dNAGLU Extends Life Span and Promotes Fitness and Stress Resistance in Drosophila
Abstract
1. Introduction
2. Results
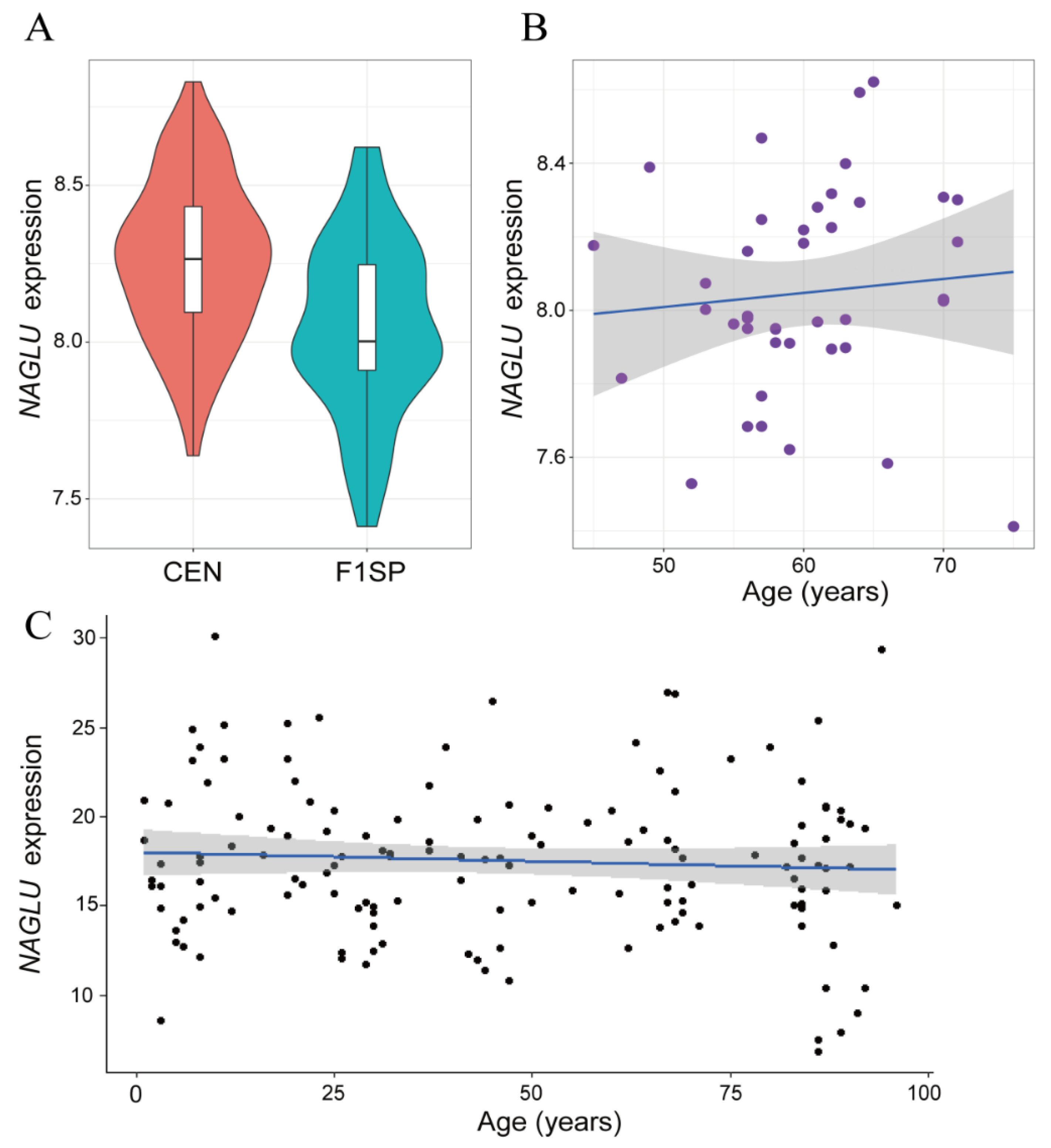
2.1. NAGLU Is Upregulated in Centenarians
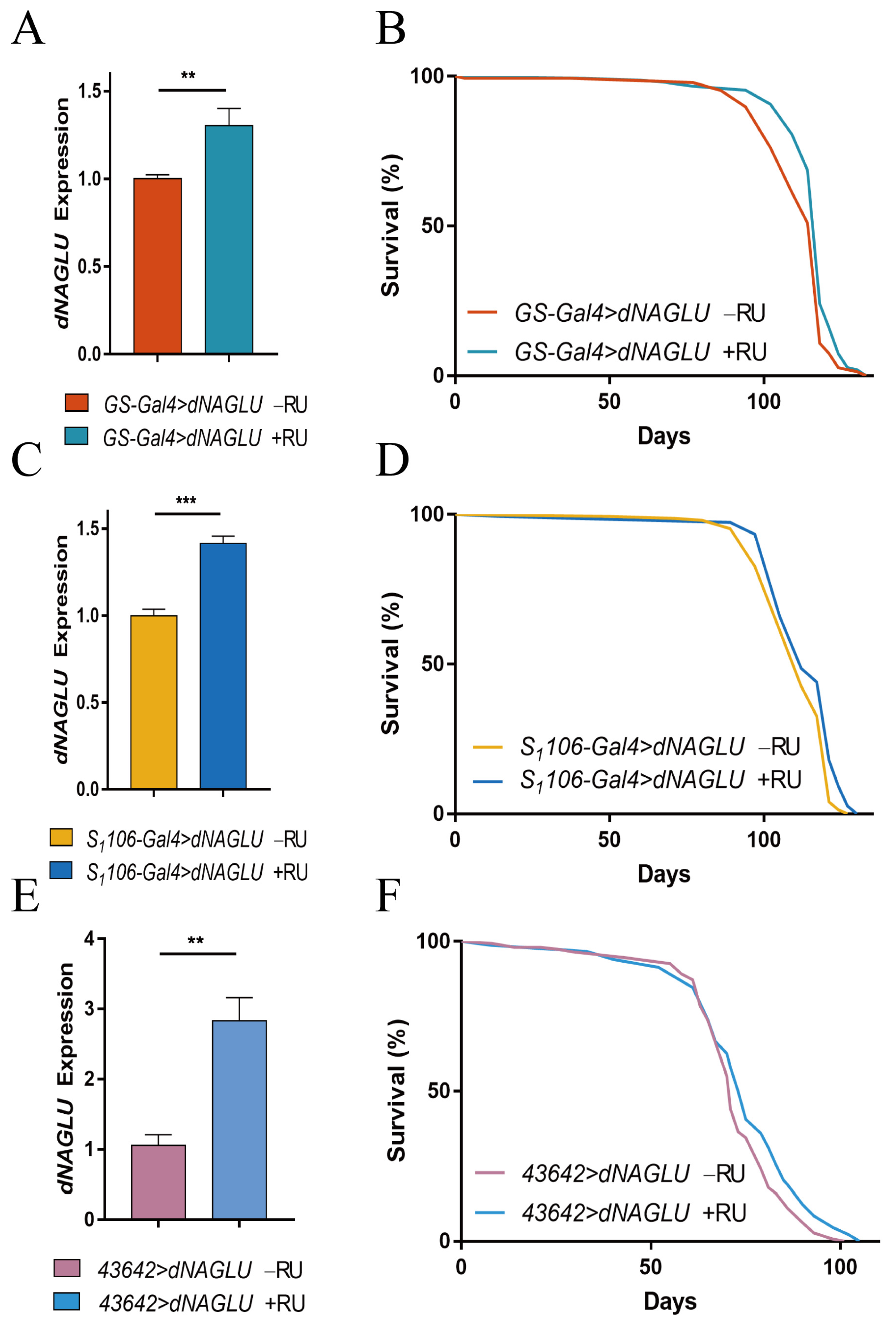
2.2. Overexpression of dNAGLU Extends Drosophila Life Span
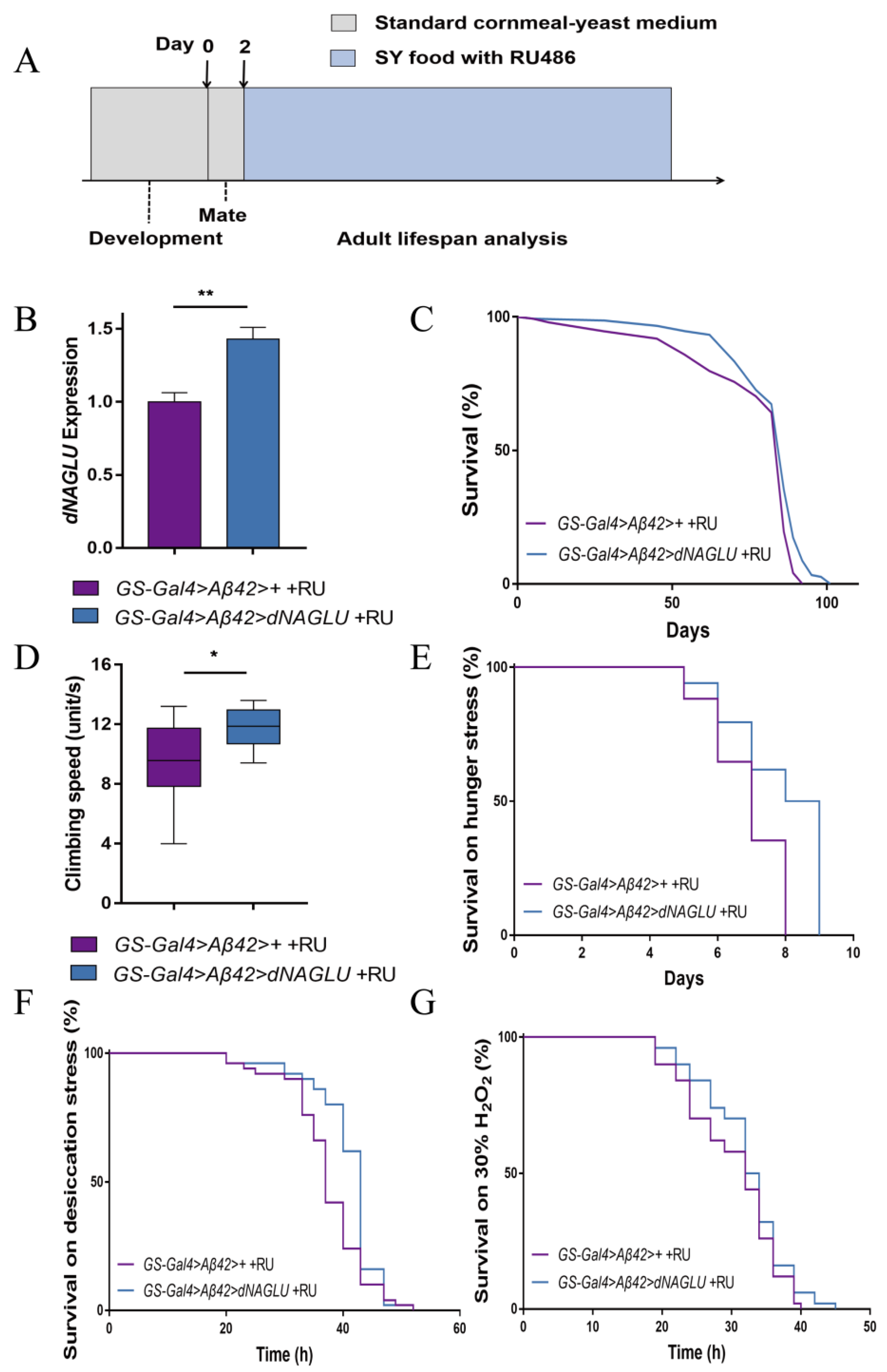
2.3. Overexpression of dNAGLU Extends Life Span and Health Span in the Drosophila AD Model
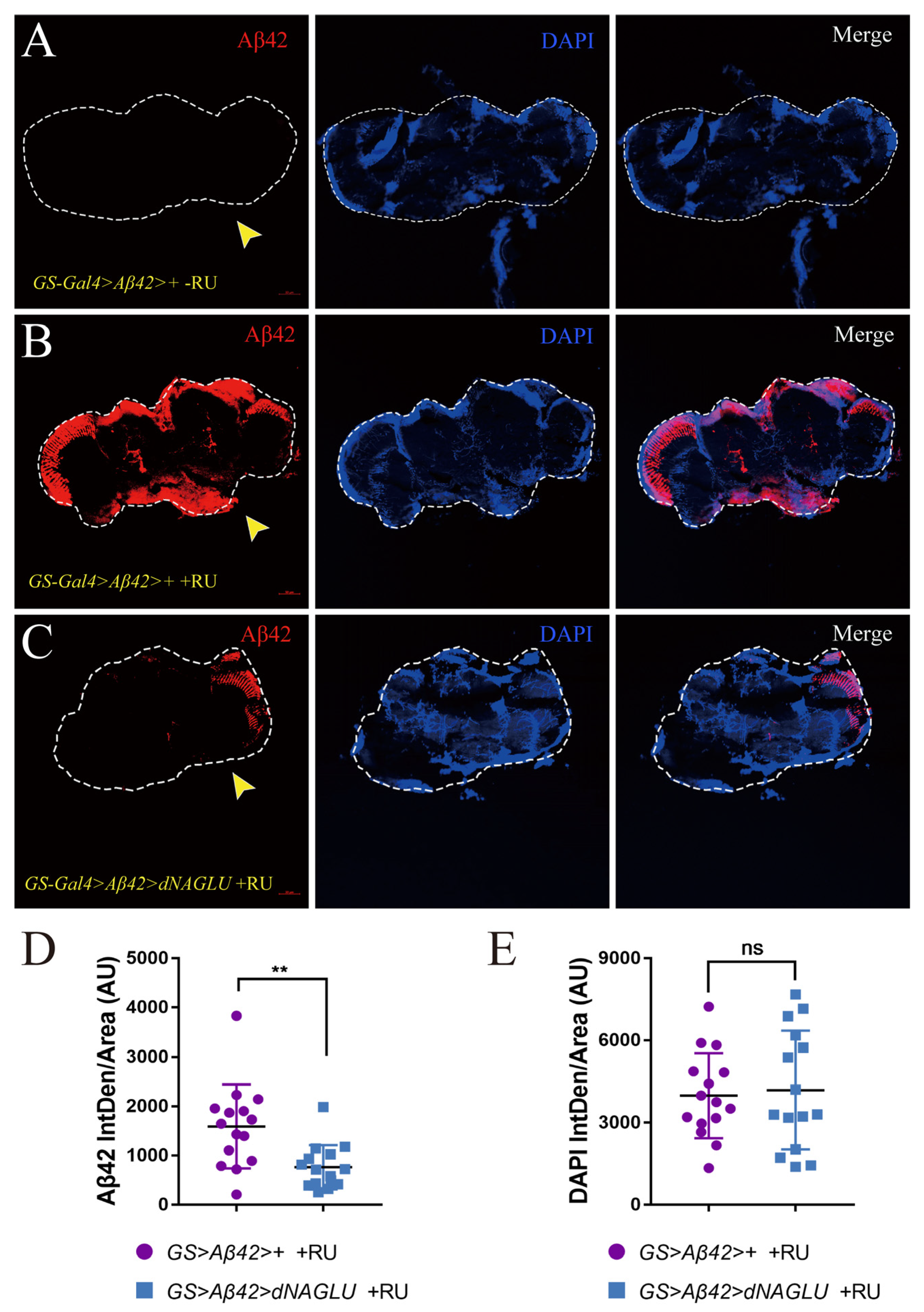
2.4. Overexpression of dNAGLU Reduces Aβ42 Deposition in the Mushroom Body in AD Drosophila
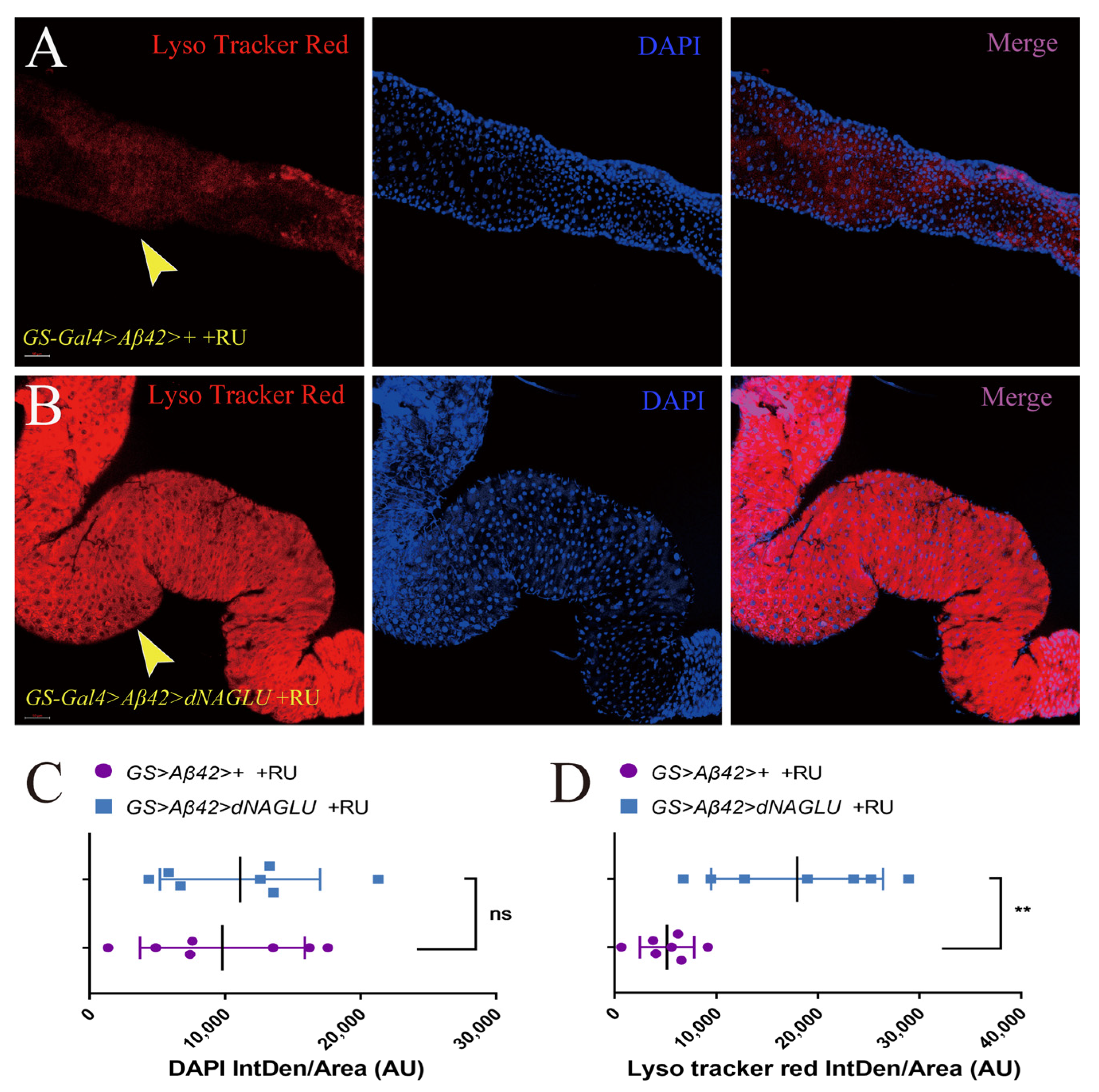
2.5. Overexpression of dNAGLU in AD Drosophila Enhances the Intestinal Lysosomal Activity
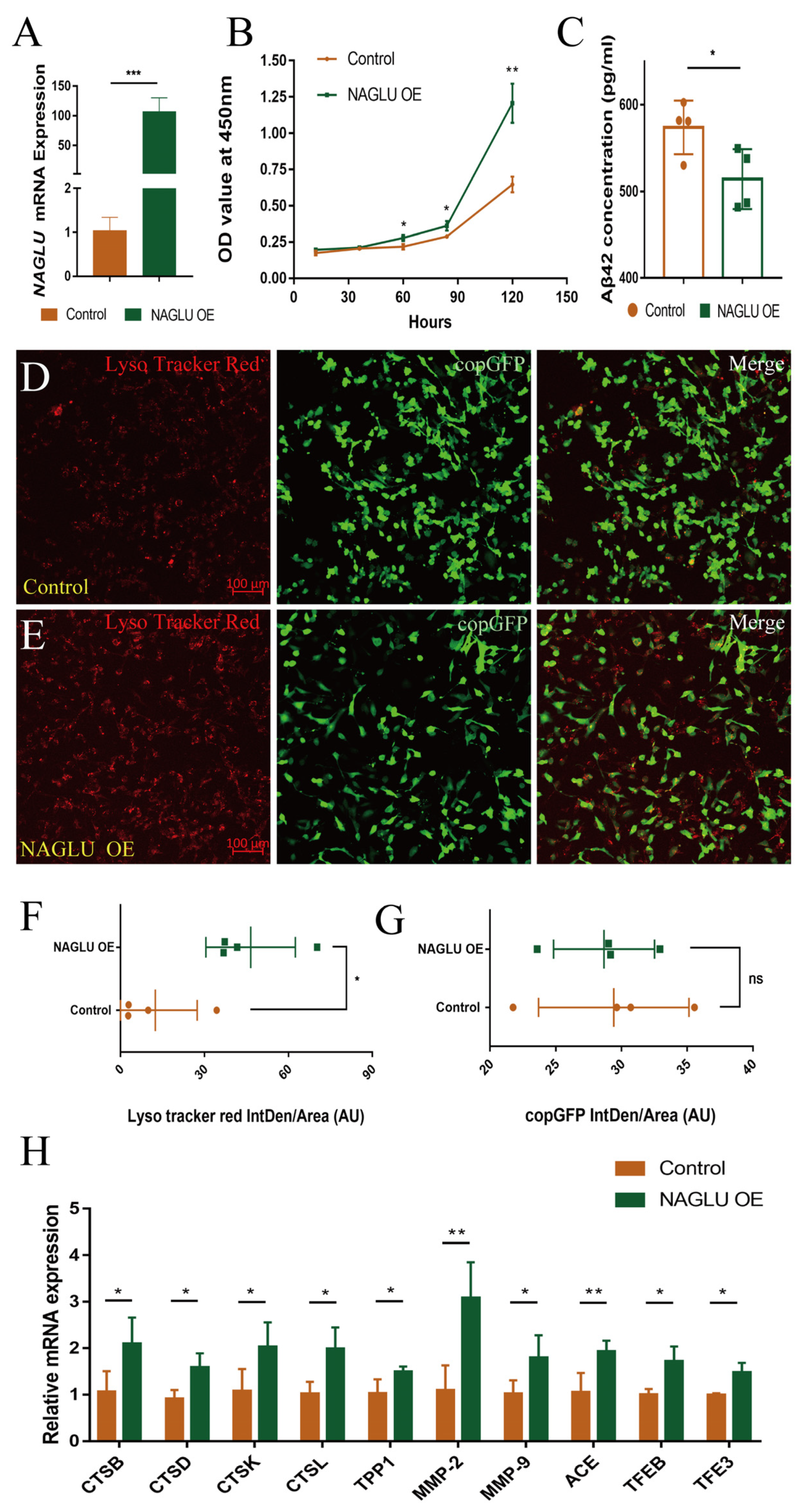
2.6. Overexpression of NAGLU Reduces Aβ42 Concentration in Human U251-APP Cells
3. Discussion
4. Materials and Methods
4.1. Drosophila Stocks and Construction of the Transgenic Flies
4.2. Husbandry and Life Span Analysis
4.3. Health Test Assays
4.4. Immunofluorescence
4.5. Autolysosome Staining
4.6. qRT-PCR
4.7. Western Blot
4.8. NAGLU Expression Analysis
4.9. Cells and Transfection
4.10. Cell Proliferation Analysis
4.11. Aβ42 ELISA Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Herskovits, A.Z.; Guarente, L. Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res. 2013, 23, 746–758. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Crews, L.; Masliah, E. Molecular mechanisms of neurodegeneration in Alzheimer’s disease. Hum. Mol. Genet. 2010, 19, R12–R20. [Google Scholar] [CrossRef]
- Hashimoto, S.; Saido, T.C. Critical review: Involvement of endoplasmic reticulum stress in the aetiology of Alzheimer’s disease. Open Biol. 2018, 8, 180024. [Google Scholar] [CrossRef]
- De Ture, M.A.; Dickson, D.W. The neuropathological diagnosis of Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 32. [Google Scholar] [CrossRef]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease_progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef]
- Mawuenyega, K.G.; Sigurdson, W.; Ovod, V.; Munsell, L.; Kasten, T.; Morris, J.C.; Yarasheski, K.E.; Bateman, R.J. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science 2010, 330, 1774. [Google Scholar] [CrossRef]
- Hitt, R.; Young-Xu, Y.; Silver, M.; Perls, T. Centenarians: The older you get, the healthier you have been. Lancet 1999, 354, 652. [Google Scholar] [CrossRef]
- Evert, J.; Lawler, E.; Bogan, H.; Perls, T. Morbidity profiles of centenarians: Survivors, delayers, and escapers. J. Gerontol. Ser. A 2003, 58, M232–M237. [Google Scholar] [CrossRef]
- Franceschi, C.; Bonafè, M. Centenarians as a model for healthy aging. Biochem. Soc. Trans. 2003, 31, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Engberg, H.; Oksuzyan, A.; Jeune, B.; Vaupel, J.W.; Christensen, K. Centenarians—A useful model for healthy aging? A 29-year follow-up of hospitalizations among 40,000 Danes born in 1905. Aging Cell 2009, 8, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.H.; Chen, X.Q.; Yu, Q.; Ye, Y.; Liu, Y.W.; Yan, D.; Yang, L.Q.; Chen, G.; Lin, R.; Yang, L.; et al. Transcriptome evidence reveals enhanced autophagy-lysosomal function in centenarians. Genome Res. 2018, 28, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Birrane, G.; Dassier, A.L.; Romashko, A.; Lundberg, D.; Holmes, K.; Cottle, T.; Norton, A.W.; Zhang, B.; Concino, M.F.; Meiyappan, M. Structural characterization of the alpha-N-acetylglucosaminidase, a key enzyme in the pathogenesis of Sanfilippo syndrome B. J. Struct. Biol. 2019, 205, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Meijer, O.; Brinke, H.T.; Ofman, R.; Ijlst, L.; Wijburg, F.; van Vlies, N. Processing of mutant N -acetyl-α-glucosaminidase in mucopolysaccharidosis type IIIB fibroblasts cultured at low temperature. Mol. Genet. Metab. 2017, 122, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Khorrami, M.; Mahdavi, M.; Fakhr, F.; Kheirollahi, M. A novel pathogenic variant in NAGLU (N-Acetyl-Alpha-Glucosaminidase) gene identified by targeted next-generation sequencing followed by in silico analysis. Iran. J. Child Neurol. 2019, 13, 173–183. [Google Scholar]
- Zhao, H.G.; Li, H.H.; Bach, G.; Schmidtchen, A.; Neufeld, E.F. The molecular basis of Sanfilippo syndrome type B. Proc. Natl. Acad. Sci. USA 1996, 93, 6101–6105. [Google Scholar] [CrossRef]
- Wang, M.C.; O’Rourke, E.J.; Ruvkun, G. Fat metabolism links germline stem cells and longevity in C. elegans. Science 2008, 322, 957–960. [Google Scholar] [CrossRef]
- Edwards, C.; Canfield, J.; Copes, N.; Rehan, M.; Lipps, D.; Bradshaw, P.C. D-beta-hydroxybutyrate extends lifespan in C. elegans. Aging 2014, 6, 621–644. [Google Scholar] [CrossRef]
- Wu, Q.; Yu, G.; Cheng, X.; Gao, Y.; Fan, X.; Yang, D.; Xie, M.; Wang, T.; Piper, M.D.W.; Yang, M. Sexual dimorphism in the nutritional requirement for adult lifespan in Drosophila melanogaster. Aging Cell 2020, 19, e13120. [Google Scholar] [CrossRef]
- Fan, X.L.; Zeng, Y.; Fan, Z.Q.; Cui, L.; Song, W.H.; Wu, Q.; Gao, Y. Dihydromyricetin promotes longevity and activates the transcription factors FOXO and AOP in Drosophila. Aging 2020, 13, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.W.; Zhao, X.; De Cecco, M.; Peterson, A.L.; Pagliaroli, L.; Manivannan, J.; Hubbard, G.B.; Ikeno, Y.; Zhang, Y.; Feng, B.; et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell 2015, 160, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.N.; Wallace, M.A.; Tomilov, A.A.; Zhou, Z.; Marcotte, G.R.; Tran, D.; Perez, G.; Gutierrez-Casado, E.; Koike, S.; Knotts, T.A.; et al. A ketogenic diet extends longevity and healthspan in adult mice. Cell Metab. 2017, 26, 539–546. [Google Scholar] [CrossRef]
- Alliance of Genome Resources. Available online: https://www.alliancegenome.org/gene/FB:FBgn0014417 (accessed on 1 July 2022).
- Scialo, F.; Sriram, A.; Stefanatos, R.; Sanz, A. Practical recommendations for the use of the GeneSwitch Gal4 system to knock-down genes in Drosophila melanogaster. PLoS ONE 2016, 11, e0161817. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef]
- Bonner, J.M.; Boulianne, G.L. Drosophila as a model to study age-related neurodegenerative disorders: Alzheimer’s disease. Exp. Gerontol. 2011, 46, 335–339. [Google Scholar] [CrossRef]
- Fleischer, J.G.; Schulte, R.; Tsai, H.H.; Tyagi, S.; Ibarra, A.; Shokhirev, M.N.; Huang, L.; Hetzer, M.W.; Navlakha, S. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 2018, 19, 221. [Google Scholar] [CrossRef]
- Arrese, E.L.; Soulages, J.L. Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef]
- Omelyanchuk, L.V.; Shaposhnikov, M.V.; Moskalev, A.A. Drosophila nervous system as a target of aging and anti-aging interventions. Front. Genet. 2015, 6, 89. [Google Scholar] [CrossRef][Green Version]
- Demontis, F.; Piccirillo, R.; Goldberg, A.L.; Perrimon, N. The influence of skeletal muscle on systemic aging and lifespan. Aging Cell 2013, 12, 943–949. [Google Scholar] [CrossRef]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef]
- Cuervo, A.M. Autophagy and aging: Keeping that old broom working. Trends Genet. 2008, 24, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, N.; Huang, X.; Hu, W.; Ma, Y.; Liang, Y.; Xie, W.; Tang, M. Atmospheric particulate matter impedes autophagic flux by impairing lysosomal milieu and integrity in human umbilical vein endothelial cells (HUVECs). Sci. Total Environ. 2021, 761, 143290. [Google Scholar] [CrossRef]
- Luo, R.; Su, L.Y.; Li, G.; Yang, J.; Liu, Q.; Yang, L.X.; Zhang, D.F.; Zhou, H.; Xu, M.; Fan, Y.; et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 2019, 16, 52–69. [Google Scholar] [CrossRef]
- Van Acker, Z.P.; Perdok, A.; Bretou, M.; Annaert, W. The microglial lysosomal system in Alzheimer’s disease: Guardian against proteinopathy. Ageing Res. Rev. 2021, 71, 101444. [Google Scholar] [CrossRef]
- Song, J.X.; Malampati, S.; Zeng, Y.; Durairajan, S.S.K.; Yang, C.B.; Tong, B.C.; Iyaswamy, A.; Shang, W.B.; Sreenivasmurthy, S.G.; Zhu, Z.; et al. A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and Tau pathology in Alzheimer’s disease models. Aging Cell 2020, 19, e13069. [Google Scholar] [CrossRef]
- Chandra, S.; Roy, A.; Patel, D.R.; Pahan, K. PPAR alpha between aspirin and plaque clearance. J. Alzheimer’s Dis. 2019, 71, 389–397. [Google Scholar] [CrossRef]
- Triolo, M.; Hood, D.A. Manifestations of age on autophagy, mitophagy and lysosomes in skeletal muscle. Cells 2021, 10, 1054. [Google Scholar] [CrossRef]
- Kuma, A.; Hatano, M.; Matsui, M.; Yamamoto, A.; Nakaya, H.; Yoshimori, T.; Ohsumi, Y.; Tokuhisa, T.; Mizushima, N. The role of autophagy during the early neonatal starvation period. Nature 2004, 432, 1032–1036. [Google Scholar] [CrossRef]
- Kan, S.H.; Aoyagi-Scharber, M.; Le, S.Q.; Vincelette, J.; Ohmi, K.; Bullens, S.; Wendt, D.J.; Christianson, T.M.; Tiger, P.M.; Brown, J.R.; et al. Delivery of an enzyme-IGFII fusion protein to the mouse brain is therapeutic for mucopolysaccharidosis type IIIB. Proc. Natl. Acad. Sci. USA 2014, 111, 14870–14875. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Lin, W.; Jiang, Y.; Lu, K.; Wei, W.; Huo, Q.; Cui, S.; Yang, X.; Li, M.; Xu, N.; et al. Electroacupuncture ameliorates beta-amyloid pathology and cognitive impairment in Alzheimer disease via a novel mechanism involving activation of TFEB (transcription factor EB). Autophagy 2021, 17, 3833–3847. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, M.; Zérah, M.; Gougeon, M.-L.; Ausseil, J.; de Bournonville, S.; Husson, B.; Zafeiriou, D.; Parenti, G.; Bourget, P.; Poirier, B.; et al. Intracerebral gene therapy in children with mucopolysaccharidosis type IIIB syndrome: An uncontrolled phase 1/2 clinical trial. Lancet Neurol. 2017, 16, 712–720. [Google Scholar] [CrossRef]
- Gougeon, M.L.; Poirier-Beaudouin, B.; Ausseil, J.; Zerah, M.; Artaud, C.; Heard, J.M.; Deiva, K.; Tardieu, M. Cell-mediated immunity to NAGLU transgene following intracerebral gene therapy in children with mucopolysaccharidosis type IIIB syndrome. Front. Immunol. 2021, 12, 655478. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Zuo, Y.; Kwan, K.M.; Liang, Y.; Ma, K.Y.; Chan, H.Y.; Huang, Y.; Yu, H.; Chen, Z.Y. Blueberry extract prolongs lifespan of Drosophila melanogaster. Exp. Gerontol. 2012, 47, 170–178. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, R.; Yang, K.; Xiao, F.; Yang, L.; Chen, G.; Li, Y.; Ye, Y.; Chen, K.; Smith, S.T.; Li, G.; et al. dNAGLU Extends Life Span and Promotes Fitness and Stress Resistance in Drosophila. Int. J. Mol. Sci. 2022, 23, 14433. https://doi.org/10.3390/ijms232214433
Xue R, Yang K, Xiao F, Yang L, Chen G, Li Y, Ye Y, Chen K, Smith ST, Li G, et al. dNAGLU Extends Life Span and Promotes Fitness and Stress Resistance in Drosophila. International Journal of Molecular Sciences. 2022; 23(22):14433. https://doi.org/10.3390/ijms232214433
Chicago/Turabian StyleXue, Rubing, Ke Yang, Fuhui Xiao, Liping Yang, Guijun Chen, Yongxuan Li, Yunshuang Ye, Kangning Chen, Sheryl T. Smith, Gonghua Li, and et al. 2022. "dNAGLU Extends Life Span and Promotes Fitness and Stress Resistance in Drosophila" International Journal of Molecular Sciences 23, no. 22: 14433. https://doi.org/10.3390/ijms232214433
APA StyleXue, R., Yang, K., Xiao, F., Yang, L., Chen, G., Li, Y., Ye, Y., Chen, K., Smith, S. T., Li, G., Kong, Q., & Zhou, J. (2022). dNAGLU Extends Life Span and Promotes Fitness and Stress Resistance in Drosophila. International Journal of Molecular Sciences, 23(22), 14433. https://doi.org/10.3390/ijms232214433





