Deep-XFCT: Deep Learning 3D-Mineral Liberation Analysis with Micro-X-ray Fluorescence and Computed Tomography
Abstract
:1. Introduction
2. Methods
2.1. Core Plug Rock Sample
Multimineral Rock Sample
2.2. Experimental Methods
2.2.1. Powder X-ray Diffraction (XRD)
2.2.2. Microcomputed Tomography (μCT)
2.2.3. Micro-X-ray Fluorescence (μXRF)
2.2.4. Raman Spectroscopy
2.3. Data Analytical Methods
2.3.1. Establishing the Training Data
K-Means Clustering
Image Registration
2.3.2. Deep Learning Segmentation
2.3.3. Establishing Ground Truth for Confirmation
3. Results
3.1. Multimodal Mineral Assignment with XRD and XRF
3.1.1. Multimodal Data
3.1.2. Mineral Assignment
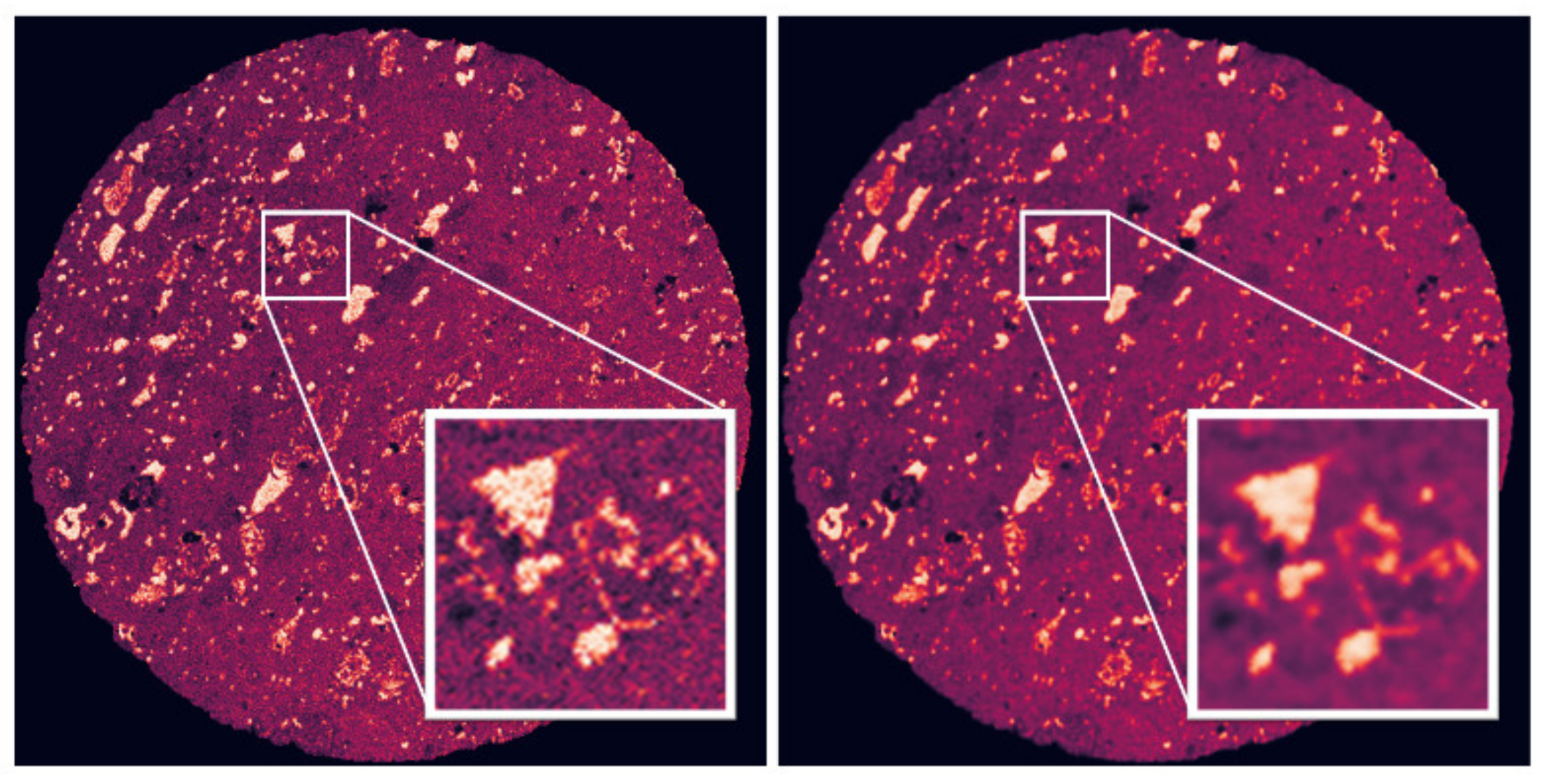
3.2. Deep Learning Segmentation of the μCT Data
3.2.1. Training Data
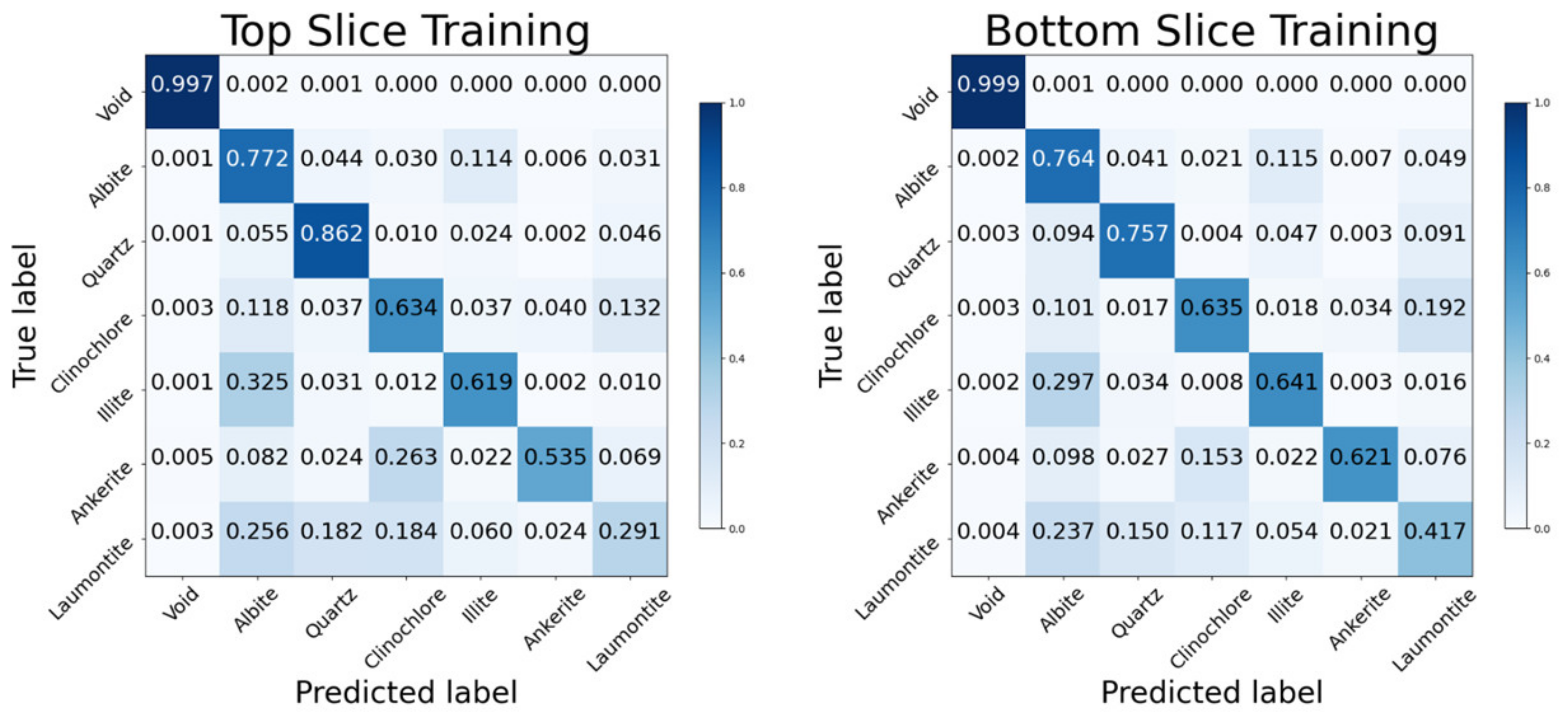
3.2.2. Validation Data
4. Discussion
4.1. Contributing Factors to Results
4.2. Segmentation Enhancement Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mariano, R.A.; Evans, C.L.; Manlapig, E. Definition of Random and Non-Random Breakage in Mineral Liberation—A review. Miner. Eng. 2016, 94, 51–60. [Google Scholar] [CrossRef]
- Lomberg, K. Best practice sampling methods, assay techniques, and quality control with reference to the platinum group elements (PGEs). J. S. Afr. Inst. Min. Metall. 2014, 114, 53–62. [Google Scholar]
- Sandmann, D. Use of Mineral Liberation Analysis (MLA) in the Characterization of Lithium-Bearing Micas. J. Miner. Mater. Charact. Eng. 2013, 1, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Heinig, T.; Bachmann, K.; Tolosana-Delgado, R.; Boogaart, G.; Gutzmer, J. Monitoring gravitational and particle shape settling effects on MLA sampling preparation. In Proceedings of the IAMG, Freiberg, Germany, 5–13 September 2015. [Google Scholar]
- Tonžetić, Ž. 5—Quantitative analysis of iron ore using SEM-based technologies. In Iron Ore; Lu, L., Ed.; Woodhead Publishing: Sawston, UK, 2015; pp. 161–189. [Google Scholar] [CrossRef]
- Babel, B.; Penz, M.; Schach, E.; Boehme, S.; Rudolph, M. Reprocessing of a Southern Chilean Zn Tailing by Flotation—A Case Study. Minerals 2018, 8, 295. [Google Scholar] [CrossRef] [Green Version]
- Schulz, B. Editorial for Special Issue “Applications of SEM Automated Mineralogy: From Ore Deposits over Processing to Secondary Resource Characterization”. Minerals 2020, 10, 1103. [Google Scholar] [CrossRef]
- Sylvester, P.J. Use of the mineral liberation analyzer (MLA) for mineralogical studies of sediments and sedimentary rocks. Mineral. Assoc. Can. 2012, 1, 1–16. [Google Scholar] [CrossRef]
- Butcher, A.; Botha, P.W.; Dunks, C. An enabling geoscience tool for automated rock typing, stratigraphic refinement, and reservoir characterisation. APPEA J. 2008, 48, 482. [Google Scholar] [CrossRef]
- Goodall, W.R.; Scales, P.J.; Butcher, A.R. The use of QEMSCAN and Diagnostic Leaching in the Characterisation of Visible Gold in Complex Ores. Miner. Eng. 2005, 18, 877–886. [Google Scholar] [CrossRef]
- Gu, Y. Automated Scanning Electron Microscope Based Mineral Liberation Analysis An Introduction to JKMRC/FEI Mineral Liberation Analyser. J. Miner. Mater. Charact. Eng. 2003, 2, 33–41. [Google Scholar] [CrossRef]
- Voytekhovsky, Y. Modal Analysis of Rocks and Ores in Thin Sections. In International Congress on Applied Mineralogy; Springer: Cham, Switzerland, 2019; pp. 162–166. [Google Scholar]
- Lemmens, H.J.; Butcher, A.R.; Botha, P.W.S.K.B. “FIB/SEM and SEM/EDX: A New Dawn for the SEM in the Core Lab?,” Petrophysics-SPWLA. J. Form. Eval. Reserv. Descr. 2011, 52, 452–456. [Google Scholar]
- Guntoro, P.I.; Ghorbani, Y.; Parian, M.; Butcher, A.R.; Kuva, J.; Rosenkranz, J. Development and experimental validation of a texture-based 3D liberation model. Miner. Eng. 2021, 164, 106828. [Google Scholar] [CrossRef]
- Mutina, A.; Bruyndonckx, P. Combined micro-X-ray tomography and micro-X-ray fluorescence study of reservoir rocks: Applicability to core analysis. Microsc. Anal. Anal. Suppl. 2013, 27, S4–S6. [Google Scholar]
- Vekemans, B.; Janssens, K.; Vincze, L.; Aerts, A.; Adams, F.; Hertogen, J. Automated segmentation of μ-XRF image sets. X-ray Spectrom. Int. J. 1997, 26, 333–346. [Google Scholar] [CrossRef]
- Arzilli, F.; Polacci, M.; Landi, P.; Giordano, D.; Baker, D.R.; Mancini, L. A novel protocol for resolving feldspar crystals in synchrotron X-ray microtomographic images of crystallized natural magmas and synthetic analogs. Am. Miner. 2016, 101, 2301–2311. [Google Scholar] [CrossRef] [Green Version]
- Juránek, R.; Výravský, J.; Kolář, M.; Motl, D.; Zemčík, P. Graph-based deep learning segmentation of EDS spectral images for automated mineral phase analysis. Comput. Geosci. 2022, 165, 105–109. [Google Scholar] [CrossRef]
- Song, Y.; Huang, Z.; Shen, C.; Shi, H.; Lange, D.A. Deep learning-based automated image segmentation for concrete petrographic analysis. Cem. Concr. Res. 2020, 135, 106118. [Google Scholar] [CrossRef]
- Tang, K.; Meyer, Q.; White, R.; Armstrong, R.T.; Mostaghimi, P.; Da Wang, Y.; Liu, S.; Zhao, C.; Regenauer-Lieb, K.; Tung, P.K.M. Deep learning for full-feature X-ray microcomputed tomography segmentation of proton electron membrane fuel cells. Comput. Chem. Eng. 2022, 161, 107768. [Google Scholar] [CrossRef]
- McCoy, J.; Auret, L. Machine learning applications in minerals processing: A review. Miner. Eng. 2018, 132, 95–109. [Google Scholar] [CrossRef]
- Bu, X.; Zhou, S.; Danstan, J.K.; Bilal, M.; Hassan, F.U.; Chao, N. Prediction of coal flotation performance using a modified deep neural network model including three input parameters from feed. Energy Sources Part A Recover. Util. Environ. Eff. 2022, 1–13. [Google Scholar] [CrossRef]
- Bu, X.; Vahed, A.T.; Ghassa, S.; Chelgani, S.C. Modelling of coal flotation responses based on operational conditions by random forest. Int. J. Oil Gas Coal Technol. 2021, 27, 457. [Google Scholar] [CrossRef]
- Chelgani, S.C.; Nasiri, H.; Alidokht, M. Interpretable modeling of metallurgical responses for an industrial coal column flotation circuit by XGBoost and SHAP-A “conscious-lab” development. Int. J. Min. Sci. Technol. 2021, 31, 1135–1144. [Google Scholar] [CrossRef]
- Tohry, A.; Yazdani, S.; Hadavandi, E.; Mahmudzadeh, E.; Chelgani, S.C. Advanced modeling of HPGR power consumption based on operational parameters by BNN: A “Conscious-Lab” development. Powder Technol. 2020, 381, 280–284. [Google Scholar] [CrossRef]
- Tohry, A.; Chelgani, S.C.; Matin, S.; Noormohammadi, M. Power-draw prediction by random forest based on operating parameters for an industrial ball mill. Adv. Powder Technol. 2020, 31, 967–972. [Google Scholar] [CrossRef]
- Sauter, E.; Sarikaya, E.; Winter, M.; Wegener, K. In-process detection of grinding burn using machine learning. Int. J. Adv. Manuf. Technol. 2021, 115, 2281–2297. [Google Scholar] [CrossRef]
- Ahmed, M.M. Effect of comminution on particle shape and surface roughness and their relation to flotation process. Int. J. Miner. Process. 2010, 94, 180–191. [Google Scholar] [CrossRef]
- Weber, C.R.; Vlahovic, W.; Wyborn, D. Validation of Hot Rock Geothermal Energy Resources; Milestone 2 Report to the Australian Greenhouse Office Under the Renewable Energy Commercialisation Program. R00032661 (GS2002/117); Geological Survey of New South Wales (DiGS): Hunter Valley, Australia, 2001.
- Philpotts, A.R.; Ague, J.J. Principles of Igneous and Metamorphic Petrology, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Lafuente, B.; Downs, R.T.; Yang, H.; Stone, N. The power of databases: The RRUFF project. In Highlights in Mineralogical Crystallography; Walter de Gruyter GmbH: Berlin, Germany, 2016; pp. 1–29. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Ron Weiss, R.; Duchesnay, E.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 2, 2825–2830. [Google Scholar]
- Dragonfly Software, 2021.2; Object Research Systems (ORS) Inc.: Montreal, QC, Canada, 2021.
- Tomasi, C.; Manduchi, R. Bilateral Filtering for Gray and Color Images. In Proceedings of the Sixth International Conference on Computer Vision (IEEE Cat. No. 98CH36271), Bombay, India, 7 January 1998; pp. 839–846. [Google Scholar]
- Maes, F.; Collignon, A.; Vandermeulen, D.; Marchal, G.; Suetens, P. Multimodality image registration by maximization of mutual information. IEEE Trans. Med. Imaging 1997, 16, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Li, X.; Sun, X.; Meng, Y.; Liang, J.; Wu, F.; Li, J. Dice Loss for Data-imbalanced NLP Tasks. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020; pp. 465–476. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Cardoso, M.J. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2017; pp. 240–248. [Google Scholar] [CrossRef] [Green Version]
- Crum, W.; Camara, O.; Hill, D. Generalized Overlap Measures for Evaluation and Validation in Medical Image Analysis. IEEE Trans. Med Imaging 2006, 25, 1451–1461. [Google Scholar] [CrossRef]
- Zeiler, M.D. Adadelta: An adaptive learning rate method. arXiv 2012, arXiv:12125701. [Google Scholar]
- Rietveld, H.M. The rietveld method. Phys. Scr. 2014, 89, 098002. [Google Scholar] [CrossRef]
- A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; Available online: https://dl.acm.org/doi/10.5555/3001460.3001507 (accessed on 15 July 2022).
- Ankerst, M.; Breunig, M.M.; Kriegel, H.; Sander, J. OPTICS: Ordering Points to Identify the Clustering Structure. ACM Sigmod Record 1999, 28, 49–60. [Google Scholar] [CrossRef]
- Hamwood, J.; Alonso-Caneiro, D.; Read, S.A.; Vincent, S.; Collins, M.J. Effect of patch size and network architecture on a convolutional neural network approach for automatic segmentation of OCT retinal layers. Biomed. Opt. Express 2018, 9, 3049–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, R.; Markert, S.M.; Britz, S.; Perschin, V.; Erbacher, C.; Stigloher, C.; Kollmannsberger, P. DeepCLEM: Automated registration for correlative light and electron microscopy using deep learning. F1000Research 2020, 9, 1275. [Google Scholar] [CrossRef]
- Li, J.; Sarma, K.V.; Chung-Ho, K.; Gertych, A.; Knudsen, B.S.; Arnold, C.W. A Multi-scale U-Net for Semantic Segmentation of Histological Images from Radical Prostatectomies. AMIA Annu. Symp. Proc. AMIA Symp. 2017, 2017, 1140–1148. [Google Scholar]
- Içek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Springer: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar] [CrossRef] [Green Version]








| Mineral Identified | Chemical Formula | Abundance (%) |
|---|---|---|
| Albite | Al1.488Ca0.491Na0.499O8Si2.506 | 50.4 (±5.6) |
| Ankerite | C2Ca0.997Fe0.676Mg0.273Mn0.054O6 | 1.4 (±0.1) |
| Clinochlore | H16Al2.884Fe0.874Mg11.126O36Si5.116 | 9.7 (±0.8) |
| Illite 2M | C2Al4K1O12Si12 | 12.0 (±1.5) |
| Laumontite | H4Al2Ca1O14Si4 | 14.0 (±1.9) |
| Quartz | SiO2 | 12.6 (±0.9) |
| Assigned Mineral | Centroid Number | Colour on Images | Al (Ct.) | Ca (Ct.) | Fe (Ct.) | K (Ct.) | Mg (Ct.) | Mn (Ct.) | Na (Ct.) | Si (Ct.) | µCT (Ct.) | Abundance (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Albite | 1 | Red | 149 | 18 | 91 | 105 | 89 | 86 | 134 | 148 | 156 | 48 |
| Ankerite | 2 | Grey | 45 | 134 | 126 | 31 | 65 | 92 | 66 | 53 | 203 | 1.98 |
| Clinochlore | 3 | Yellow | 72 | 13 | 191 | 20 | 203 | 220 | 67 | 73 | 187 | 4.06 |
| Illite | 4 | Light blue | 163 | 16 | 74 | 167 | 81 | 75 | 136 | 159 | 150 | 24.8 |
| Laumontite | 5 | White | 121 | 16 | 118 | 44 | 131 | 139 | 113 | 132 | 155 | 7.06 |
| Quartz | 6 | Dark blue | 182 | 12 | 36 | 27 | 72 | 52 | 182 | 179 | 120 | 14 |
| Assigned Mineral | Label in Figure 2 | Raman Detection | Al Mass % | Ca Mass % | Fe Mass % | K Mass % | Mg Mass % | Mn Mass % | Na Mass % | Si Mass % |
|---|---|---|---|---|---|---|---|---|---|---|
| Albite | F | Detected | 16.60 | 2.61 | 11.93 | 4.88 | 1.66 | 0.24 | 8.82 | 51.82 |
| Ankerite | A | Undetected | 16.12 | 3.23 | 18.39 | 3.57 | 4.69 | 0.56 | 6.33 | 45.54 |
| Clinochlore | C | Detected | 15.88 | 3.13 | 13.23 | 4.94 | 2.59 | 0.33 | 7.24 | 51.10 |
| Illite | D | Undetected | 16.59 | 3.14 | 10.30 | 6.77 | 1.09 | 0.23 | 7.61 | 52.74 |
| Laumontite | E | Undetected | 16.29 | 3.28 | 10.33 | 6.41 | 1.42 | 0.22 | 7.70 | 52.73 |
| Quartz | B | Detected | 16.35 | 2.03 | 8.72 | 2.37 | 1.96 | 0.24 | 8.79 | 58.64 |
| Mineral | Deep-XFCT Segmentation Abundance (%) |
|---|---|
| Albite | 50.48 |
| Ankerite | 1.85 |
| Clinochlore | 5.18 |
| Illite | 20.71 |
| Laumontite | 7.06 |
| Quartz | 14.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tung, P.K.M.; Halim, A.Y.; Wang, H.; Rich, A.; Marjo, C.; Regenauer-Lieb, K. Deep-XFCT: Deep Learning 3D-Mineral Liberation Analysis with Micro-X-ray Fluorescence and Computed Tomography. Energies 2022, 15, 5326. https://doi.org/10.3390/en15155326
Tung PKM, Halim AY, Wang H, Rich A, Marjo C, Regenauer-Lieb K. Deep-XFCT: Deep Learning 3D-Mineral Liberation Analysis with Micro-X-ray Fluorescence and Computed Tomography. Energies. 2022; 15(15):5326. https://doi.org/10.3390/en15155326
Chicago/Turabian StyleTung, Patrick Kin Man, Amalia Yunita Halim, Huixin Wang, Anne Rich, Christopher Marjo, and Klaus Regenauer-Lieb. 2022. "Deep-XFCT: Deep Learning 3D-Mineral Liberation Analysis with Micro-X-ray Fluorescence and Computed Tomography" Energies 15, no. 15: 5326. https://doi.org/10.3390/en15155326
APA StyleTung, P. K. M., Halim, A. Y., Wang, H., Rich, A., Marjo, C., & Regenauer-Lieb, K. (2022). Deep-XFCT: Deep Learning 3D-Mineral Liberation Analysis with Micro-X-ray Fluorescence and Computed Tomography. Energies, 15(15), 5326. https://doi.org/10.3390/en15155326






