Silica/Lignin Carrier as a Factor Increasing the Process Performance and Genetic Diversity of Microbial Communities in Laboratory-Scale Anaerobic Digesters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substrates and Inoculum
2.2. Experimental Procedure
2.2.1. Batch Preparation
2.2.2. Carriers Preparation
2.2.3. Bacillus Amyloliquefaciens Biomass
2.2.4. Anaerobic Digestion
2.3. Analysis Techniques
2.3.1. Physicochemical Analysis
2.3.2. Microbiological and Biochemical Analysis
2.3.3. DNA Extraction and Next-Generation Sequencing (NGS)
2.3.4. Statistical and Bioinformatics Analyses
3. Results and Discussion
3.1. Substrate Characteristics
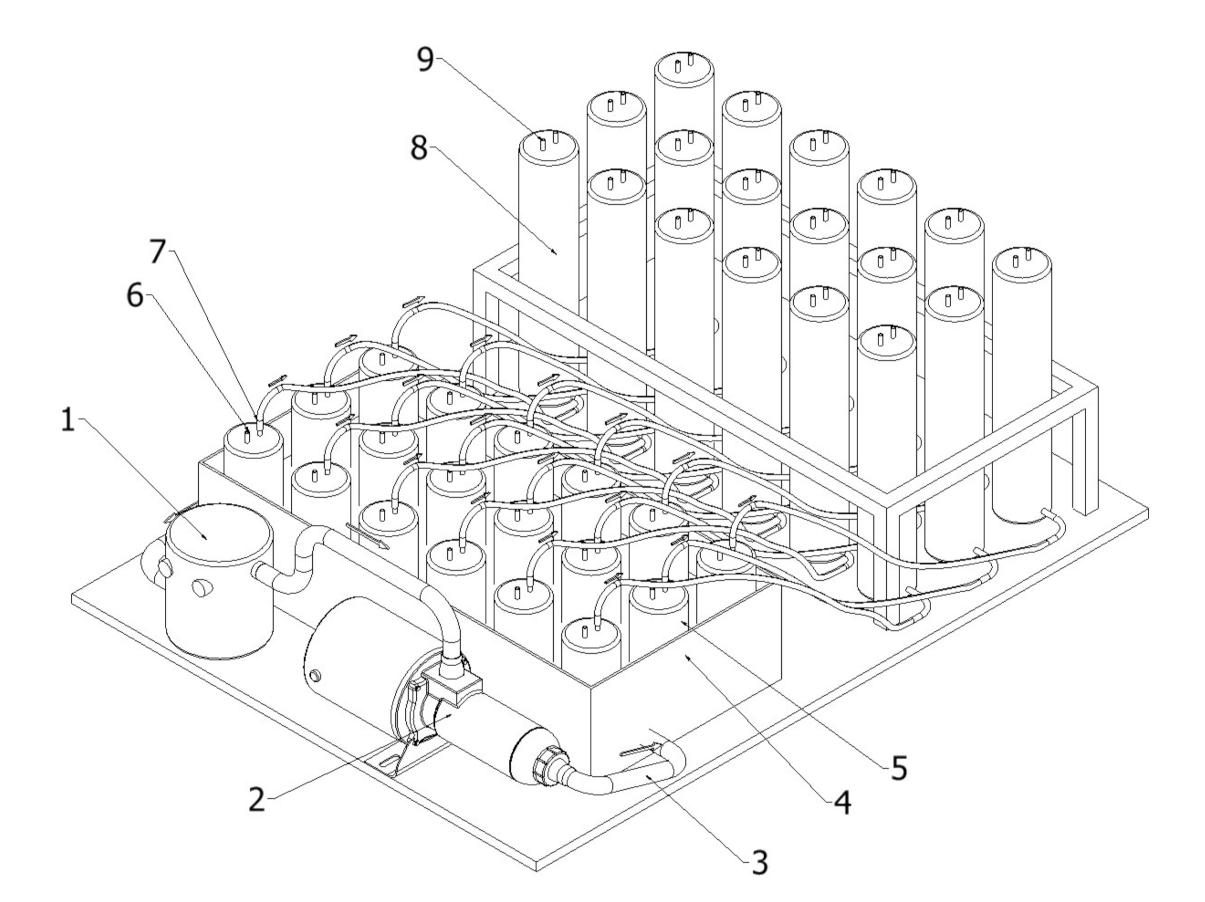
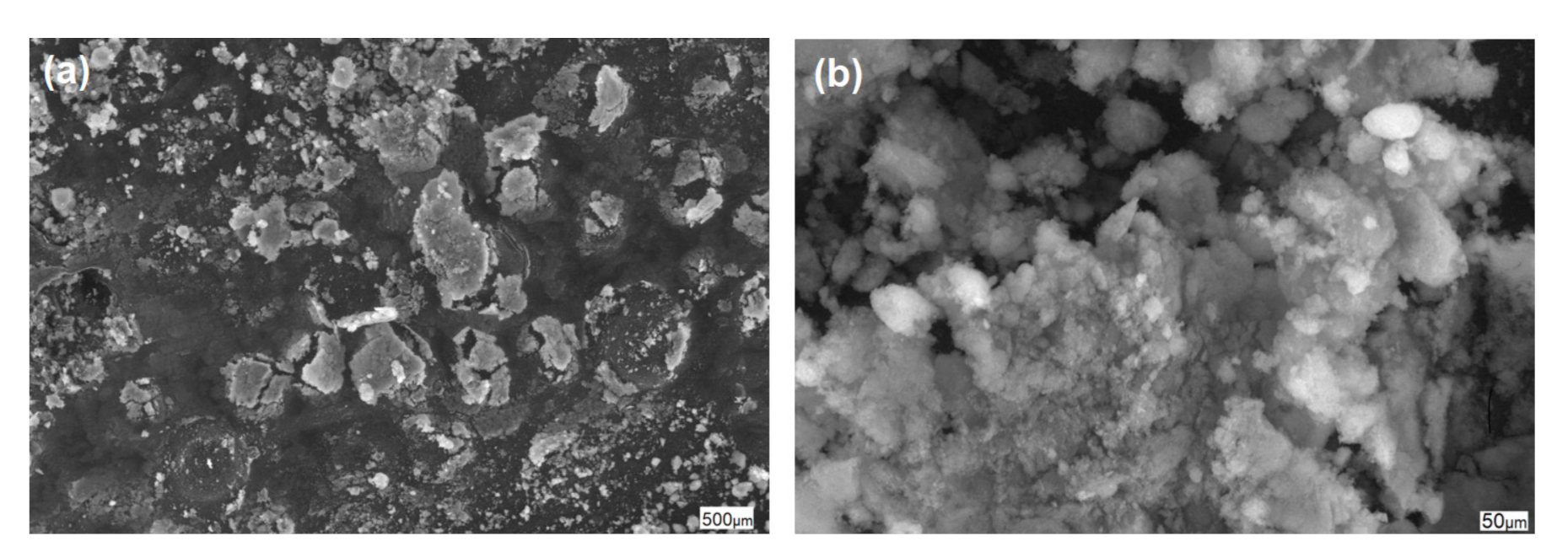
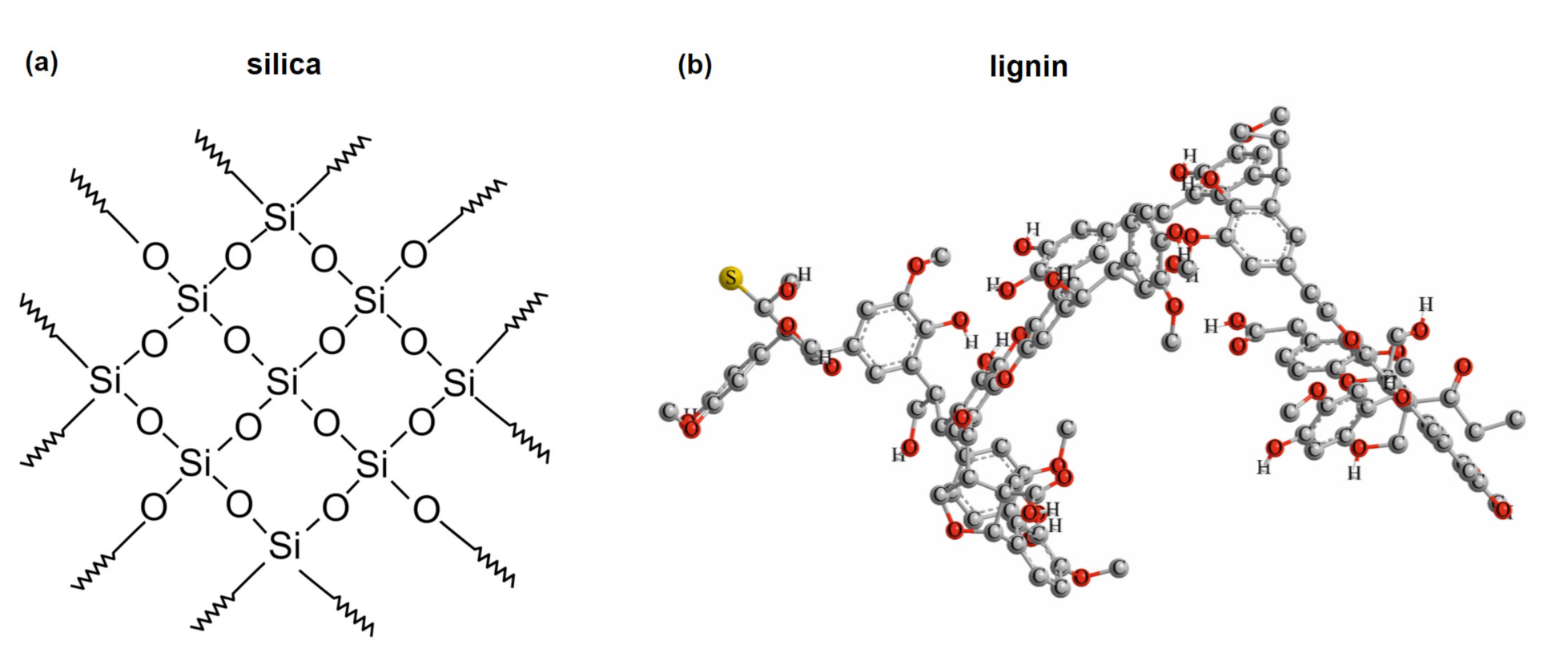
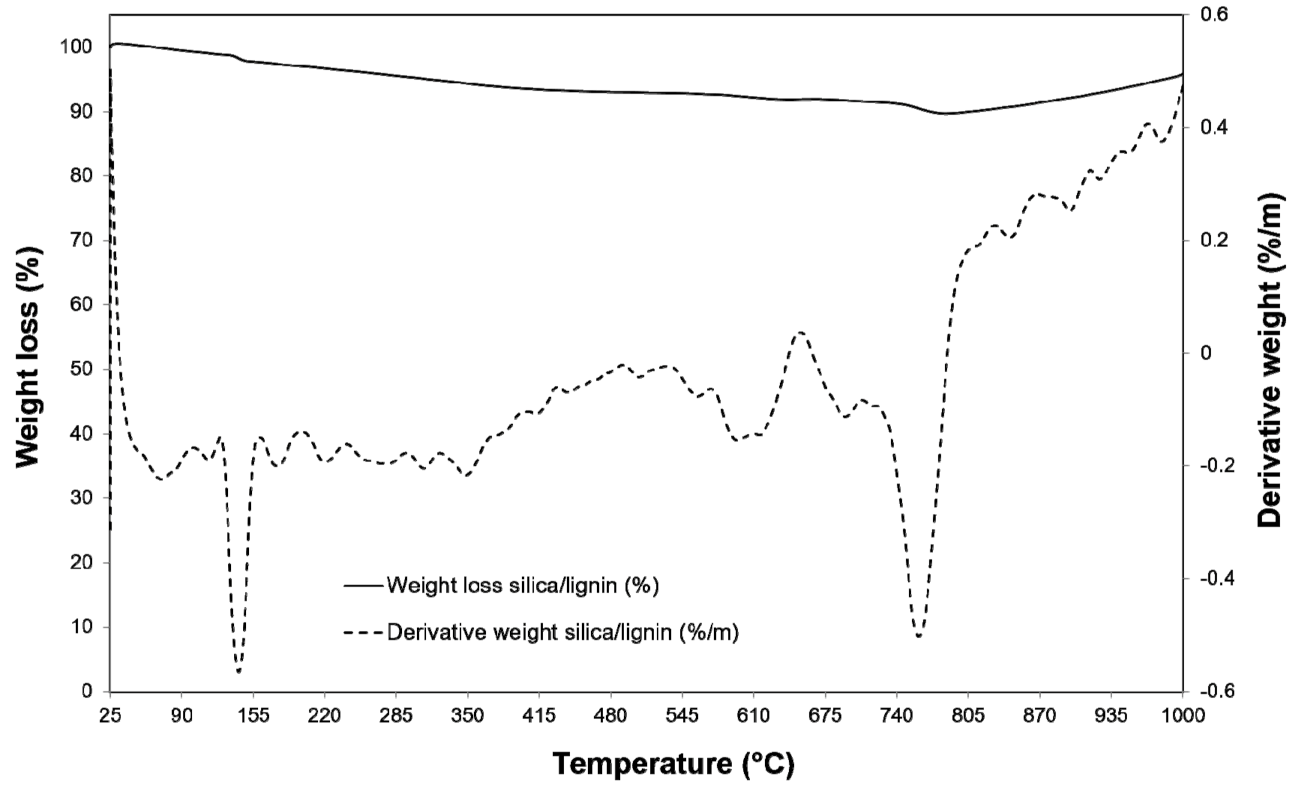
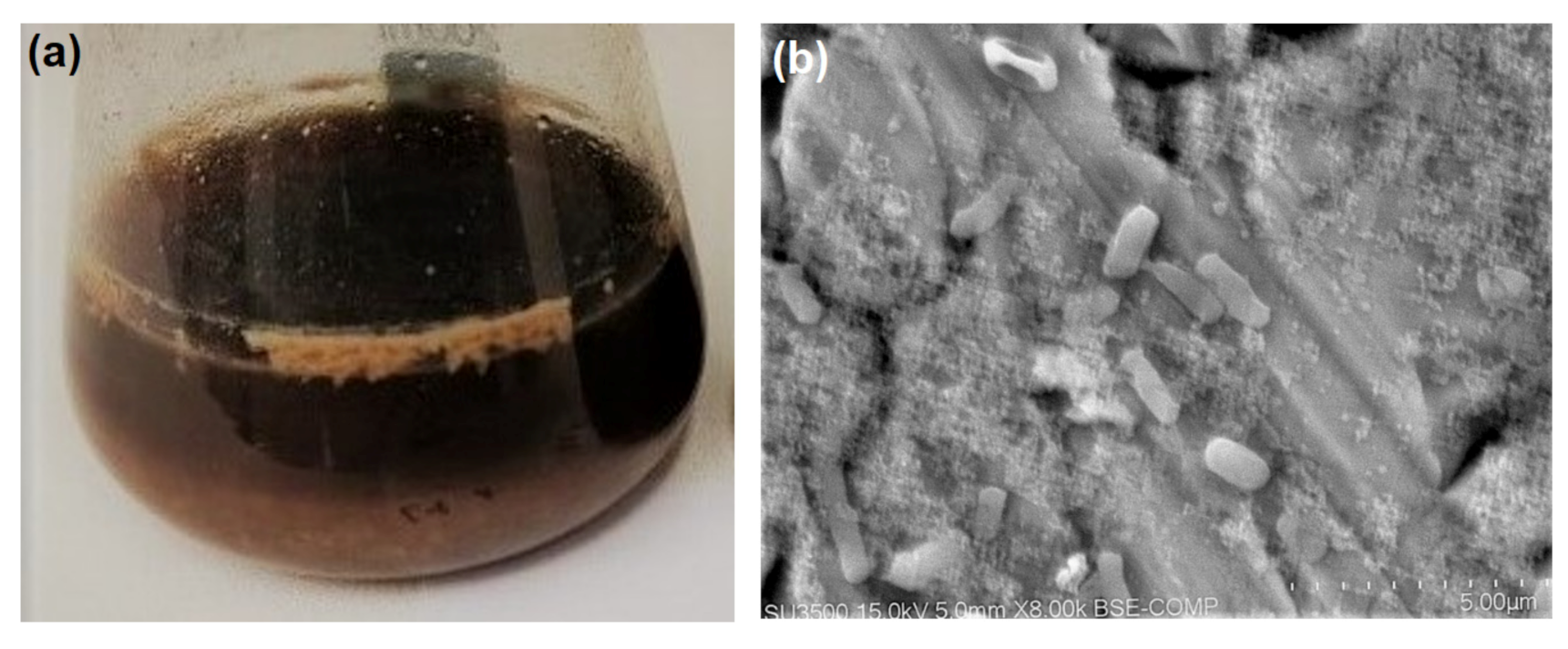
3.2. Properties and Efficiency of a Carrier
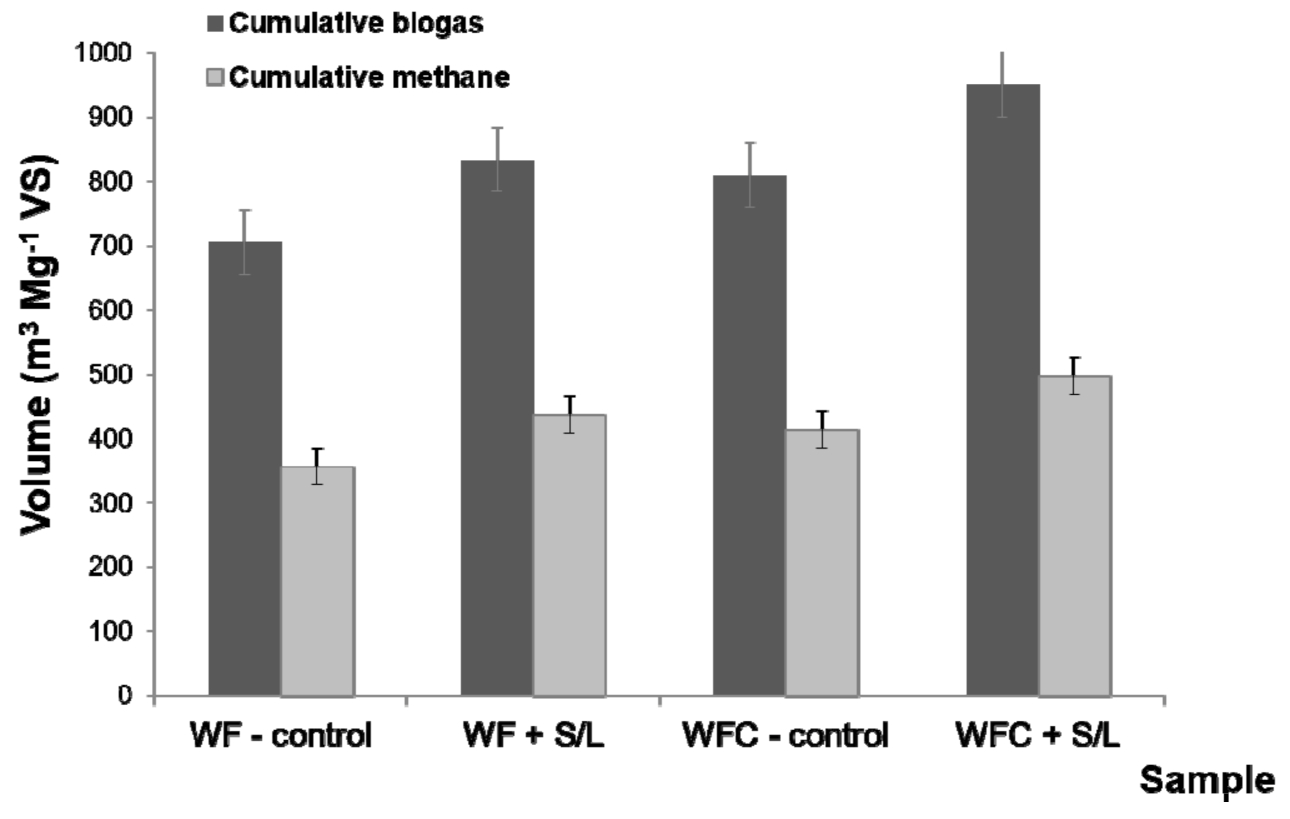
3.3. Stability and Performance of Anaerobic Digestion
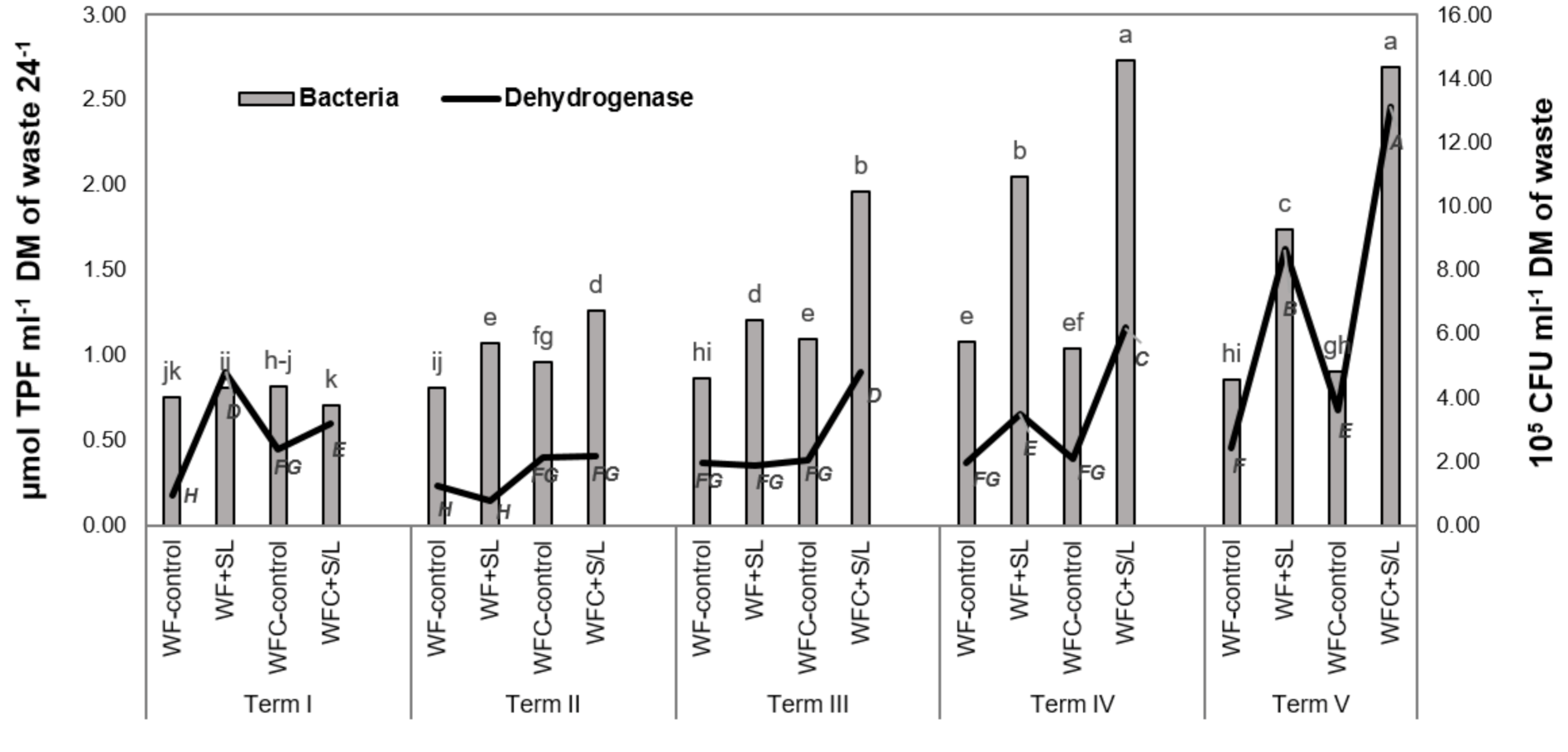
3.4. Total Bacterial Count and Dehydrogenase Activity in Digested Samples
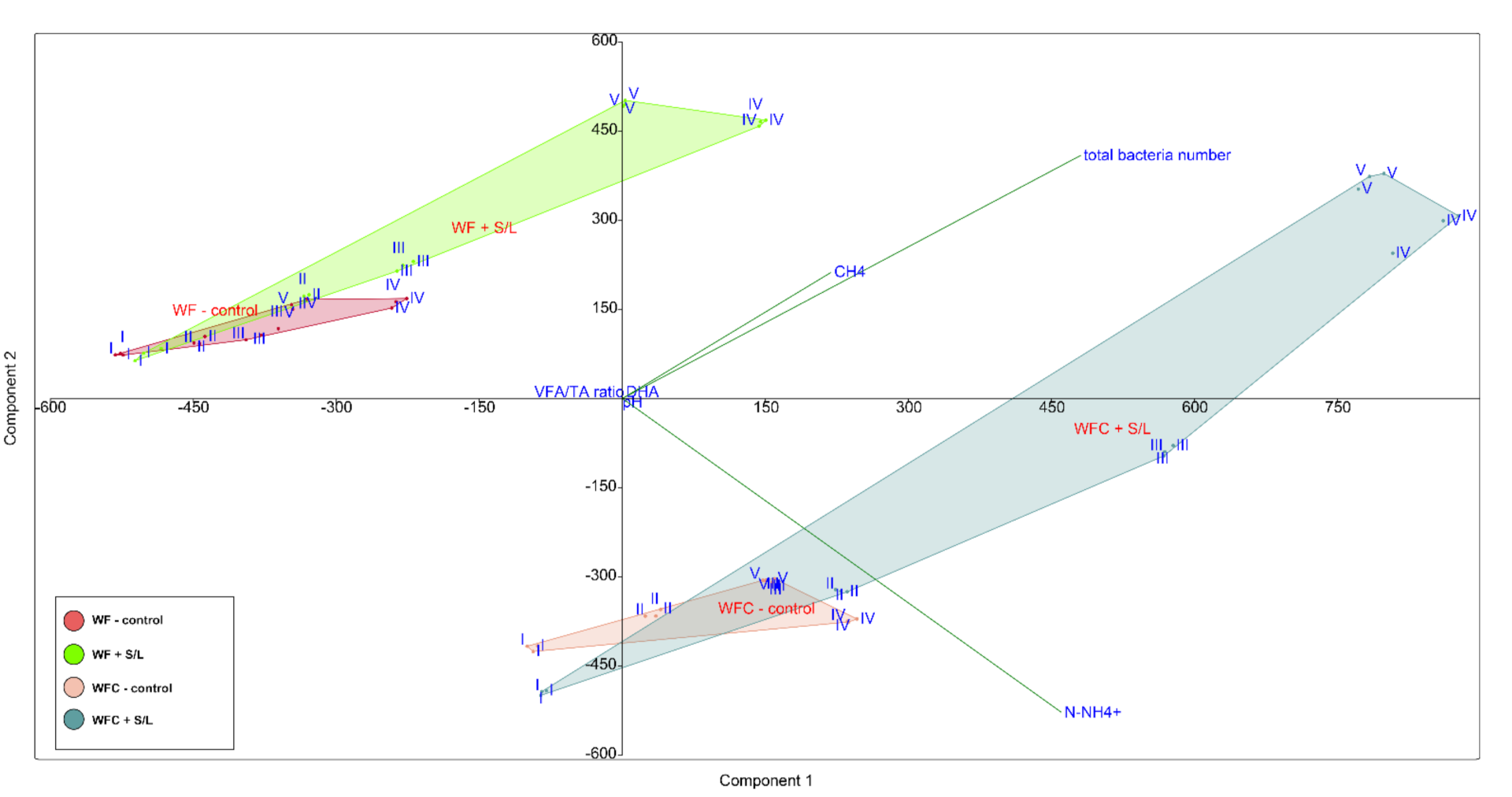
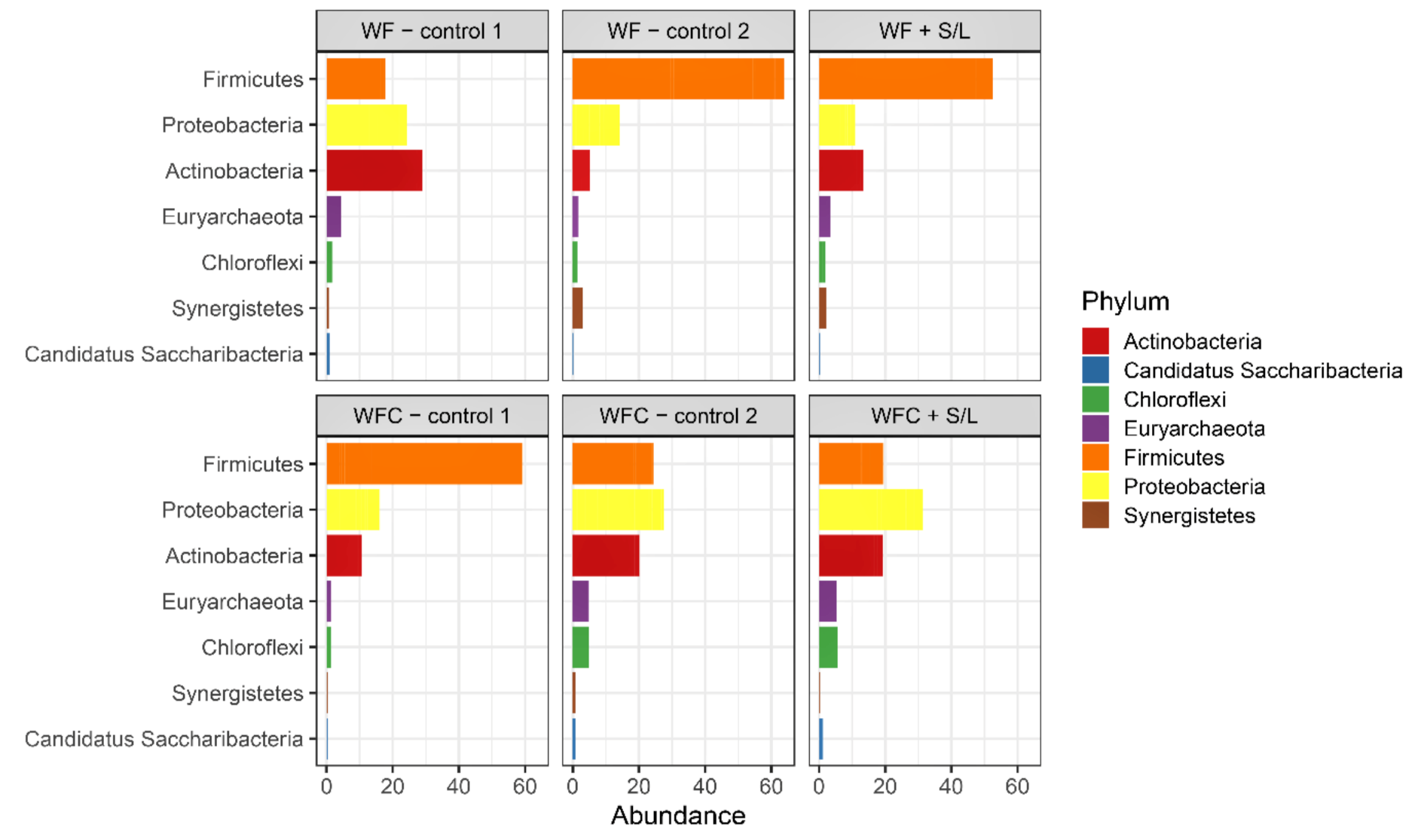
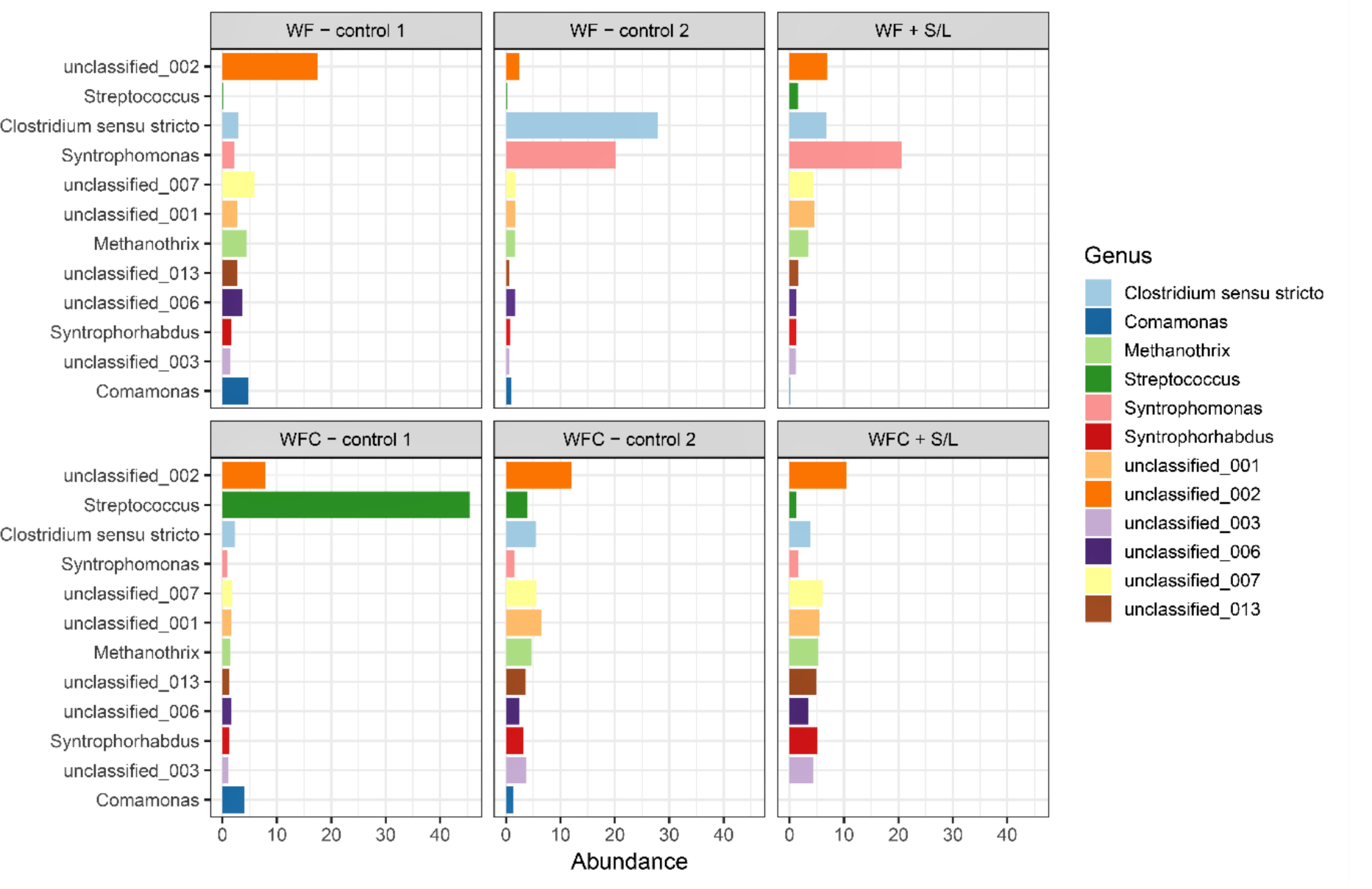
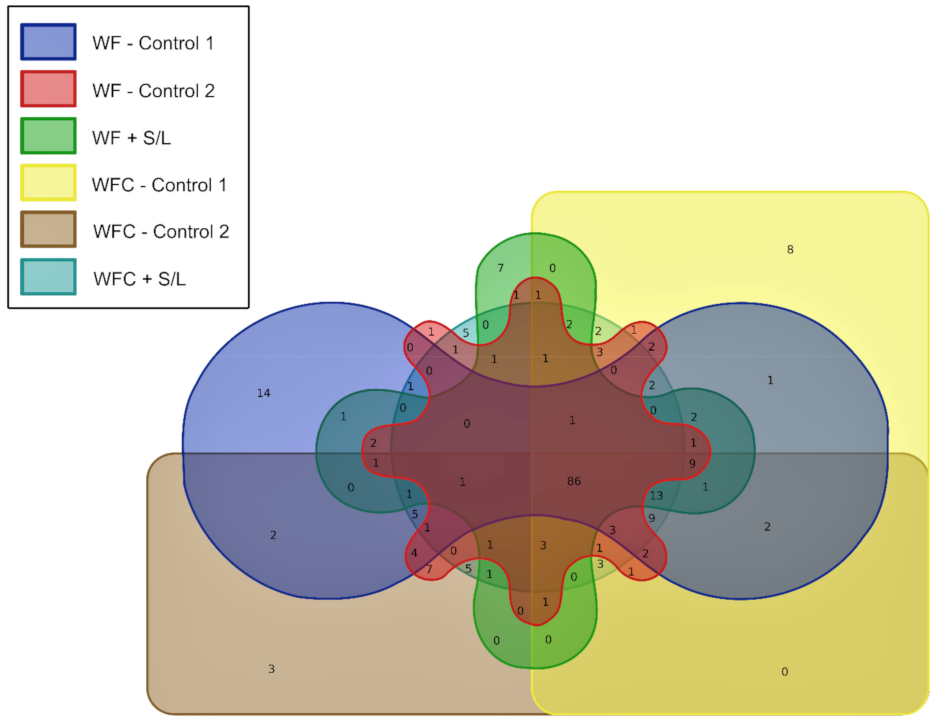
3.5. Bacterial Community Abundance and Composition
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pilarska, A.A.; Pilarski, K.; Witaszek, K.; Waliszewska, H.; Zborowska, M.; Waliszewska, B.; Kolasiński, M.; Szwarc-Rzepka, K. Treatment of dairy waste by anaerobic digestion with sewage sludge. Ecol. Chem. Eng. 2016, 23, 99–115. [Google Scholar] [CrossRef] [Green Version]
- Pilarska, A.A. Anaerobic co-digestion of waste wafers from the confectionery production with sewage sludge. Pol. J. Environ. Stud. 2018, 27, 237–245. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Waliszewska, B.; Zborowska, M.; Witaszek, K.; Waliszewska, H.; Kolasiński, M.; Szwarc-Rzepka, K. Evaluation of bio-methane yields for high-energy organic waste and sewage sludge: A pilot-scale study for a wastewater treatment plant. Environ. Eng. Manag. J. 2019, 18, 2019–2030. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Pilarski, K.; Ryniecki, A.; Tomaszyk, K.; Dach, J.; Wolna-Maruwka, A. Utilization of vegetable dumplings waste from industrial production by anaerobic digestion. Int. Agrophys. 2017, 31, 93–102. [Google Scholar] [CrossRef]
- Witaszek, K.; Pilarski, K.; Niedbała, G.; Pilarska, A.A.; Herkowiak, M. Energy efficiency of comminution and extrusion of maize substrates subjected to methane fermentation. Energies 2020, 13, 1887. [Google Scholar] [CrossRef]
- Pilarski, K.; Pilarska, A.A.; Boniecki, P.; Niedbała, G.; Durczak, K.; Witaszek, K.; Mioduszewska, N.; Kowalik, I. The efficiency of industrial and laboratory anaerobic digesters of organic substrates: The use of the biochemical methane potential correction coefficient. Energies 2020, 13, 1280. [Google Scholar] [CrossRef] [Green Version]
- Mioduszewska, N.; Pilarska, A.A.; Pilarski, K.; Adamski, M. The influence of the process of sugar beet storage on its biochemical methane potential. Energies 2020, 13, 5104. [Google Scholar] [CrossRef]
- Manzanares, P. The role of biorefinering research in the development of a modern bioeconomy. Acta Innov. 2020, 37, 47–56. [Google Scholar] [CrossRef]
- Lopes, T.F.; Łukasik, R.M. Economic, social and environmental impacts attained by the use of the effluents generated within a small-scale biorefinery concept. Acta Innov. 2020, 36, 57–63. [Google Scholar] [CrossRef]
- Rusín, J.; Kašáková, K.; Chamrádová, K. Anaerobic digestion of waste wafer material from the confectionery production. Energy 2015, 85, 194–199. [Google Scholar] [CrossRef]
- Ximenes, J.; Siqueira, A.; Kochańska, E.; Łukasik, R.M. Valorisation of Agri- and Aquaculture Residues via Biogas Production for Enhanced Industrial Application. Energies 2021, 14, 2519. [Google Scholar] [CrossRef]
- Dzionek, A.; Wojcieszyńska, D.; Guzik, U. Natural carriers in bioremediation: A review. Electron. J. Biotechnol. 2016, 23, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Pilarska, A.A.; Pilarski, K.; Wolna-Maruwka, A. Cell immobilization on lignin–polyvinylpyrrolidone material used for anaerobic digestion of waste wafers and sewage sludge. Environ. Eng. Sci. 2019, 36, 478–490. [Google Scholar] [CrossRef]
- Gong, W.; Ran, Z.; Ye, F.; Zhao, G. Lignin from bamboo shoot shells as an activator and novel immobilizing support for α-amylase. Food Chem. 2017, 228, 455–462. [Google Scholar] [CrossRef]
- Weiß, S.; Zankel, A.; Lebuhn, M.; Petrak, S.; Somitsch, W.; Guebitz, G.M. Investigation of microorganisms colonising activated zeolites during anaerobic biogas production from grass silage. Bioresour. Technol. 2011, 102, 4353–4359. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, G.; Rákhely, G.; Kovács, K.L. Hydrogen production from biopolymers by Caldicellulosiruptor saccharolyticus and stabilization of the system by immobilization. Int. J. Hydrogen Energy 2008, 33, 6953–6961. [Google Scholar] [CrossRef]
- Purnomo, C.W.; Mellyanawaty, M.; Budhijanto, W. Simulation and experimental study on iron impregnated microbial immobilization in zeolite for production of biogas. Waste Biomass Valor. 2017, 8, 2413–2421. [Google Scholar] [CrossRef]
- Jesionowski, T. Characterisation of pigments obtained by adsorption of C.I. Basic Blue 9 and C.I. Acid Orange 52 dyes onto silica particles precipitated via the emulsion route. Dyes Pigm. 2005, 67, 81–92. [Google Scholar] [CrossRef]
- Clemments, A.M.; Botella, P.; Landry, C.C. Protein adsorption from biofluids on silica nanoparticles: Corona analysis as a function of particle diameter and porosity. ACS Appl. Mater. Interfaces 2015, 7, 21682–21689. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Xu, Y.; Dong, B. Effect of the micron-sized silica particles (MSSP) on biogas conversion of sewage sludge. Water Res. 2017, 115, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Dong, B.; Yang, D.; Li, N.; Dai, X. Micron-sized silica particles in wastewater influenced the distribution of organic matters in sludge and their anaerobic degradation. J. Hazard. Mater. 2020, 393, 122340. [Google Scholar] [CrossRef]
- Ge, Y.; Qin, L.; Li, Z. Lignin microspheres: An effective and recyclable natural polymer-based adsorbent for lead ion removal. Mater. Design 2016, 95, 141–147. [Google Scholar] [CrossRef]
- Pilarska, A.A.; Wolna-Maruwka, A.; Pilarski, K. Kraft lignin grafted with polyvinylpyrrolidone as a novel microbial carrier in biogas production. Energies 2018, 11, 3246. [Google Scholar] [CrossRef] [Green Version]
- Pilarska, A.A.; Wolna-Maruwka, A.; Pilarski, K.; Janczak, D.; Przybył, K.; Gawrysiak-Witulska, M. The use of lignin as a microbial carrier in the co-digestion of cheese and wafer waste. Polymers 2019, 11, 2073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilarska, A.A.; Wolna-Maruwka, A.; Niewiadomska, A.; Pilarski, K.; Olesienkiewicz, A. A Comparison of the influence of kraft lignin and the kraft lignin/silica system as cell carriers on the stability and efficiency of the anaerobic digestion process. Energies 2020, 13, 5803. [Google Scholar] [CrossRef]
- Aguado, J.; Arsuaga, J.M.; Arencibi, A.; Lindo, M.; Gascón, V. Aqueous heavy metals removal by adsorption on amine-functionalized mesoporous silica. J. Hazard. Mater. 2009, 163, 213–221. [Google Scholar] [CrossRef]
- Saikia, J.; Yazdimamaghani, M.; Moghaddam, S.P.H.; Ghandehari, H. Differential protein adsorption and cellular uptake of silica nanoparticles based on size and porosity. ACS Appl. Mater. Interfaces 2016, 8, 34820–34832. [Google Scholar] [CrossRef] [Green Version]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; van Braekel, J.; Fu, Q.; Roosens, N.H.C.; de Keersmaecker, S.C.J.; Vanneste, K. Targeting the 16S rRNA gene for bacterial identification in complex mixed samples: Comparative evaluation of second (Illumina) and third (Oxford Nanopore Technologies) generation sequencing technologies. J. Mol. Sci. 2020, 21, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norm VDI 4630. Fermentation of Organic Materials Characterization of the Substrate, Sampling, Collection of Material Data, Fermentation Tests; German Engineers Club: Düsseldorf, Germany, 2006. [Google Scholar]
- DIN Guideline 38 414-S8. Characterisation of the Substrate, Sampling, Collection of Material Data, Fermentation Tests; German Institute for Standardization: Berlin, Germany, 1985. [Google Scholar]
- Pilarska, A.A.; Pilarski, K.; Wolna-Maruwka, A.; Boniecki, P.; Zaborowicz, M. Use of confectionery waste in biogas production by the anaerobic digestion process. Molecules 2019, 24, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camiña, F.; Trasar-Cepeda, C.; Gil-Sotres, F.; Leirós, C. Measurement of dehydrogenase activity in acid soilsrich in organic matter. Soil Biol. Biochem. 1998, 30, 1005–1011. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; Vienna Insurance Group AG: Vienna, Austria, 2016; Available online: https://www.R-project.org/ (accessed on 9 March 2021).
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. cPhyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef]
- Wright, E.S. RDP v16 Modified Training Set for 16S rRNA Classification. 2019. Available online: http://www2.decipher.codes/Classification/TrainingSets/RDP_v16-mod_March2018.RData (accessed on 9 March 2018).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.L.; Ortiz, R.; Steele, T.W.J.; Stuckey, D.C. Toxicants inhibiting anaerobic digestion: A review. Biotechnol. Adv. 2014, 32, 1523–1534. [Google Scholar] [CrossRef]
- Kupiec, K.; Konieczka, P. Charakterystyka, procesy chemicznej modyfikacji oraz zastosowanie krzemionki. Ecol. Chem. Eng. 2007, 14, 473–487. [Google Scholar]
- Pilarska, A.; Bula, K.; Myszka, K.; Rozmanowski, T.; Szwarc-Rzepka, K.; Pilarski, K.; Chrzanowski, Ł.; Czaczyk, K.; Jesionowski, T. Functional polypropylene composites filled with ultra-fine magnesium hydroxide. Open Chem. 2015, 13, 161–171. [Google Scholar] [CrossRef]
- Pilarska, A.; Markiewicz, E.; Ciesielczyk, F.; Jesionowski, T. The influence of spray drying on dispersive the and physicochemical properties of magnesium oxide. Dry. Technol. 2011, 29, 1210–1218. [Google Scholar] [CrossRef]
- Klapiszewski, L.; Rzemieniecki, T.; Krawczyk, M.; Malina, D.; Norman, M.; Zdarta, J.; Majchrzak, I.; Dobrowolska, A.; Czaczyk, K.; Jesionowski, T. Kraft lignin/silica–AgNPs as a functional material with antibacterial activity. Colloids Surf. B Biointerfaces 2015, 134, 220–228. [Google Scholar] [CrossRef]
- Ralph, J.; Lundguist, K.; Brunow, G.; Lu, F.; Kim, H.; Schatz, P.F. Marita, J.M.; Hatfield, R.D.; Ralph, S.A.; Christensen, J.H. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl- propanoids. Phytochem. Rev. 2004, 3, 29–60. [Google Scholar] [CrossRef]
- Yamamoto, T.; Yabushita, S.; Irisawa, T.; Tanabe, Y. Enhancement of bending strength, thermal stability and recyclability of carbon-fiber-reinforced thermoplastics by using silica colloids. Compos. Sci. Technol. 2019, 18, 107665. [Google Scholar] [CrossRef]
- Bhatt, B.; Prajapati, V.; Patel, K.; Trivedi, U. Kitchen waste for economical amylase production using Bacillus amyloliquefaciens KCP2. Biocatal. Agric. Biotechnol. 2020, 26, 101654. [Google Scholar] [CrossRef]
- Karunakaran, G.; Suriyaprabha, R.; Manivasakan, P.; Yuvakkumar, R.; Rajendran, V.; Prabu, P.; Kannan, N. Effect of nanosilica and silicon sources on plant growth promoting rhizobacteria, soil nutrients and maize seed germination. IET Nanobiotechnol. 2013, 7, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Krakat, N.; Demirel, B.; Anjum, R.; Dietz, D. Methods of ammonia removal in anaerobic digestion: A review. Water Sci. Technol. 2017, 76, 1925–1938. [Google Scholar] [CrossRef] [PubMed]
- Suschka, J.; Grübel, K. Nitrogen in the process of waste activated sludge anaerobic digestion. Arch. Environ. Prot. 2014, 40, 123–136. [Google Scholar] [CrossRef] [Green Version]
- Tapia-Olivares, V.R.; Vazquez-Bello, E.A.; Aguilar-Garnica, E.; Escalante, F.M. Valorization of lignin as an immobilizing agent for bioinoculant production using Azospirillum brasilense as a model bacteria. Molecules 2019, 24, 4613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Xie, X.; Wang, Z.; Tao, Y.; Niu, X.; Huang, X.; Liu, L.; Li, Z. Immobilization of Lactobacillus rhamnosus in mesoporous silica-based material: An efficiency continuous cell-recycle fermentation system for lactic acid production. J. Biosci. Bioeng. 2016, 121, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, R.; He, Q.; Ji, B.; Wang, H.; Yang, K. Adaptation to salinity: Response of biogas production and microbial communities in anaerobic digestion of kitchen waste to salinity stress. J. Biosci. Bioeng. 2020, 130, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Yenigün, O.; Demirel, B. Ammonia inhibition in anaerobic digestion: A review. Process. Biochem. 2013, 48, 901–911. [Google Scholar] [CrossRef]
- Bhatta, R.; Tajima, K.; Kurihara, M. Influence of temperature and pH on fermentation pattern and methane production in the rumen simulating fermenter (RUSITEC). Asian Aust. J. Animal Sci. 2006, 19, 376–380. [Google Scholar] [CrossRef]
- Rawat, N.; Joshi, G.K. Bacterial community structure analysis of a hot spring soil by next generation sequencing of ribosomal RNA. Genomics 2019, 111, 1053–1058. [Google Scholar] [CrossRef]
- Furtak, K.; Grządziel, J.; Gałązka, A.; Niedźwiecki, J. Prevalence of unclassified bacteria in the soil bacterial community from floodplain meadows (fluvisols) under simulated flood conditions revealed by a metataxonomic approachss. Catena 2020, 188, 104448. [Google Scholar] [CrossRef]
- Banach, A.; Ciesielski, S.; Bacza, T.; Pieczykolan, M.; Ziembińska-Buczyńska, A. Microbial community composition and methanogens’ biodiversity during a temperature shift in a methane fermentation chamber. Environ. Technol. 2019, 40, 3252–3263. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Knapp, B.A.; Farbmacher, T.; Ebner, C.; Insam, H.; Franke-Whittle, I.H. Searching for links in the biotic characteristics and abiotic parameters of nine different biogas plants. Microb. Biotechnol. 2012, 5, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Bedard, D.L.; Ritalahti, K.M.; Löffler, F.E. The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl. Environ. Microbiol. 2007, 73, 2513–2521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Doesburg, W.; van Eekert, M.H.A.; Middeldorp, P.J.M.; Balk, M.; Schraa, G.; Stams, A.J.M. Reductive dechlorination of beta-hexachlorocyclohexane (beta-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 2005, 54, 87–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, M.S.; Clark, C.M.; Ómarsdóttir, S.; Sanchez, L.M.; Murphy, B.T. Minimizing taxonomic and natural product redundancy in microbial libraries using MALDI-TOF MS and the bioinformatics pipeline IDBac. J. Nat. Prod. 2019, 82, 2167–2173. [Google Scholar] [CrossRef] [PubMed]
- Gärtner, A.; Wiese, J.; Imhoff, J.F. Diversity of Micromonospora strains from the deep Mediterranean Sea and their potential to produce bioactive compounds. AIMS Microbiol. 2016, 2, 205–221. [Google Scholar] [CrossRef]
- Yamada, T.; Imachi, H.; Ohashi, A.; Harada, H.; Hanada, S.; Kamagata, Y.; Sekiguchi, Y. Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int. J. Syst. Evol. Microbiol. 2007, 57, 2299–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, S.; Pan, X.C.; Mei, R.; Wang, Y.N.; Sun, J.Q.; Liu, X.Y.; Tang, Y.Q.; Wu, X.L. Ottowia shaoguanensis sp. nov., isolated from coking wastewater. Curr. Microbiol. 2014, 68, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Teramoto, M.; Harayama, S. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 1999, 65, 2813–2819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, Y.; Tamaki, H.; Matsuzawa, H.; Nigaya, M.; Mori, K.; Kamagata, Y. Microbial community analysis in the roots of aquatic plants and isolation of novel microbes including an organism of the candidate phylum OP10. Microbes Environ. 2012, 27, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.L.; Hanada, S.; Ohashi, A.; Harada, H.; Kamagata, Y.; Sekiguchi, Y. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl. Environ. Microbial. 2008, 74, 2051–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, T.; Bian, S.; Ko, J.H.; Liu, J.; Shi, X.; Xu, Q. Exploring the roles of zero-valent iron in two-stage food waste anaerobic digestion. Waste Manag. 2020, 107, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, Y. Application of ethanol-type fermentation in establishmentof direct interspecies electron transfer: A practical engineering case study. Renew. Energy 2019, 136, 846–855. [Google Scholar] [CrossRef]
- Laothanachareon, T.; Kanchanasuta, S.; Mhuanthong, W.; Phalakornkule, C.; Pisutpaisal, N.; Champreda, V. Analysis of microbial community adaptation in mesophilic hydrogen fermentation from food waste by tagged 16S rRNA gene pyrosequencing. J. Environ. Manag. 2014, 144, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.A.; Sobczak, A.; Dziembowski, A.; Lipinski, L. Genomic analysis of γ-hexachlorocyclohexane-degrading Sphingopyxis lindanitolerans WS5A3p strain in the context of the pangenome of Sphingopyxis. Genes 2019, 10, 688. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.Q.; Shigematsu, T.; Morimura, S.; Kida, K. Dynamics of the microbial community during continuous methane fermentation in continuously stirred tank reactors. J. Biosci. Bioeng. 2015, 119, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Pindi, P.K.; Ashwitha, K.; Rani, A.S. Chryseomicrobium palamuruense sp. nov., a haloalkalitolerant bacterium isolated from a sediment sample. Int. J. Syst. Evol. Microbiol. 2016, 66, 3731–3736. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Lai, Y.; Xue, F.; Sun, L.; Wang, J. Paracandidimonas caeni sp. nov., isolated from sludge. Int. J. Syst. Evol. Microbiol. 2019, 69, 3332–3337. [Google Scholar] [CrossRef] [PubMed]
| Samples | WF (g) | CE (g) | S/L (g) | Inoculum (g) | pH | TS (%) | VS (%) |
|---|---|---|---|---|---|---|---|
| WF-control | 9.8 | - | - | 830.2 | 7.15 | 4.01 | 65.64 |
| WF + S/L | 9.8 | - | 20.0 | 830.2 | 7.08 | 4.00 | 64.62 |
| WFC-control | 5.5 | 2.9 | - | 832.6 | 6.96 | 3.76 | 65.57 |
| WFC + S/L | 5.5 | 2.9 | 20.0 | 832.6 | 6.75 | 3.76 | 64.55 |
| Waste | pH | Cond. | TS | VS | C/N Ratio | C | N | N-NH4 |
|---|---|---|---|---|---|---|---|---|
| (mS cm−1) | (wt %) | (wt %TS) | (wt %TS) | (wt %TS) | (wt %TS) | |||
| Wafers | 6.92 | 1.88 | 71.62 | 99.53 | 46.22 | 43.45 | 0.94 | 0.32 |
| Cheese | 4.59 | 74.36 | 32.15 | 94.86 | 3.48 | 49.64 | 14.25 | 0.49 |
| Inoc. | 7.03 | 32.16 | 3.21 | 65.24 | 3.30 | 26.37 | 7.99 | 3.97 |
| Samples | Fermentation Time (Days) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 6 | 9 | 12 | 15 | 18 | 21 | |
| pH (-) | ||||||||
| WF-control | 6.92 | 7.05 | 7.13 | 7.28 | 7.16 | 7.09 | 7.21 | 7.29 |
| WF + S/L | 6.89 | 6.98 | 7.05 | 7.17 | 7.23 | 7.14 | 7.17 | 7.25 |
| WFC-control | 7.05 | 7.12 | 7.31 | 7.44 | 7.38 | 7.50 | 7.46 | 7.53 |
| WFC + S/L | 6.85 | 6.94 | 7.10 | 718 | 7.32 | 7.29 | 7.14 | 7.16 |
| VFA/TA ratio (-) | ||||||||
| WF-control | 0.36 | 0.43 | 0.38 | 0.31 | 0.29 | 0.25 | 0.26 | 0.24 |
| WF + S/L | 0.42 | 0.37 | 0.36 | 0.33 | 0.40 | 0.29 | 0.25 | 0.22 |
| WFC-control | 0.38 | 0.40 | 0.43 | 0.42 | 0.39 | 0.34 | 0.28 | 0.25 |
| WFC + S/L | 0.41 | 0.39 | 0.42 | 0.38 | 0.35 | 0.37 | 0.29 | 0.23 |
| N–NH4+ (mg L−1) | ||||||||
| WF-control | 148 | 164 | 173 | 189 | 222 | 278 | 249 | 216 |
| WF + S/L | 165 | 169 | 214 | 225 | 238 | 296 | 264 | 183 |
| WFC-control | 805 | 812 | 875 | 892 | 936 | 998 | 917 | 881 |
| WFC + S/L | 871 | 939 | 986 | 964 | 905 | 893 | 815 | 792 |
| Parameter | Term | Combination | Interaction |
|---|---|---|---|
| Bacteria | 2225.42 *** | 3915.19 *** | 600.59 *** |
| Dehydrogenase | 1882.39 *** | 1924.33 *** | 452.00 *** |
| Unclassified Symbol (in This Research) | NCBI Accession Numbers (% of Sequence Identity) | Source/Environment | References (If Available) | Closest Relative |
|---|---|---|---|---|
| unclassified_001 | EF059533 (97.2%) | PCB-dechlorinating enrichment culture | Bedard et al. (2007) [62] | Sedimentibacter sp |
| AY766467 (96.5%) | Anaerobic coculture enriched with a hexachlorocyclohexane (HCH) polluted soil. | Wim van Doesburg et al. (2005) [63] | Sedimentibacter sp | |
| unclassified_002 | MK143173 (98.8%) | Algae (Iceland) | Costa et al. (2019) [64] | Knoellia sp. |
| KX256211 (98.8%) | Eastern Mediterranean Sea Sediment | Gärtner et al. (2016) [65] | Intrasporangium sp. | |
| unclassified_003 | NR_041354 (97%) | Thermophilic digester sludge, methanogenic propionate-degrading consortia | Yamada et al. (2007) [66] | Bellilinea caldifistulae |
| KX261406 (93.8%) | Sludge and beet sugar industrial wastewater | - | Levilinea saccharolytica | |
| unclassified_006 | KC252871 (100%) | Activated sludge | - | Comamonadaceae bacterium |
| KF751647 (99.8%) | Wastewater treatment system | - | Diaphorobacter sp. | |
| NR_125656 (99.3%) | Coking wastewater | Geng et al. (2014) [67] | Ottowia sp. | |
| unclassified_007 | AB021325 (98%) | Activated sludge with phenol as the sole carbon source | Watanebe et al. (1999) [68] | Uncultured/unclassified |
| JQ899231 (97.5%) | Marine soil sediment | - | Streptomyces aomiensis | |
| unclassified_013 | AB529706 (98%) | Rhizoplane | Tanaka et al. (2012) [69] | Uncultured/unclassified |
| HM124367 (96.8%) | Lake sediment | - | Hyphomicrobium sp. |
| Sample | Observed | Shannon (H`) | Simpson (1/D) |
|---|---|---|---|
| WF-control 1 | 167 | 3.869 | 0.9513 |
| WF-control 2 | 137 | 3.089 | 0.8741 |
| WF + S/L | 138 | 3.541 | 0.9345 |
| WFC-control 1 | 161 | 2.779 | 0.7803 |
| WFC-control 2 | 166 | 3.896 | 0.9620 |
| WFC + S/L | 152 | 3.835 | 0.9635 |
| Abbreviation | Meaning |
|---|---|
| AD | anaerobic digestion |
| NGS | next-generation sequencing |
| 16S rRNA | prokaryotic 16S ribosomal RNA gene |
| TS | total solids |
| VS | volatile solids |
| WF | wafer waste |
| CE | cheese waste |
| WFC | wafer and cheese co-substrates |
| S/L | silica/lignin carrier |
| PBS | phosphate buffered saline |
| DIN | Deutsches Institut für Normung |
| VDI | Verein Deutscher Ingenieure |
| HRT | hydraulic retention time |
| VFA | volatile fatty acids |
| VFA/TA ratio | volatile fatty acids-to-total alkalinity ratio |
| C/N ratio | carbon/nitrogen ratio |
| TTC | 2,3,5-triphenyltetrazolium chloride |
| TPF | triphenylformazan |
| DHA | dehydrogenase activity |
| CFU | colony-forming units |
| AnB | anaerobic bacteria |
| DM | dry matter |
| PE | paired-end |
| PCR | polymerase chain reaction |
| RDP | ribosomal database project |
| IDTAXA | taxonomic classification algorithm |
| DADA2 | software package that models and corrects Illumina-sequenced amplicon errors |
| PCA | principal component analysis |
| SEM | scanning electron microscope |
| BET surface | Brunauer-Emmett-Teller |
| TG | thermogravimetry |
| OTUs | operational taxonomic units |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
A. Pilarska, A.; Wolna-Maruwka, A.; Niewiadomska, A.; Pilarski, K.; Adamski, M.; Grzyb, A.; Grządziel, J.; Gałązka, A. Silica/Lignin Carrier as a Factor Increasing the Process Performance and Genetic Diversity of Microbial Communities in Laboratory-Scale Anaerobic Digesters. Energies 2021, 14, 4429. https://doi.org/10.3390/en14154429
A. Pilarska A, Wolna-Maruwka A, Niewiadomska A, Pilarski K, Adamski M, Grzyb A, Grządziel J, Gałązka A. Silica/Lignin Carrier as a Factor Increasing the Process Performance and Genetic Diversity of Microbial Communities in Laboratory-Scale Anaerobic Digesters. Energies. 2021; 14(15):4429. https://doi.org/10.3390/en14154429
Chicago/Turabian StyleA. Pilarska, Agnieszka, Agnieszka Wolna-Maruwka, Alicja Niewiadomska, Krzysztof Pilarski, Mariusz Adamski, Aleksandra Grzyb, Jarosław Grządziel, and Anna Gałązka. 2021. "Silica/Lignin Carrier as a Factor Increasing the Process Performance and Genetic Diversity of Microbial Communities in Laboratory-Scale Anaerobic Digesters" Energies 14, no. 15: 4429. https://doi.org/10.3390/en14154429
APA StyleA. Pilarska, A., Wolna-Maruwka, A., Niewiadomska, A., Pilarski, K., Adamski, M., Grzyb, A., Grządziel, J., & Gałązka, A. (2021). Silica/Lignin Carrier as a Factor Increasing the Process Performance and Genetic Diversity of Microbial Communities in Laboratory-Scale Anaerobic Digesters. Energies, 14(15), 4429. https://doi.org/10.3390/en14154429










