Lactic Fermentation Spectral Analysis of Target Substrates and Food and Feed Wastes for Energy Applications
Abstract
1. Introduction
1.1. Feed and Food Waste and Methods of Its Processing
- -
- Incineration;
- -
- Methane fermentation;
- -
- Gasification;
- -
- Pyrolysis.
1.2. The Role of Lactic Acid Fermentation in the Anaerobic Stabilization of Waste
1.3. Evaluation of Technological Parameters of Lactic Fermentation
2. Materials and Methods
2.1. General Characteristics of the Research Material
2.2. Research Equipment
2.2.1. Marking the Weight of the Laboratory Sample
2.2.2. Determining the Sample Volume
2.2.3. Ambient Temperature
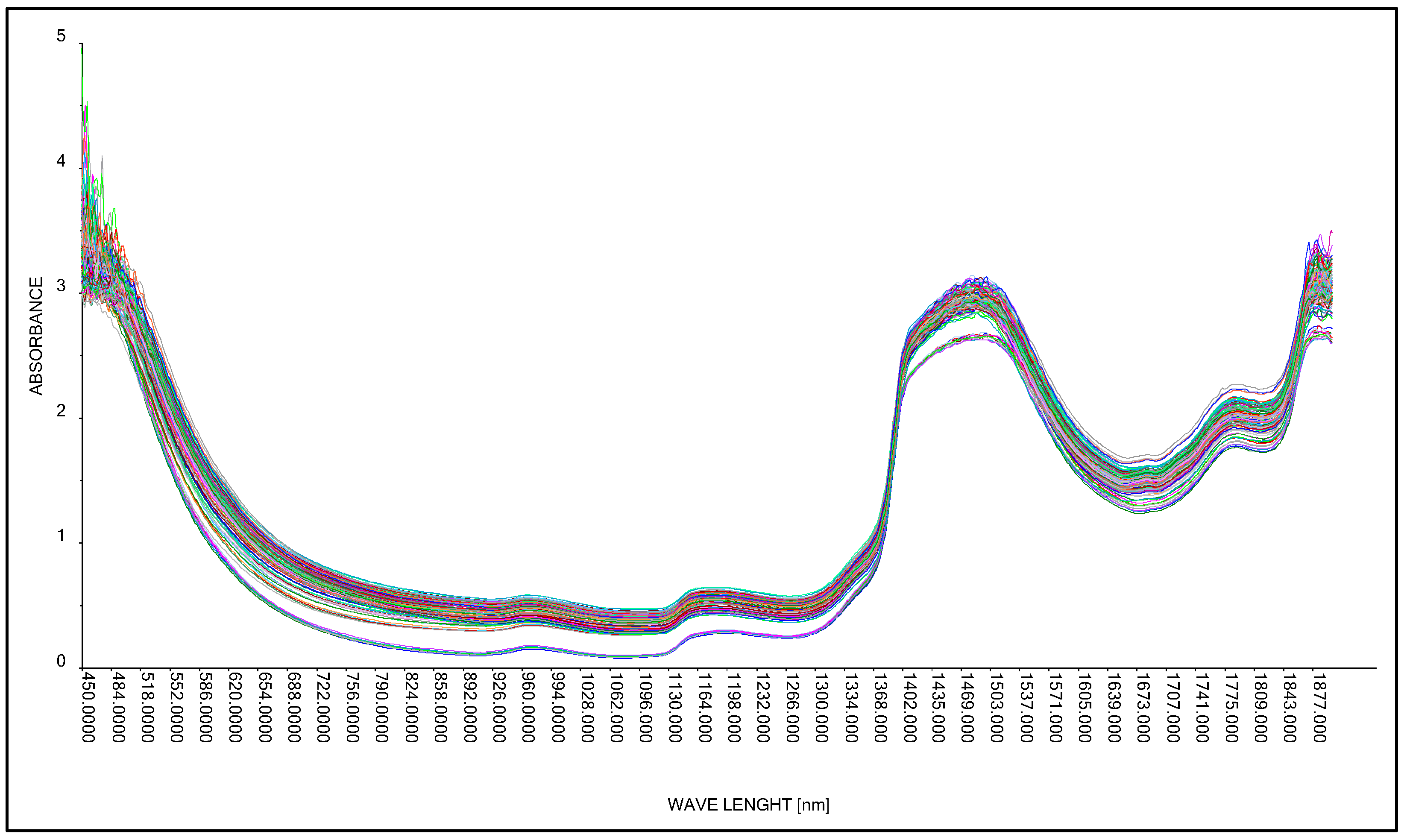
2.2.4. Research Station in the Process of Acquiring Spectral Spectra in the near Infrared Range
2.2.5. Statistical Analysis of Research Results
- Null hypothesis (H0): there is no statistically significant difference in the mean values between the analyzed data groups;
- Alternative hypothesis (H1): there exists a statistically significant difference in the mean values between the analyzed data groups.
3. Results
3.1. Acquisition of Spectral Spectra of Concentrated Lactic Acid
3.2. Acquisition of Spectral Spectra of an Aqueous Solution of Lactic Acid
3.2.1. Spectral Spectrum of an Aqueous Solution of Lactic Acid Using a Niron Head
3.2.2. Spectral Spectrum of an Aqueous Solution of Lactic Acid Using the A40 Head
3.3. Acquisition of Spectral Spectra of Aqueous Solutions of Lactic Acid and Molasses
3.3.1. Spectral Spectra of an Aqueous Solution of Lactic Acid and Molasses Using a Niron Head
3.3.2. Spectral Spectra of an Aqueous Solution of Lactic Acid and Molasses Using an A40 Head
4. Discussion
- −
- For grass silage (dry material): 4.9 (Lac), 2.0 (HAc), 3.7 (pH), 3.1 (NH3-N);
- −
- For grass silage (wet material): 3.3 (EtOH);
- −
- For corn silage: 4.7 (Lac), 1.9 (HAc), 2.4 (pH), 2.9 (NH3-N), 4.0 (EtOH).
5. Conclusions
- ◦
- First, the possibility of determining the concentration of lactic acid in liquid mixtures using the near-infrared (NIR) method in the analyzed wavelength range from 400 nm to 2250 nm was confirmed.
- ◦
- Identifying spectral noise clearly outlines the limitation of the data range (in the model up to 1900 nm) and improves the quality of the built calibration models.
- ◦
- Identification of optimal wavelength sub-ranges and gradual modifications of spectral spectra allow the areas of most valuable data for the calibration model to be determined.
- ◦
- The collection of data with increased size (150 NIR scans each time) contributed to the preparation of correct models of lactic acid concentration both in aqueous solution and in a mixture with molasses.
- ◦
- It has been demonstrated that Niron and A40 detection heads (absorption and reflectance) in the range of 450–1900 nm enable the creation of a high-quality database through the effective activation of lactic acid chemical bonds in an environment with limited optical transparency.
- ◦
- The results of the quality parameters of the calibration models confirmed that the proprietary method of spectral acquisition using 50 mL samples in a glass Petri dish is correct. The radiation waves penetrated the sample and reflected off the surface in a manner that allowed spectra to be obtained and changes in lactic acid concentration to be identified.
- ◦
- Building an effective neural network-based calibration model that recognizes the presence of lactic acid in a water-molasses mixture requires successive model iterations with wavelength range control to effectively improve the model quality parameters.
- ◦
- Mathematical transformations, such as baseline, moving average, SGolay filter, and outlier elimination, play a key role. This is an effective strategy for achieving high correlation between the model and the predictive and validation data.
- ◦
- It was found that the addition of 25% molasses—despite significantly reducing the transparency of the solution—does not prevent the construction of a high-quality NIR model. The spectral response of lactic acid is recognizable even in the difficult molasses medium, thanks to the excitation of chemical bonds in the NIR and partially MIR ranges.
- ◦
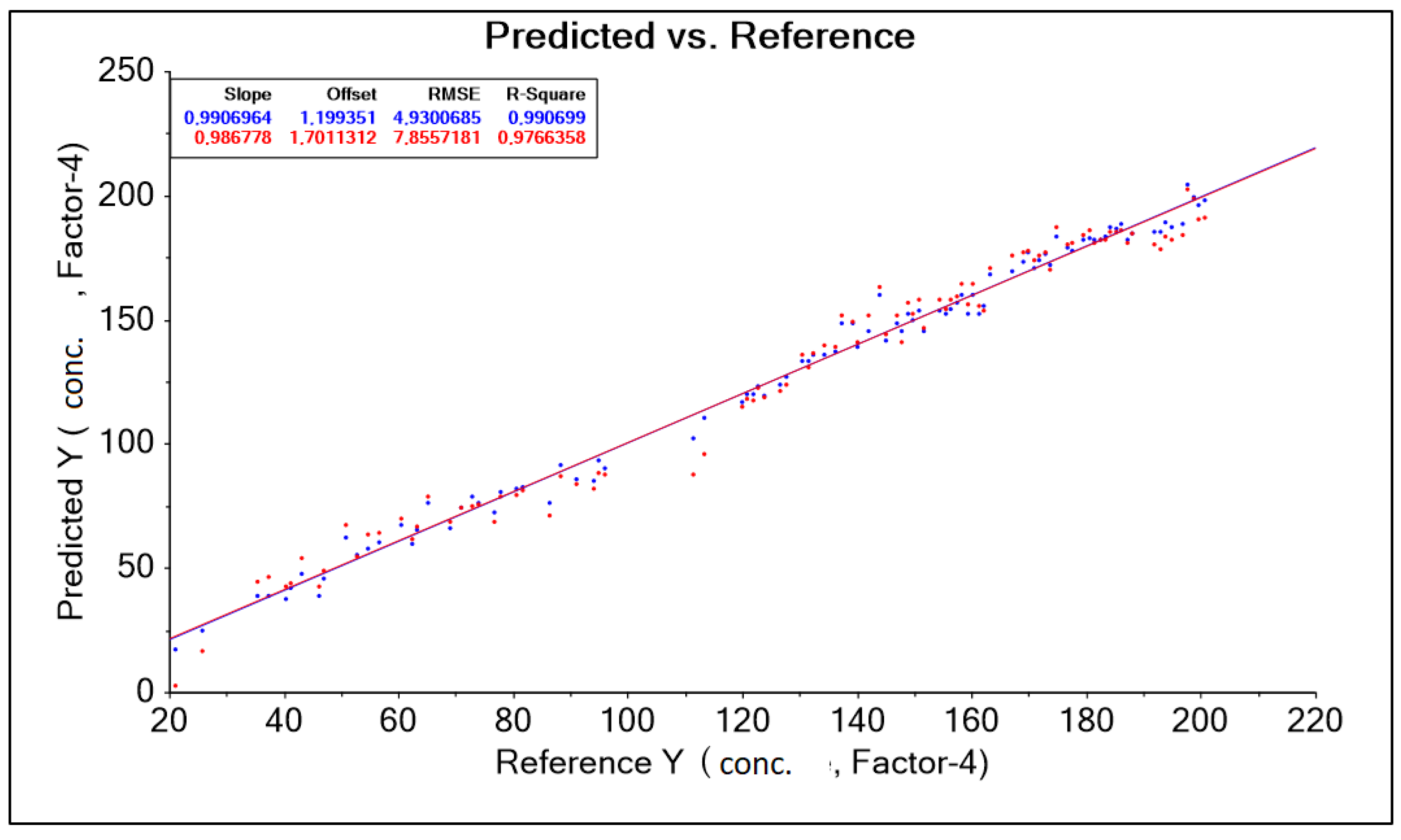
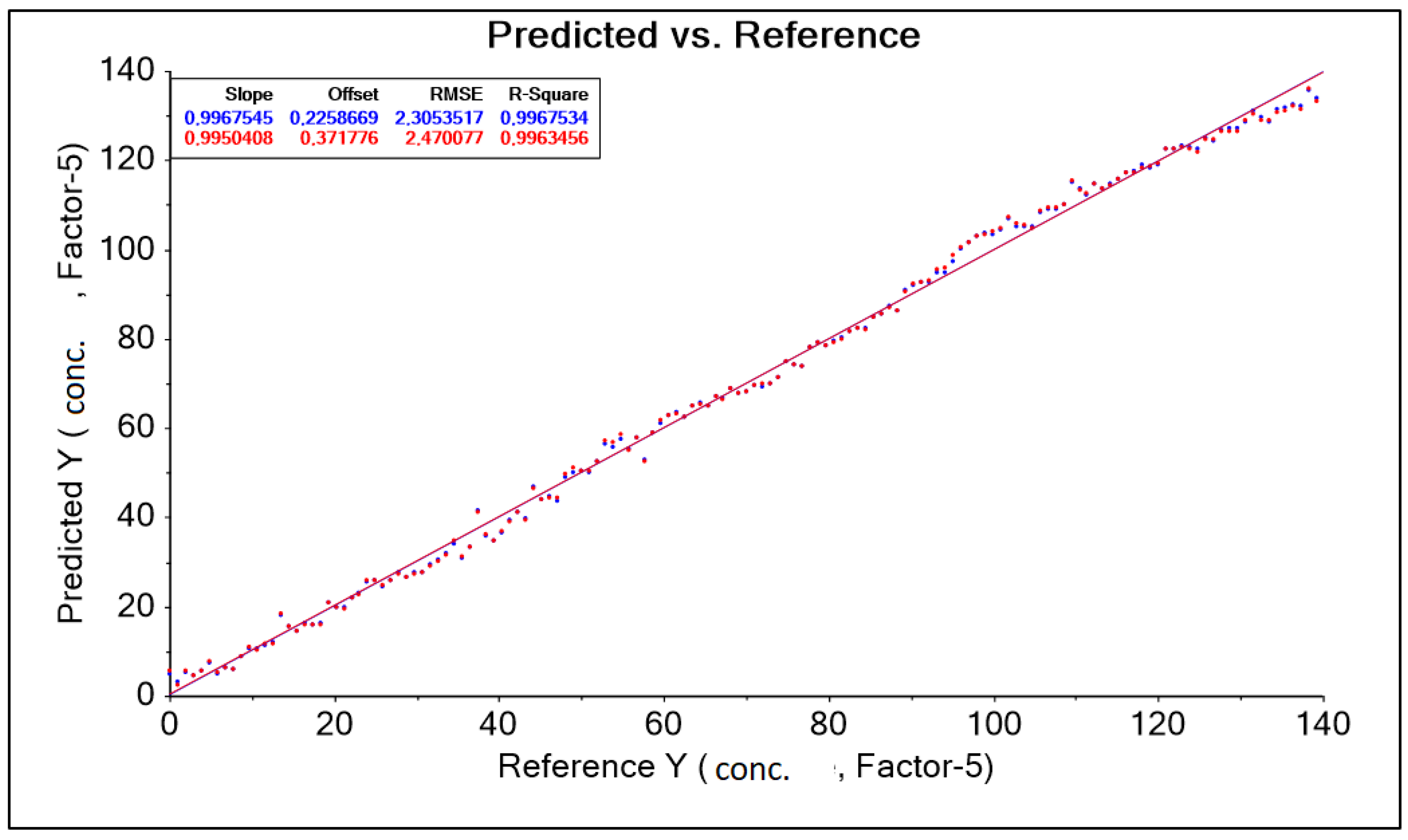
- The models were characterized by low RMSE errors and high R2, confirming the effectiveness of the method in determining lactic acid concentration in aquatic environments and in food/feed mixtures.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhao, X.F.; Yuan, Y.Q.; Chen, Q.K.; Li, Q.; Huang, Y.; Wu, D.; Li, L. Effect of total solids contents on the performance of anaerobic digester treating food waste and kinetics evaluation. In Proceedings of the E3S Web of Conferences 272, 01026 ICEPG 2021, Xiamen, China, 21–23 May 2021. [Google Scholar] [CrossRef]
- Li, Y.; Jin, Y.; Borrion, A.; Li, H.; Li, J. Effects of Organic Composition on the Anaerobic Biodegradability of Food Waste. Bioresour. Technol. 2017, 243, 836–845. [Google Scholar] [CrossRef]
- Yasin, N.H.M.; Mumtaz, T.; Hassan, M.A. Food waste and food processing waste for biohydrogen production: A review. J. Environ. Manag. 2013, 130, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Kibler, K.M.; Reinhart, D.; Hawkins, C.; Motlagh, A.M.; Wright, J. Food waste and the food-energy-water nexus: A review of food waste management alternatives. Waste Manag. 2018, 74, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, O. Energy and Waste Management. Energies 2017, 10, 1072. [Google Scholar] [CrossRef]
- Czekała, W.; Pulka, J.; Jasiński, T.; Szewczyk, P.; Bojarski, W.; Jasiński, J. Waste as substrates for agricultural biogas plants: A case study from Poland. J. Water Land Dev. 2023, 56, 45–50. [Google Scholar] [CrossRef]
- Wałowski, G. Assessment of technological simulation of an agricultural biogas installation using integration mechanisms. J. Water Land Dev. 2023, 59, 283–290. [Google Scholar] [CrossRef]
- Kupryś-Caruk, M.; Lisowski, A.; Chomontowski, C. The effect of silage additive on the kinetics of biogas production from lignocellulosic perennial crops. J. Water Land Dev. 2023, 56, 58–66. [Google Scholar] [CrossRef]
- Tezer, Ö.; Karabağ, N.; Öngen, A.; Çolpan, C.Ö.; Ayol, A. Biomass gasification for sustainable energy production: A review. Int. J. Hydrogen Energy 2022, 47, 15419–15433. [Google Scholar] [CrossRef]
- Modak, S.; Katiyar, P.; Talukdar, D.; Gole, B. Pyrolytic evaluation of essential oil industry waste: Effect of pyrolysis temperature on bio-oil composition. J. Environ. Manag. 2025, 392, 126757. [Google Scholar] [CrossRef]
- Mieldažys, R.; Jotautienė, E.; Pocius, A.; Jasinskas, A. Analysis of organic agricultural waste usage for fertilizer production. Agron. Res. 2016, 14, 143–149. [Google Scholar]
- Chojnacka, K.; Mikula, K.; Skrzypczak, D.; Izydorczyk, G.; Gorazda, K.; Kulczycka, J.; Kominko, H.; Moustakas, K.; Witek-Krowiak, A. Practical aspects of biowastes conversion to fertilizers. Biomass Convers. Biorefin. 2024, 14, 1515–1533. [Google Scholar] [CrossRef]
- Huang, J.; Wang, J.; Liu, S. Advanced Fermentation Techniques for Lactic Acid Production from Agricultural Waste. Fermentation 2023, 9, 765. [Google Scholar] [CrossRef]
- Walters, K.A.; Myers, K.S.; Ingle, A.T.; Donohue, T.J.; Noguera, D.R. Effect of Temperature and pH on Microbial Communities Fermenting a Dairy Coproduct Mixture. Fermentation 2024, 10, 422. [Google Scholar] [CrossRef]
- Adamski, M.; Czechlowski, M.; Durczak, K.; Garbowski, T. Determination of the Concentration of Propionic Acid in an Aqueous Solution by POD-GP Model and Spectroscopy. Energies 2021, 14, 8288. [Google Scholar] [CrossRef]
- Piątek, M.; Bartkowiak, A.M. Effectiveness of using physical pretreatment of lignocellulosic biomass. J. Water Land Dev. 2023, 58, 62–69. [Google Scholar] [CrossRef]
- Zaccariello, L.; Mastellone, M.L.; D’Amelia, L.I.; Catauro, M.; Morrone, B. Assessment of Integration between Lactic Acid, Biogas and Hydrochar Production in OFMSW Plants. Energies 2020, 13, 6593. [Google Scholar] [CrossRef]
- Curtin, L.V. Molasses—General Considerations; National Feed Ingredients Association: Iowa, IA, USA, 1983. [Google Scholar]
- Al-Amin, K.; Kawsar, M.; Mamun, M.T.R.B.; Hossain, M.S. Fourier transform infrared spectroscopic technique for analysis of inorganic materials: A review. Nanoscale Adv. 2025, 7, 6677–6702. [Google Scholar] [CrossRef]
- Šarić, L.Ć.; Filipčev, B.V.; Šimurina, O.D.; Plavšić, D.V.; Šarić, B.M.; Lazarević, J.M.; Milovanović, I.L. Sugar beet molasses: Properties and applications in osmotic dehydration of fruits and vegetables. Food Feed Res. 2016, 43, 135–144. [Google Scholar] [CrossRef]
- Bogoczek, R.; Napierała, W. Kwas mlekowy jakość, właściwości i kierunki zastosowań. Przemysł Spożywczy Rocz. 1998, 52, 43–47. [Google Scholar]
- van Lieshout, G.P. The Physical Properties of Lactic Acid and Derivatives; A Literature Review; TU Delft: Delft, The Netherlands, 1992. [Google Scholar]
- Czekała, W.; Nowak, M.; Bojarski, W. Characteristics of Substrates Used for Biogas Production in Terms of Water Content. Fermentation 2023, 9, 449. [Google Scholar] [CrossRef]
- Tekin, Y.; Kuang, B.; Mouazen, A.M. Potential of on-line visible and near infrared spectroscopy for measurement of pH for deriving variable rate lime recommendations. Sensors 2013, 13, 10177–10190. [Google Scholar] [CrossRef]
- Atelge, M.R.; Atabani, A.E.; Banu, J.R.; Krisa, D.; Kaya, M.; Eskicioglu, C.; Kumar, G.; Lee, C.; Yildiz, Y.Ş.; Unalan, S.; et al. A critical review of pretreatment technologies to enhance anaerobic digestion and energy recovery. Fuel 2020, 270, 117494. [Google Scholar] [CrossRef]
- Kintl, A.; Vítěz, T.; Huňady, I.; Sobotková, J.; Hammerschmiedt, T.; Vítězová, M.; Brtnický, M.; Holátko, J.; Elbl, J. Effect of Mycotoxins in Silage on Biogas Production. Bioengineering 2023, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Kupryaniuk, K.; Witaszek, K.; Vaskina, I.; Filipek-Kaźmierczak, S.; Kupryaniuk, J.; Sołowiej, P.; Dach, J. The Effect of Corn Ensiling Methods on Digestibility and Biogas Yield. Energies 2025, 18, 188. [Google Scholar] [CrossRef]
- Xue, X.; Tian, H.; Zhao, K.; Yu, Y.; Xiao, Z.; Zhuo, C.; Sun, J. Rapid Lactic Acid Content Detection in Secondary Fermentation of Maize Silage Using Colorimetric Sensor Array Combined with Hyperspectral Imaging. Agriculture 2024, 14, 1653. [Google Scholar] [CrossRef]
- Moll, V.; Beć, K.B.; Grabska, J.; Huck, C.W. Investigation of Water Interaction with Polymer Matrices by Near-Infrared (NIR) Spectroscopy. Molecules 2022, 27, 5882. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Roger, J.M.; Mallet, A.; Marini, F. Preprocessing NIR Spectra for Aquaphotomics. Molecules 2022, 27, 6795. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saager, R.B.; Cuccia, D.J.; Durkin, A.J. Determination of optical properties of turbid media spanning visible and near-infrared regimes via spatially modulated quantitative spectroscopy. J. Biomed. Opt. 2010, 15, 017012. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gál, L.; Paračková, P.; Kaliňáková, B.; Šimonová, S.; Reháková, M.; Čeppan, M. Microbial contaminated paper substrate: UV–Vis–NIR spectra of model systems. Chem. Pap. 2024, 78, 2603–2611. [Google Scholar] [CrossRef]
- Macedo, M.G.; Laporte, M.F.; Lacroix, C. Quantification of exopolysaccharide, lactic acid, and lactose concentrations in culture broth by near-infrared spectroscopy. J. Agric. Food Chem. 2002, 50, 1774–1779. [Google Scholar] [CrossRef] [PubMed]
- Păucean, A.; Vodnar, D.C.; Mureșan, V.; Fetea, F.; Ranga, F.; Man, S.M.; Muste, S.; Socaciu, C. Monitoring lactic acid concentrations by infrared spectroscopy: A new developed method for Lactobacillus fermenting media with potential food applications. Acta Aliment. 2017, 46, 420–427. [Google Scholar] [CrossRef]
- Sørensen, L.K. Prediction of Fermentation Parameters in Grass and Corn Silage by Near Infrared Spectroscopy. J. Dairy Sci. 2004, 87, 3826–3835. [Google Scholar] [CrossRef]









| Ingredients | Molasses | Beet Molasses [13] |
|---|---|---|
| Energy (kcal) | 290 | 232 |
| Protein [g] | 0 | 6 |
| Fats [g] | 0 | 0 |
| Simple sugars [g] | 76 | 48 |
| Sucrose [g] | 29 | - |
| Glucose [g] | 12 | - |
| Fructose [g] | 13 | - |
| Fiber [g] | 0 | 0 |
| Sodium [mg] | 37 | 200 |
| Potassium [mg] | 1472 | 4700 |
| Calcium [mg] | 205 | 200 |
| Phosphorus [mg] | 31 | 30 |
| Magnesium [mg] | 242 | - |
| Iron [mg] | 4.7 | 11.7 |
| Zinc [mg] | 0.4 | 4 |
| Cooper [mg] | 0.6 | 1.3 |
| Manganese [mg] | 1.5 | 1 |
| Selenium [mg] | 17.8 | - |
| Vitamin B1 [mg] | 0.04 | 0 |
| Vitamin B2 [mg] | 0.002 | - |
| Vitamin PP [mg] | 0.93 | 0.14 |
| Vitamin B6 [mg] | 0.67 | - |
| A40 | Aqueous Molasses Solution | ||||||
|---|---|---|---|---|---|---|---|
| Dose No. | Concentration [g·dm−3] | Dose No. | Concentration [g·dm−3] | Dose No. | Concentration [g·dm−3] | Dose No. | Concentration [g·dm−3] |
| 000 | 0 | 037 | 35.52 | 074 | 71.04 | 111 | 106.56 |
| 001 | 0.96 | 038 | 36.48 | 075 | 72 | 112 | 107.52 |
| 002 | 1.92 | 039 | 37.44 | 076 | 72.96 | 113 | 108.48 |
| 003 | 2.88 | 040 | 38.4 | 077 | 73.92 | 114 | 109.44 |
| 004 | 3.84 | 041 | 39.36 | 078 | 74.88 | 115 | 110.4 |
| 005 | 4.8 | 042 | 40.32 | 079 | 75.84 | 116 | 111.36 |
| 006 | 5.76 | 043 | 41.28 | 080 | 76.8 | 117 | 112.32 |
| 007 | 6.72 | 044 | 42.24 | 081 | 77.76 | 118 | 113.28 |
| 008 | 7.68 | 045 | 43.2 | 082 | 78.72 | 119 | 114.24 |
| 009 | 8.64 | 046 | 44.16 | 083 | 79.68 | 120 | 115.2 |
| 010 | 9.6 | 047 | 45.12 | 084 | 80.64 | 121 | 116.16 |
| 011 | 10.56 | 048 | 46.08 | 085 | 81.6 | 122 | 117.12 |
| 012 | 11.52 | 049 | 47.04 | 086 | 82.56 | 123 | 118.08 |
| 013 | 12.48 | 050 | 48 | 087 | 83.52 | 124 | 119.04 |
| 014 | 13.44 | 051 | 48.96 | 088 | 84.48 | 125 | 120 |
| 015 | 14.4 | 052 | 49.92 | 089 | 85.44 | 126 | 120.96 |
| 016 | 15.36 | 053 | 50.88 | 090 | 86.4 | 127 | 121.92 |
| 017 | 16.32 | 054 | 51.84 | 091 | 87.36 | 128 | 122.88 |
| 018 | 17.28 | 055 | 52.8 | 092 | 88.32 | 129 | 123.84 |
| 019 | 18.24 | 056 | 53.76 | 093 | 89.28 | 130 | 124.8 |
| 020 | 19.2 | 057 | 54.72 | 094 | 90.24 | 131 | 125.76 |
| 021 | 20.16 | 058 | 55.68 | 095 | 91.2 | 132 | 126.72 |
| 022 | 21.12 | 059 | 56.64 | 096 | 92.16 | 133 | 127.68 |
| 023 | 22.08 | 060 | 57.6 | 097 | 93.12 | 134 | 128.64 |
| 024 | 23.04 | 061 | 58.56 | 098 | 94.08 | 135 | 129.6 |
| 025 | 24 | 062 | 59.52 | 099 | 95.04 | 136 | 130.56 |
| 026 | 24.96 | 063 | 60.48 | 100 | 96 | 137 | 131.52 |
| 027 | 25.92 | 064 | 61.44 | 101 | 96.96 | 138 | 132.48 |
| 028 | 26.88 | 065 | 62.4 | 102 | 97.92 | 139 | 133.44 |
| 029 | 27.84 | 066 | 63.36 | 103 | 98.88 | 140 | 134.4 |
| 030 | 28.8 | 067 | 64.32 | 104 | 99.84 | 141 | 135.36 |
| 031 | 29.76 | 068 | 65.28 | 105 | 100.8 | 142 | 136.32 |
| 032 | 30.72 | 069 | 66.24 | 106 | 101.76 | 143 | 137.28 |
| 033 | 31.68 | 070 | 67.2 | 107 | 102.72 | 144 | 138.24 |
| 034 | 32.64 | 071 | 68.16 | 108 | 103.68 | 145 | 139.2 |
| 035 | 33.6 | 072 | 69.12 | 109 | 104.64 | 146 | 140.16 |
| 036 | 34.56 | 073 | 70.08 | 110 | 105.6 | ||
| Niron | Aqueous Molasses Solution | ||||||
|---|---|---|---|---|---|---|---|
| Dose No. | Concentration [g·dm−3] | Dose No. | Concentration [g·dm−3] | Dose No. | Concentration [g·dm−3] | Dose No. | Concentration [g·dm−3] |
| 000 | 0 | 037 | 35.52 | 074 | 71.04 | 111 | 106.56 |
| 001 | 0.96 | 038 | 36.48 | 075 | 72 | 112 | 107.52 |
| 002 | 1.92 | 039 | 37.44 | 076 | 72.96 | 113 | 108.48 |
| 003 | 2.88 | 040 | 38.4 | 077 | 73.92 | 114 | 109.44 |
| 004 | 3.84 | 041 | 39.36 | 078 | 74.88 | 115 | 110.4 |
| 005 | 4.8 | 042 | 40.32 | 079 | 75.84 | 116 | 111.36 |
| 006 | 5.76 | 043 | 41.28 | 080 | 76.8 | 117 | 112.32 |
| 007 | 6.72 | 044 | 42.24 | 081 | 77.76 | 118 | 113.28 |
| 008 | 7.68 | 045 | 43.2 | 082 | 78.72 | 119 | 114.24 |
| 009 | 8.64 | 046 | 44.16 | 083 | 79.68 | 120 | 115.2 |
| 010 | 9.6 | 047 | 45.12 | 084 | 80.64 | 121 | 116.16 |
| 011 | 10.56 | 048 | 46.08 | 085 | 81.6 | 122 | 117.12 |
| 012 | 11.52 | 049 | 47.04 | 086 | 82.56 | 123 | 118.08 |
| 013 | 12.48 | 050 | 48 | 087 | 83.52 | 124 | 119.04 |
| 014 | 13.44 | 051 | 48.96 | 088 | 84.48 | 125 | 120 |
| 015 | 14.4 | 052 | 49.92 | 089 | 85.44 | 126 | 120.96 |
| 016 | 15.36 | 053 | 50.88 | 090 | 86.4 | 127 | 121.92 |
| 017 | 16.32 | 054 | 51.84 | 091 | 87.36 | 128 | 122.88 |
| 018 | 17.28 | 055 | 52.8 | 092 | 88.32 | 129 | 123.84 |
| 019 | 18.24 | 056 | 53.76 | 093 | 89.28 | 130 | 124.8 |
| 020 | 19.2 | 057 | 54.72 | 094 | 90.24 | 131 | 125.76 |
| 021 | 20.16 | 058 | 55.68 | 095 | 91.2 | 132 | 126.72 |
| 022 | 21.12 | 059 | 56.64 | 096 | 92.16 | 133 | 127.68 |
| 023 | 22.08 | 060 | 57.6 | 097 | 93.12 | 134 | 128.64 |
| 024 | 23.04 | 061 | 58.56 | 098 | 94.08 | 135 | 129.6 |
| 025 | 24 | 062 | 59.52 | 099 | 95.04 | 136 | 130.56 |
| 026 | 24.96 | 063 | 60.48 | 100 | 96 | 137 | 131.52 |
| 027 | 25.92 | 064 | 61.44 | 101 | 96.96 | 138 | 132.48 |
| 028 | 26.88 | 065 | 62.4 | 102 | 97.92 | 139 | 133.44 |
| 029 | 27.84 | 066 | 63.36 | 103 | 98.88 | 140 | 134.4 |
| 030 | 28.8 | 067 | 64.32 | 104 | 99.84 | 141 | 135.36 |
| 031 | 29.76 | 068 | 65.28 | 105 | 100.8 | 142 | 136.32 |
| 032 | 30.72 | 069 | 66.24 | 106 | 101.76 | 143 | 137.28 |
| 033 | 31.68 | 070 | 67.2 | 107 | 102.72 | 144 | 138.24 |
| 034 | 32.64 | 071 | 68.16 | 108 | 103.68 | 145 | 139.2 |
| 035 | 33.6 | 072 | 69.12 | 109 | 104.64 | 146 | 140.16 |
| 036 | 34.56 | 073 | 70.08 | 110 | 105.6 | ||
| Slope | Free Term | Mean Square Error RMSE [g·dm−3] | Coefficient of Determination R2 | |||||
|---|---|---|---|---|---|---|---|---|
| Name | Model | Validation | Model | Validation | Name | Model | Validation | Model |
| PLS Nitron 450–1900 nm | 0.970 | 0.948 | 2.968 | 4.6000 | 10.011 | 17.614 | 0.970 | 0.909 |
| PLS Nitron 450–1900 nm no out aut | 0.977 | 0.969 | 2.271 | 2.645 | 8.839 | 10.722 | 0.977 | 0.967 |
| PLS Nitron 450–950 nm raw | 0.890 | 0.881 | 11.029 | 11.812 | 19.295 | 21.638 | 0.890 | 0.862 |
| PLS Nitron 450–950 nm no out aut | 0.929 | 0.914 | 7.191 | 8.799 | 16.057 | 19.227 | 0.929 | 0.899 |
| PLS Nitron 950–1900 nm raw | 0.977 | 0.959 | 2.276 | 3.338 | 8.767 | 12.941 | 0.977 | 0.951 |
| PLS Nitron 950–1900 nm no out aut | 0.978 | 0.955 | 2.252 | 4.921 | 8.273 | 10.794 | 0.978 | 0.964 |
| PLS Nitron 950–1900 nm baseline | 0.978 | 0.959 | 2.150 | 4.210 | 8.519 | 10.728 | 0.978 | 0.966 |
| PLS Nitron 950–1900 nm baseline no out aut | 0.984 | 0.967 | 2.049 | 3.74 | 6.317 | 9.582 | 0.984 | 0.965 |
| PLS Nitron 950–1900 nm baseline Moving Average 9 | 0.972 | 0.951 | 2.823 | 4.787 | 9.763 | 12.312 | 0.972 | 0.955 |
| PLS Nitron 950–1900 nm baseline Moving Average 9 no out aut | 0.991 | 0.987 | 1.199 | 1.701 | 4.930 | 7.855 | 0.990 | 0.976 |
| Slope | Free Term | Mean Square Error RMSE [g·dm−3] | Coefficient of Determination R2 | |||||
|---|---|---|---|---|---|---|---|---|
| Name | Model | Validation | Model | Validation | Name | Model | Validation | Model |
| PLS A40 450–1900 | 0.991 | 0.991 | 0.861 | 0.991 | 5.391 | 5.694 | 0.991 | 0.990 |
| PLS A40 920–1900 | 0.994 | 0.994 | 0.623 | 0.575 | 4.584 | 4.784 | 0.994 | 0.993 |
| PLS A40 920–1900 no out aut | 0.994 | 0.994 | 0.623 | 0.575 | 4.584 | 4.784 | 0.994 | 0.993 |
| PLS A40 920–1900 baseline | 0.994 | 0.994 | 0.589 | 0.519 | 4.460 | 4.622 | 0.994 | 0.994 |
| PLS A40 920–1900 baseline no out, aut | 0.994 | 0.994 | 0.592 | 0.530 | 4.354 | 4.601 | 0.994 | 0.993 |
| PLS A40 920–1900 baseline no out, by hand | 0.994 | 0.994 | 0.570 | 0.671 | 4.186 | 4.355 | 0.994 | 0.994 |
| PLS A40 1400–1900 baseline | 0.988 | 0.987 | 1.163 | 1.210 | 6.266 | 6.311 | 0.988 | 0.988 |
| PLS A40 1400–1900 baseline no out, by hand | 0.988 | 0.986 | 1.241 | 1.472 | 6.206 | 6.303 | 0.988 | 0.988 |
| PLS A40 920–1900 baseline SGolay | 0.984 | 0.971 | 1.593 | 2.616 | 7.335 | 10.202 | 0.984 | 0.969 |
| Slope | Free Term | Mean Square Error RMSE [g·dm−3] | Coefficient of Determination R2 | |||||
|---|---|---|---|---|---|---|---|---|
| Name | Model | Validation | Model | Validation | Model | Validation | Model | Validation |
| PLS Niron | 0.948 | 0.887 | 3.644 | 8.459 | 9.288 | 12.938 | 0.948 | 0.900 |
| PLS Niron baseline | 0.948 | 0.885 | 3.676 | 8.721 | 9.329 | 14.894 | 0.947 | 0.869 |
| PLS Niron baseline no out | 0.958 | 0.897 | 3.268 | 8.000 | 7.962 | 11.039 | 0.957 | 0.920 |
| PLS Niron 500–1850 nm | 0.959 | 0.882 | 2.832 | 8.995 | 8.189 | 14.004 | 0.959 | 0.883 |
| PLS Niron baseline 500–1850 nm | 0.951 | 0.893 | 3.414 | 7.055 | 8.990 | 11.899 | 0.951 | 0.915 |
| PLS Niron baseline 850–1850 nm | 0.955 | 0.905 | 3.160 | 6.158 | 8.651 | 12.471 | 0.955 | 0.908 |
| PLS Niron baseline 550–950 nm | 0.867 | 0.807 | 9.330 | 14.793 | 14.864 | 19.216 | 0.867 | 0.784 |
| PLS Niron 500–1850 nm no out aut | 0.962 | 0.889 | 2.920 | 0.196 | 7.547 | 10.463 | 0.962 | 0.931 |
| PLS Niron 500–1850 nm no out by hand 1 | 0.964 | 0.917 | 2.749 | 6.600 | 6.962 | 10.406 | 0.964 | 0.921 |
| PLS Niron 500–1850 nm no out by hand 2 | 0.979 | 0.880 | 1.583 | 10.627 | 5.249 | 11.081 | 0.979 | 0.913 |
| PLS Niron 500–1850 nm no out by hand 3 | 0.984 | 0.908 | 1.268 | 7.320 | 4.687 | 8.471 | 0.983 | 0.948 |
| PLS Niron 500–1850 nm no out by hand 4 | 0.985 | 0.928 | 1.175 | 5.887 | 4.455 | 8.062 | 0.985 | 0.951 |
| PLS Niron 500–1850 nm bez out by hand 5 | 0.985 | 0.918 | 1.171 | 6.588 | 4.435 | 7.918 | 0.985 | 0.953 |
| Slope | Free Term | Mean Square Error RMSE [g·dm−3] | Coefficient of Determination R2 | |||||
|---|---|---|---|---|---|---|---|---|
| Name | Model | Validation | Model | Validation | Model | Validation | Model | Validation |
| PLS A40 450–1900 nm | 0.991 | 0.973 | 0.616 | 2.294 | 3.806 | 4.727 | 0.991 | 0.986 |
| PLS A40 450–1900 nm baseline | 0.985 | 0.971 | 1.034 | 2.159 | 4.932 | 5.436 | 0.985 | 0.982 |
| PLS A40 550–1850 nm | 0.994 | 0.983 | 0.431 | 1.355 | 3.184 | 3.991 | 0.994 | 0.990 |
| PLS A40 550–1850 nm baseline | 0.997 | 0.995 | 0.226 | 0.371 | 2.305 | 2.470 | 0.997 | 0.996 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamski, M.; Herkowiak, M.; Marek, P.; Dzida, K.; Kapłan, M.; Klimek, K.E. Lactic Fermentation Spectral Analysis of Target Substrates and Food and Feed Wastes for Energy Applications. Energies 2025, 18, 6360. https://doi.org/10.3390/en18236360
Adamski M, Herkowiak M, Marek P, Dzida K, Kapłan M, Klimek KE. Lactic Fermentation Spectral Analysis of Target Substrates and Food and Feed Wastes for Energy Applications. Energies. 2025; 18(23):6360. https://doi.org/10.3390/en18236360
Chicago/Turabian StyleAdamski, Mariusz, Marcin Herkowiak, Przemysław Marek, Katarzyna Dzida, Magdalena Kapłan, and Kamila E. Klimek. 2025. "Lactic Fermentation Spectral Analysis of Target Substrates and Food and Feed Wastes for Energy Applications" Energies 18, no. 23: 6360. https://doi.org/10.3390/en18236360
APA StyleAdamski, M., Herkowiak, M., Marek, P., Dzida, K., Kapłan, M., & Klimek, K. E. (2025). Lactic Fermentation Spectral Analysis of Target Substrates and Food and Feed Wastes for Energy Applications. Energies, 18(23), 6360. https://doi.org/10.3390/en18236360






