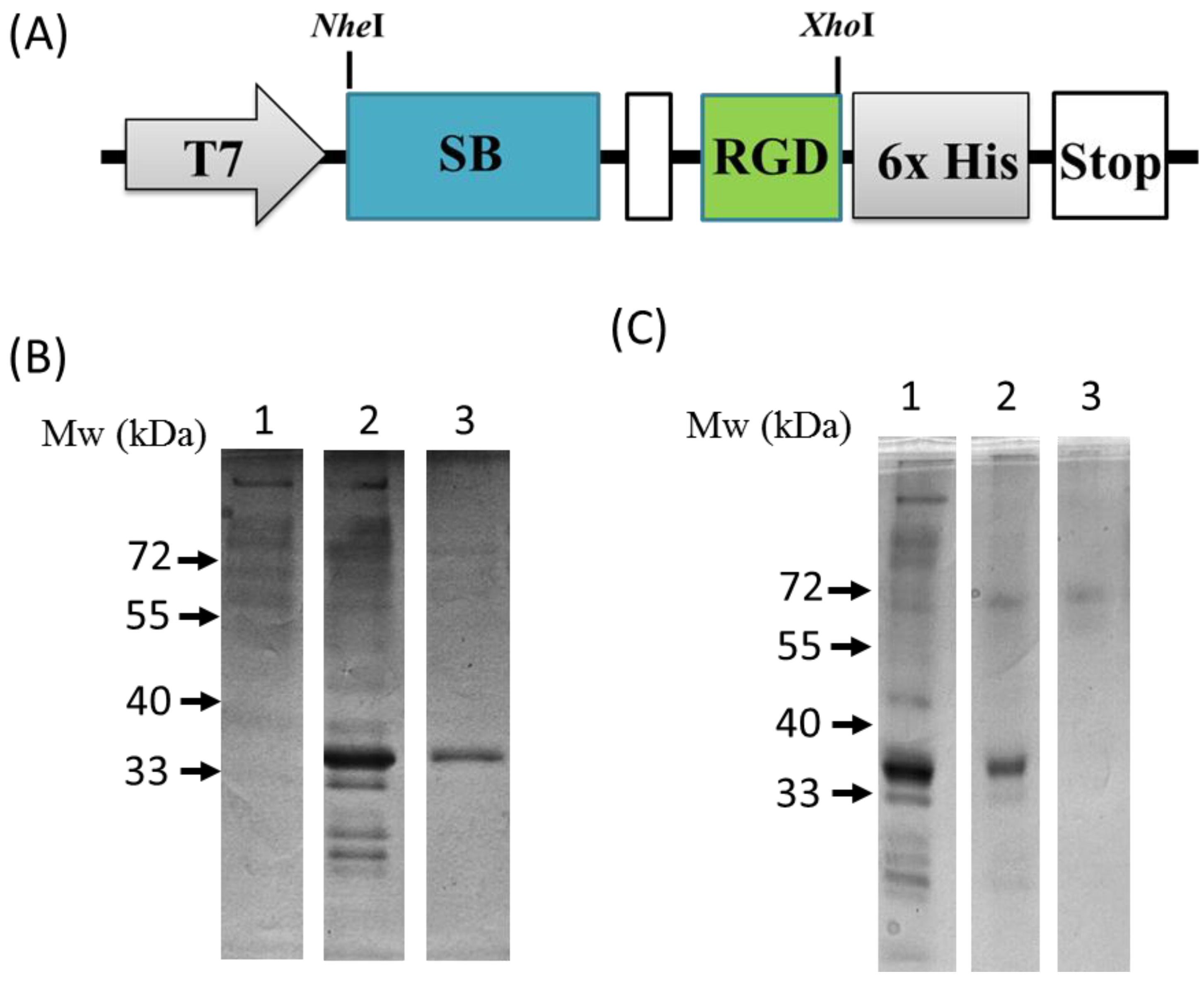
2.3. Plasmid Construction and the Expression and Purification of the SB-RGD Protein
2.4. Dissociation Testing of SB-RGD from Silica Particles
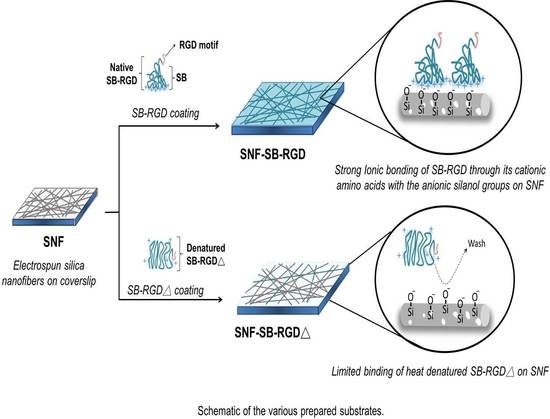
2.5. Preparation of SNF-SB-RGD and the SNF Coated with Heat-Denatured SB-RGD (SNF-SB-RGD∆)
2.6. Examining the Immobilization of Fusion Protein on SNF-SB-RGD and SNF-SB-RGD∆
2.7. In Vitro Culture of PC12 Cells
2.8. Cell Viability Assay
2.9. Immunocytochemistry Staining
2.10. Cell Morphology Study
2.11. Statistical Analysis
3. Results and Discussion
3.1. Genetic Engineering and Expression of the Recombinant SB-RGD Fusion Protein.
3.2. Purification and Functional Testing of SB-RGD
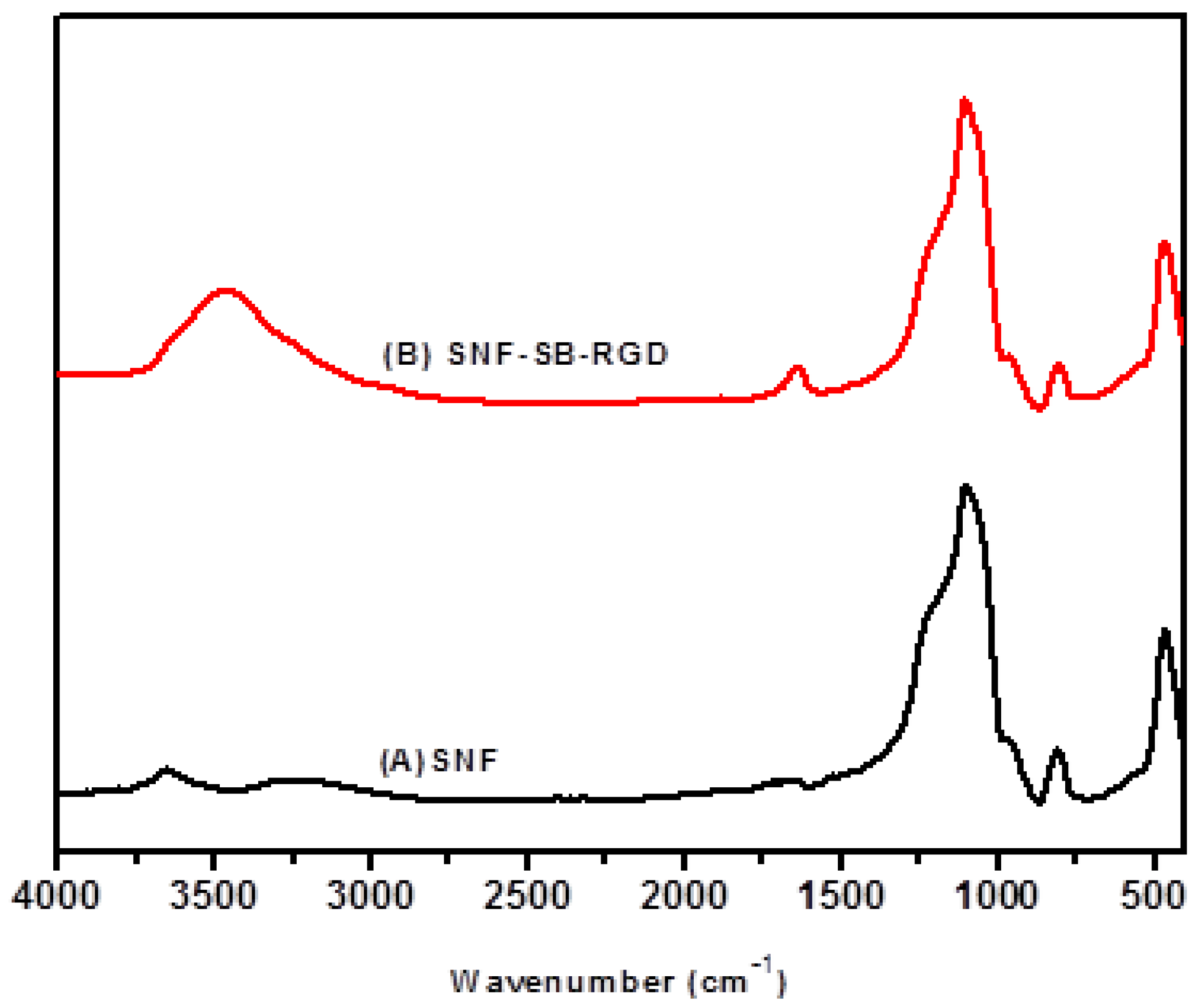
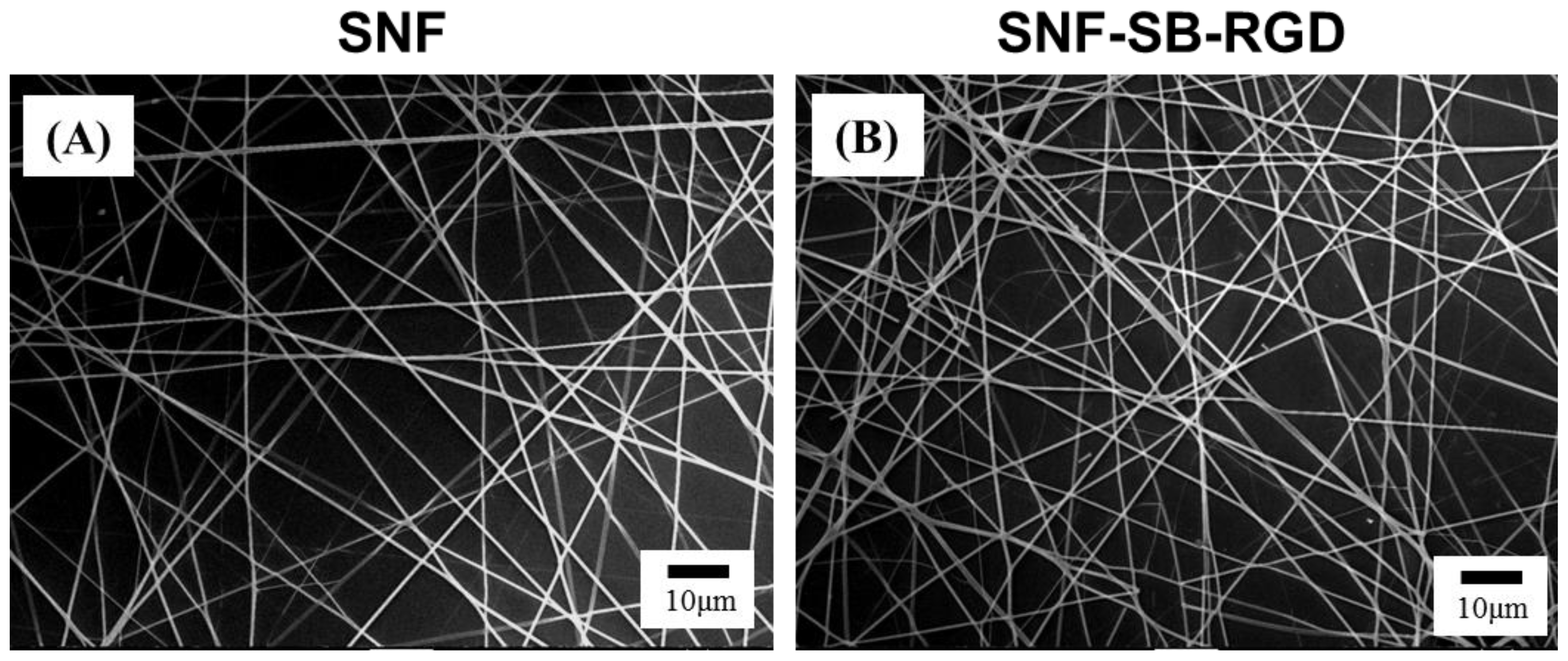
3.3. Preparation and Characterization of SNF-SB-RGD
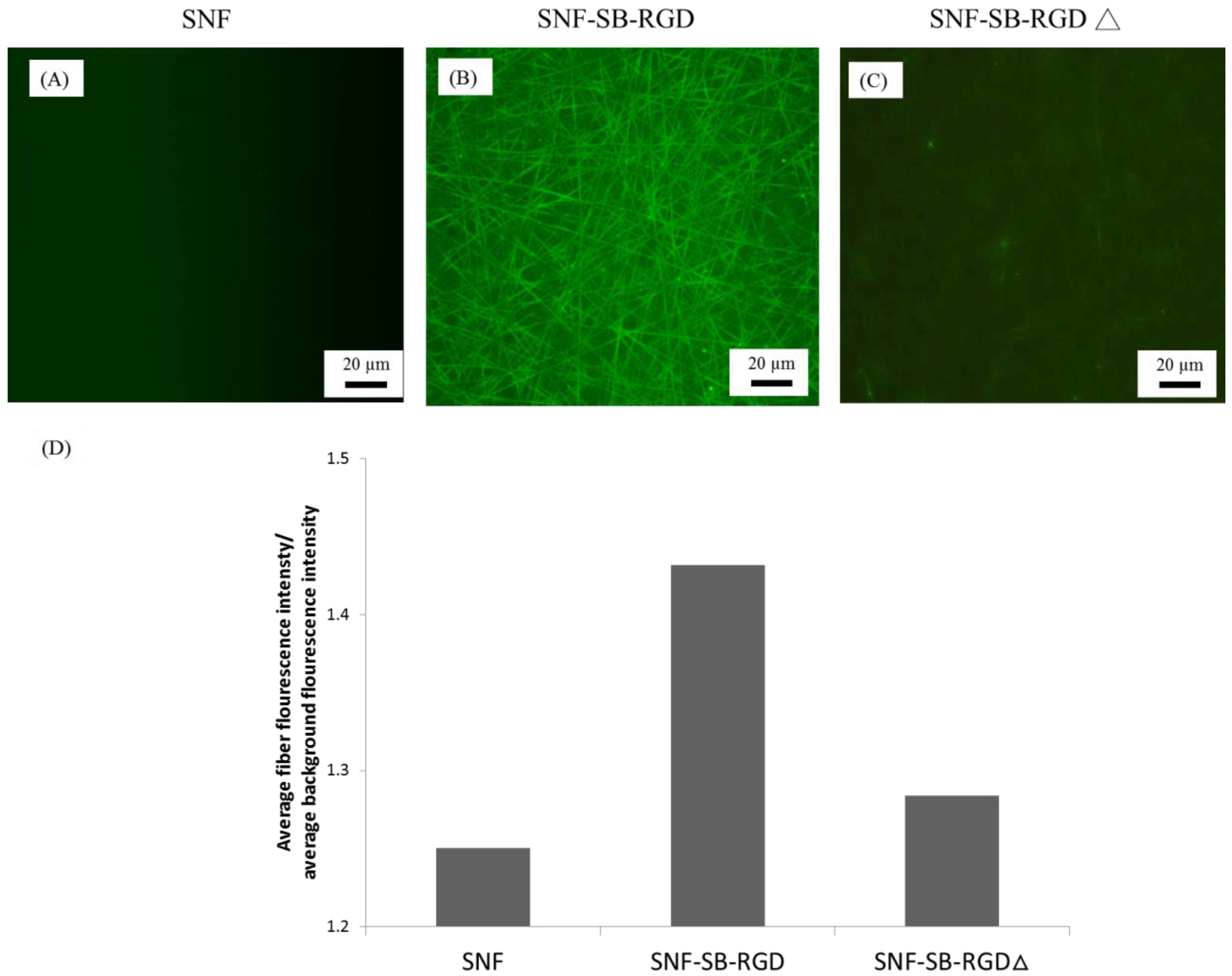
3.4. Binding of SB-RGD to the SNF Substrate’s Surface
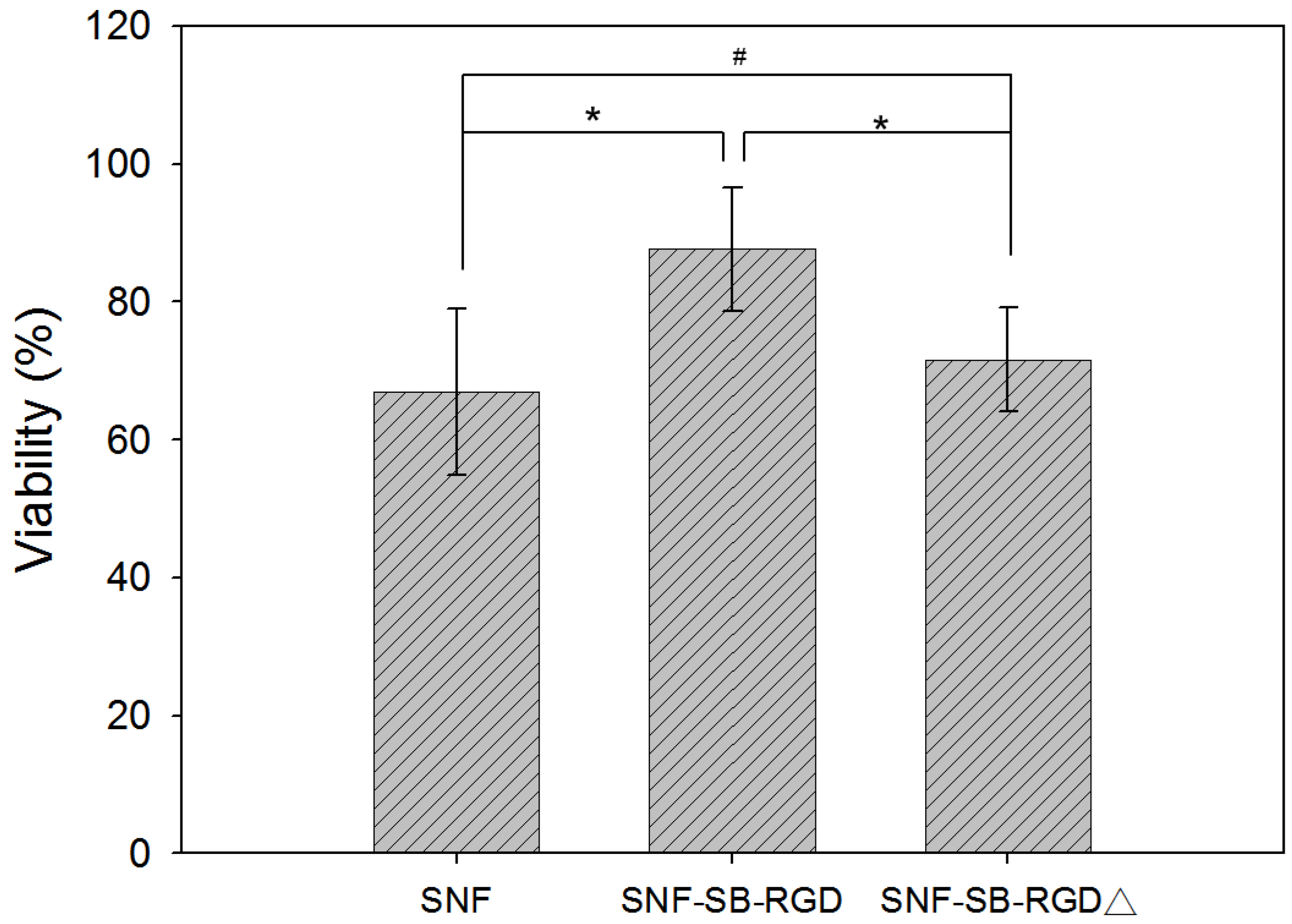
3.5. Effect of Fusion-Protein-Modified SNF on Cell Viability
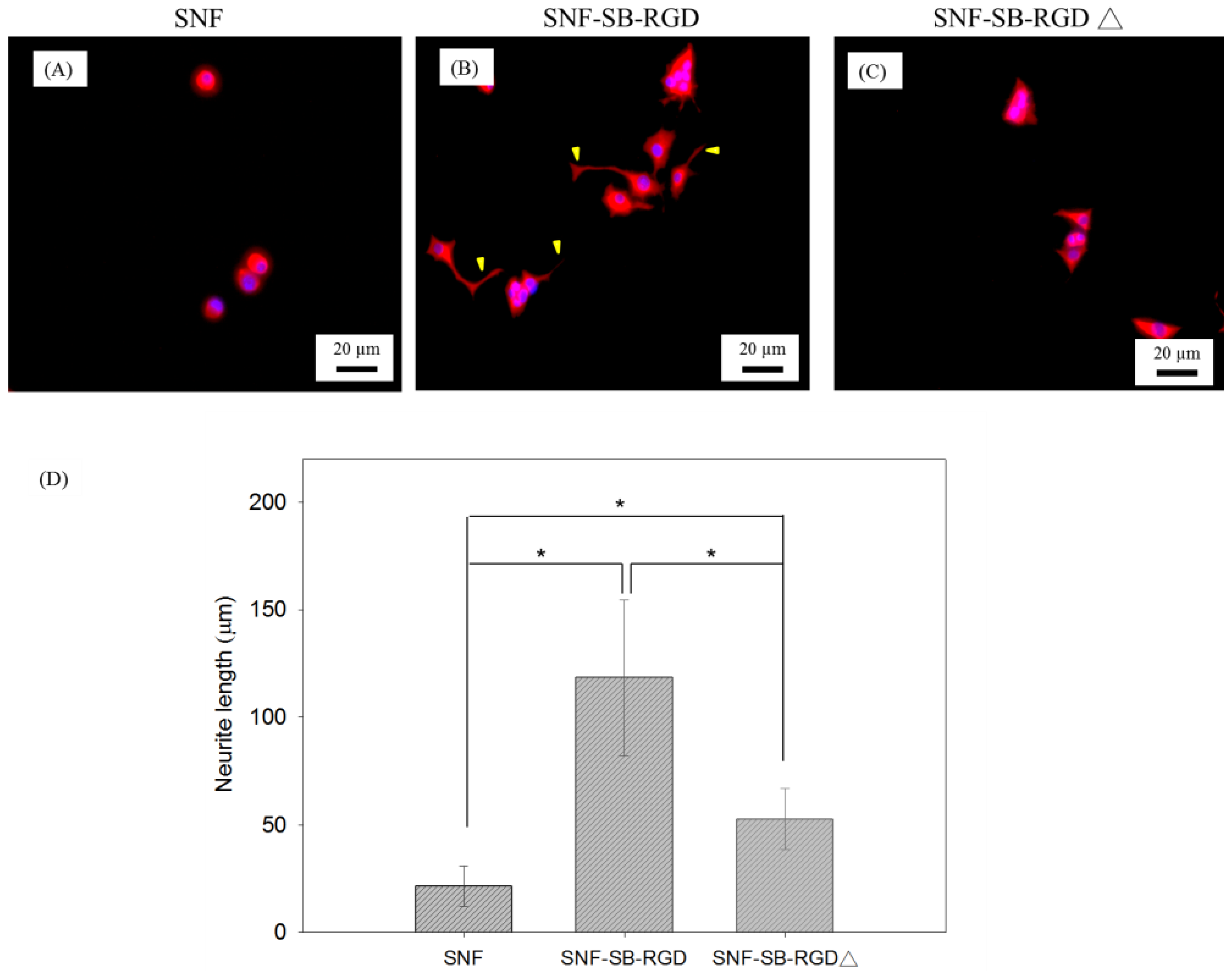
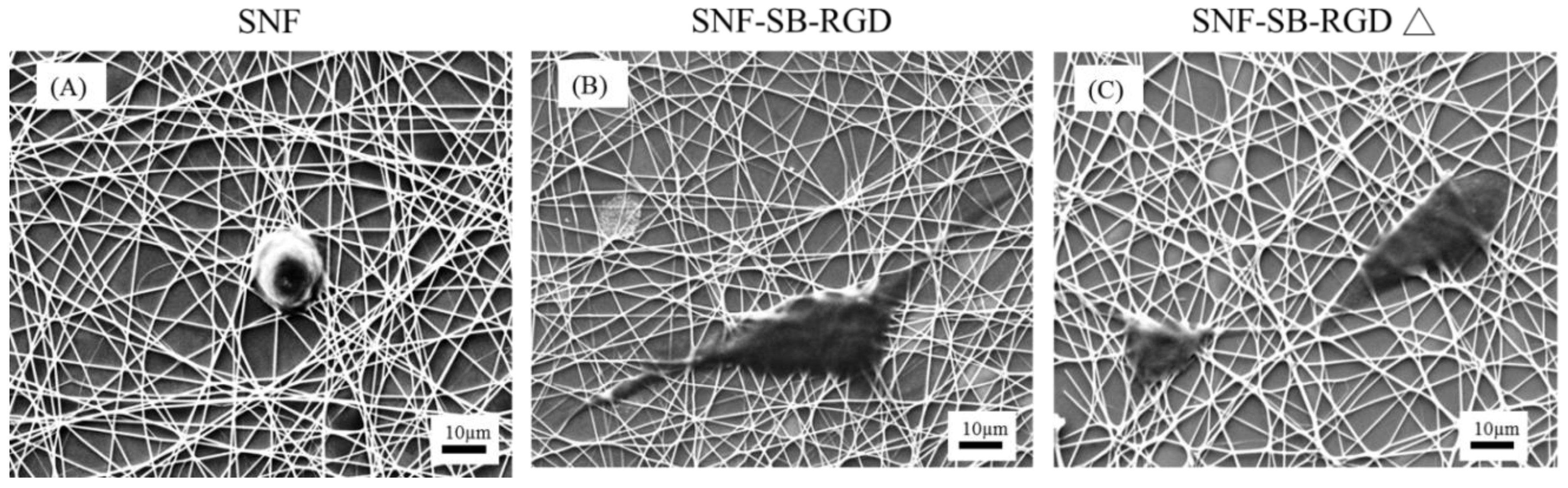
3.6. Effect of the Various Substrates on the Neurite Extension of PC12 Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dou, Q.Q.; Teng, C.P.; Ye, E.; Loh, X.J. Effective near-infrared photodynamic therapy assisted by upconversion nanoparticles conjugated with photosensitizers. Int. J. Nanomed. 2015, 10, 419–432. [Google Scholar]
- Peela, N.; Truong, D.; Saini, H.; Chu, H.; Mashaghi, S.; Ham, S.L.; Singh, S.; Tavana, H.; Mosadegh, B.; Nikkhah, M. Advanced biomaterials and microengineering technologies to recapitulate the stepwise process of cancer metastasis. Biomaterials 2017, 133, 176–207. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Oo, N.N.L.; Lee, J.P.; Li, Z.; Loh, X.J. Recent development of synthetic nonviral systems for sustained gene delivery. Drug Discov. Today 2017, 22, 1318–1335. [Google Scholar] [CrossRef] [PubMed]
- Loh, X.J.; Lee, T.-C.; Dou, Q.; Deen, G.R. Utilising inorganic nanocarriers for gene delivery. Biomater. Sci. 2016, 4, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarayanan, R.; Sridhar, R.; Loh, X.J.; Nandhakumar, M.; Barathi, V.A.; Kalaipriya, M.; Kwan, J.L.; Liu, S.P.; Beuerman, R.W.; Ramakrishna, S. Interaction of gelatin with polyenes modulates antifungal activity and biocompatibility of electrospun fiber mats. Int. J. Nanomed. 2014, 9, 2439–2458. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Oves, M.; Almeelbi, T.; Al-Makishah, N.H.; Barakat, M.A. Hybrid chitosan/polyaniline-polypyrrole biomaterial for enhanced adsorption and antimicrobial activity. J. Colloid Interface Sci. 2017, 490, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Kai, D.; Tan, M.J.; Prabhakaran, M.P.; Chan, B.Q.Y.; Liow, S.S.; Ramakrishna, S.; Loh, X.J. Biocompatible electrically conductive nanofibers from inorganic-organic shape memory polymers. Colloids Surf. B Biointerfaces 2016, 148, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Sensharma, P.; Madhumathi, G.; Jayant, R.D.; Jaiswal, A.K. Biomaterials and cells for neural tissue engineering: Current choices. Mater. Sci. Eng. C 2017, 77, 1302–1315. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, F.J. Biomaterials & scaffolds for tissue engineering. Mater. Today 2011, 14, 88–95. [Google Scholar]
- Lohmeyer, J.A.; Siemers, F.; Machens, H.G.; Mailander, P. The clinical use of artificial nerve conduits for digital nerve repair: A prospective cohort study and literature review. J. Reconstr. Microsurg. 2009, 25, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Bertleff, M.J.O.E.; Meek, M.F.; Nicolai, J.-P.A. A prospective clinical evaluation of biodegradable neurolac nerve guides for sensory nerve repair in the hand. J. Hand Surg. Am. 2005, 30, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Liverani, L.; Boccaccini, A. Versatile production of poly(epsilon-caprolactone) fibers by electrospinning using benign solvents. Nanomaterials 2016, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Sundaramurthi, D.; Jaidev, L.R.; Ramana, L.N.; Sethuraman, S.; Krishnan, U.M. Osteogenic differentiation of stem cells on mesoporous silica nanofibers. RSC Adv. 2015, 5, 69205–69214. [Google Scholar] [CrossRef]
- Chen, W.S.; Hsieh, P.H.; Yang, W.N.; Fan-Jen, P.Z.; Yang, M.-L.; Yeh, J.M.; Wei, Y.; Chin, T.Y.; Chen-Yang, Y.W. Chemically modified electrospun silica nanofibers for promoting growth and differentiation of neural stem cells. J. Mater. Chem. B 2014, 2, 1205–1215. [Google Scholar] [CrossRef]
- Feng, Z.V.; Chen, W.S.; Keratithamkul, K.; Stoick, M.; Kapala, B.; Johnson, E.; Huang, A.-C.; Chin, T.Y.; Chen-Yang, Y.W.; Yang, M.-L. Degradation of the electrospun silica nanofiber in a biological medium for primary hippocampal neuron–effect of surface modification. Int. J. Nanomed. 2016, 11, 729–741. [Google Scholar]
- Garcia Cruz, D.M.; Gomez Ribelles, J.L.; Salmeron Sanchez, M. Blending polysaccharides with biodegradable polymers. I. Properties of chitosan/polycaprolactone blends. J. Biomed. Mater. Res. B Appl. Biomater. 2008, 85, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, L.A.; Rebaza, L.M.; Derzic, S.; Schwartz, P.H.; Monuki, E.S. Regulation of human neural precursor cells by laminin and integrins. J. Neurosci. Res. 2006, 83, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M. Adhesive recognition sequences. J. Biol. Chem. 1991, 266, 12809–12812. [Google Scholar] [PubMed]
- Andrade, F.K.; Moreira, S.M.; Domingues, L.; Gama, F.M. Improving the affinity of fibroblasts for bacterial cellulose using carbohydrate-binding modules fused to RGD. J. Biomed. Mater. Res. A 2010, 92, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, P.; Chen, C.B.; Wang, Z.H.; Ma, P.; Chen, G.Q. The improvement of fibroblast growth on hydrophobic biopolyesters by coating with polyhydroxyalkanoate granule binding protein PhaP fused with cell adhesion motif RGD. Biomaterials 2010, 31, 8921–8930. [Google Scholar] [CrossRef] [PubMed]
- Bilem, I.; Chevallier, P.; Plawinski, L.; Sone, E.D.; Durrieu, M.C.; Laroche, G. RGD and BMP-2 mimetic peptide crosstalk enhances osteogenic commitment of human bone marrow stem cells. Acta Biomater. 2016, 36, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Meroni, D.; Lo Presti, L.; Di Liberto, G.; Ceotto, M.; Acres, R.G.; Prince, K.C.; Bellani, R.; Soliveri, G.; Ardizzone, S. A close look at the structure of the TiO2-APTES interface in hybrid nanomaterials and its degradation pathway: An experimental and theoretical study. J. Phys. Chem. C 2017, 121, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Dettin, M.; Bagno, A.; Gambaretto, R.; Iucci, G.; Conconi, M.T.; Tuccitto, N.; Menti, A.M.; Grandi, C.; Di Bello, C.; Licciardello, A.; et al. Covalent surface modification of titanium oxide with different adhesive peptides: Surface characterization and osteoblast-like cell adhesion. J. Biomed. Mater. Res. A 2009, 90, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Neff, J.A.; Tresco, P.A.; Caldwell, K.D. Surface modification for controlled studies of cell–ligand interactions. Biomaterials 1999, 20, 2377–2393. [Google Scholar] [CrossRef]
- Hersel, U.; Dahmen, C.; Kessler, H. RGD modified polymers: Biomaterials for stimulated cell adhesion and beyond. Biomaterials 2003, 24, 4385–4415. [Google Scholar] [CrossRef]
- Moreira, S.M.; Andrade, F.K.; Domingues, L.; Gama, M. Development of a strategy to functionalize a dextrin-based hydrogel for animal cell cultures using a starch-binding module fused to RGD sequence. BMC Biotechnol. 2008, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Li, J.; Li, L.; Dong, Y.; Chen, G.-Q.; Chen, K.C. Enhanced proliferation and differentiation of neural stem cells grown on PHA films coated with recombinant fusion proteins. Acta Biomater. 2013, 9, 7845–7854. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kwon, C.; Jeon, H. Genetically engineered phage induced selective H9c2 cardiomyocytes patterning in PDMS microgrooves. Materials 2017, 10, 973. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.S.; Guo, L.Y.; Tang, C.C.; Tsai, C.K.; Huang, H.H.; Chin, T.Y.; Yang, M.L. The effect of laminin surface modification of electrospun silica nanofiber substrate on neuronal tissue engineering. Nanomaterials 2018, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Nomura, K.; Hata, Y.; Nishimura, T.; Asami, Y.; Kuroda, A. The Si-tag for immobilizing proteins on a silica surface. Biotechnol. Bioeng. 2007, 96, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.E.; Shastri, V.R.; Vacanti, J.P.; Langer, R. Stimulation of neurite outgrowth using an electrically conducting polymer. Proc. Natl. Acad. Sci. USA 1997, 94, 8948–8953. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.A.; Tischler, A.S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 1976, 73, 2424–2428. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.Y.; Au, N.P.; Ma, C.H. The association between laminin and microglial morphology in vitro. Sci. Rep. 2016, 6, 28580. [Google Scholar] [CrossRef] [PubMed]
- Amritraj, A.; Wang, Y.; Revett, T.J.; Vergote, D.; Westaway, D.; Kar, S. Role of cathepsin d in u18666a-induced neuronal cell death: Potential implication in niemann-pick type c disease pathogenesis. J. Biol. Chem. 2013, 288, 3136–3152. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.-J.; Zhang, H.; Huang, H.; Wang, X.-M.; Zhou, Z.-Y.; Cui, F.-Z.; An, Y.-H. In vitro behavior of neural stem cells in response to different chemical functional groups. Biomaterials 2009, 30, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Jacob, M.; Sarria, J.C.; Steiner, P.; Hirling, H.; Unser, M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. A 2004, 58, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Ninomiya, K.; Hirota, R.; Kuroda, A. Single-step affinity purification of recombinant proteins using the silica-binding Si-tag as a fusion partner. Protein Expr. Purif. 2010, 71, 91–95. [Google Scholar] [CrossRef] [PubMed]
- De Campos Vidal, B.; Mello, M.L.S. Collagen type i amide i band infrared spectroscopy. Micron 2011, 42, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Kim, M.H.; Park, S.H.; Lee, H.J.; Kim, I.K.; Chung, H.M. Synthesis of Arg-Gly-Asp (RGD) sequence conjugated thermo-reversible gel via the PEG spacer arm as an extracellular matrix for a pheochromocytoma cell (PC12) culture. Biosci. Biotechnol. Biochem. 2004, 68, 2224–2229. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Jia, T.T.; Huang, Q.X.; Qiu, Y.Y.; Xu, J.; Yin, P.H.; Liu, T. Mesoporous silica nanoparticles (MSNs)-based organic/inorganic hybrid nanocarriers loading 5-fluorouracil for the treatment of colon cancer with improved anticancer efficacy. Colloids Surf. B Biointerfaces 2017, 159, 375–385. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.S.; Guo, L.Y.; Masroujeh, A.M.; Augustine, A.M.; Tsai, C.K.; Chin, T.Y.; Chen-Yang, Y.W.; Yang, M.-L. A Single-Step Surface Modification of Electrospun Silica Nanofibers Using a Silica Binding Protein Fused with an RGD Motif for Enhanced PC12 Cell Growth and Differentiation. Materials 2018, 11, 927. https://doi.org/10.3390/ma11060927
Chen WS, Guo LY, Masroujeh AM, Augustine AM, Tsai CK, Chin TY, Chen-Yang YW, Yang M-L. A Single-Step Surface Modification of Electrospun Silica Nanofibers Using a Silica Binding Protein Fused with an RGD Motif for Enhanced PC12 Cell Growth and Differentiation. Materials. 2018; 11(6):927. https://doi.org/10.3390/ma11060927
Chicago/Turabian StyleChen, Wen Shuo, Ling Yu Guo, Amien Mohamed Masroujeh, Anna Morgan Augustine, Cheng Kang Tsai, Ting Yu Chin, Yui Whei Chen-Yang, and Mong-Lin Yang. 2018. "A Single-Step Surface Modification of Electrospun Silica Nanofibers Using a Silica Binding Protein Fused with an RGD Motif for Enhanced PC12 Cell Growth and Differentiation" Materials 11, no. 6: 927. https://doi.org/10.3390/ma11060927
APA StyleChen, W. S., Guo, L. Y., Masroujeh, A. M., Augustine, A. M., Tsai, C. K., Chin, T. Y., Chen-Yang, Y. W., & Yang, M.-L. (2018). A Single-Step Surface Modification of Electrospun Silica Nanofibers Using a Silica Binding Protein Fused with an RGD Motif for Enhanced PC12 Cell Growth and Differentiation. Materials, 11(6), 927. https://doi.org/10.3390/ma11060927