Ecological Aerobic Ammonia and Methane Oxidation Involved Key Metal Compounds, Fe and Cu
Abstract
1. Introduction
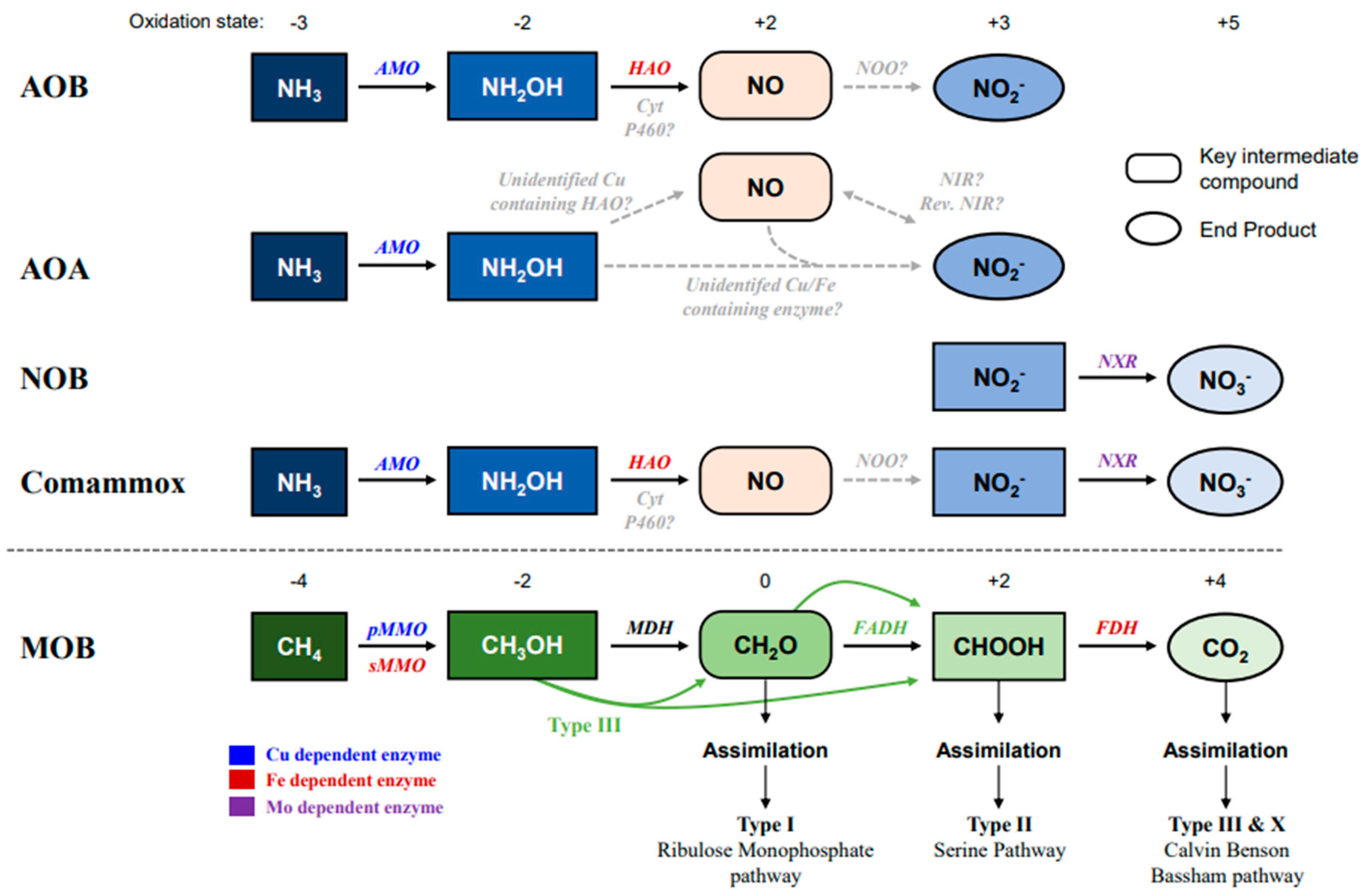
2. Ammonia and Methane Oxidation Pathways Mediated by Metal Compounds
2.1. Copper and Iron in the Ammonia-Oxidizing Pathway
2.2. Copper and Iron in the Methane-Oxidizing Pathways
2.3. Ammonia Oxidation by MOB and Methane Oxidation by AOM
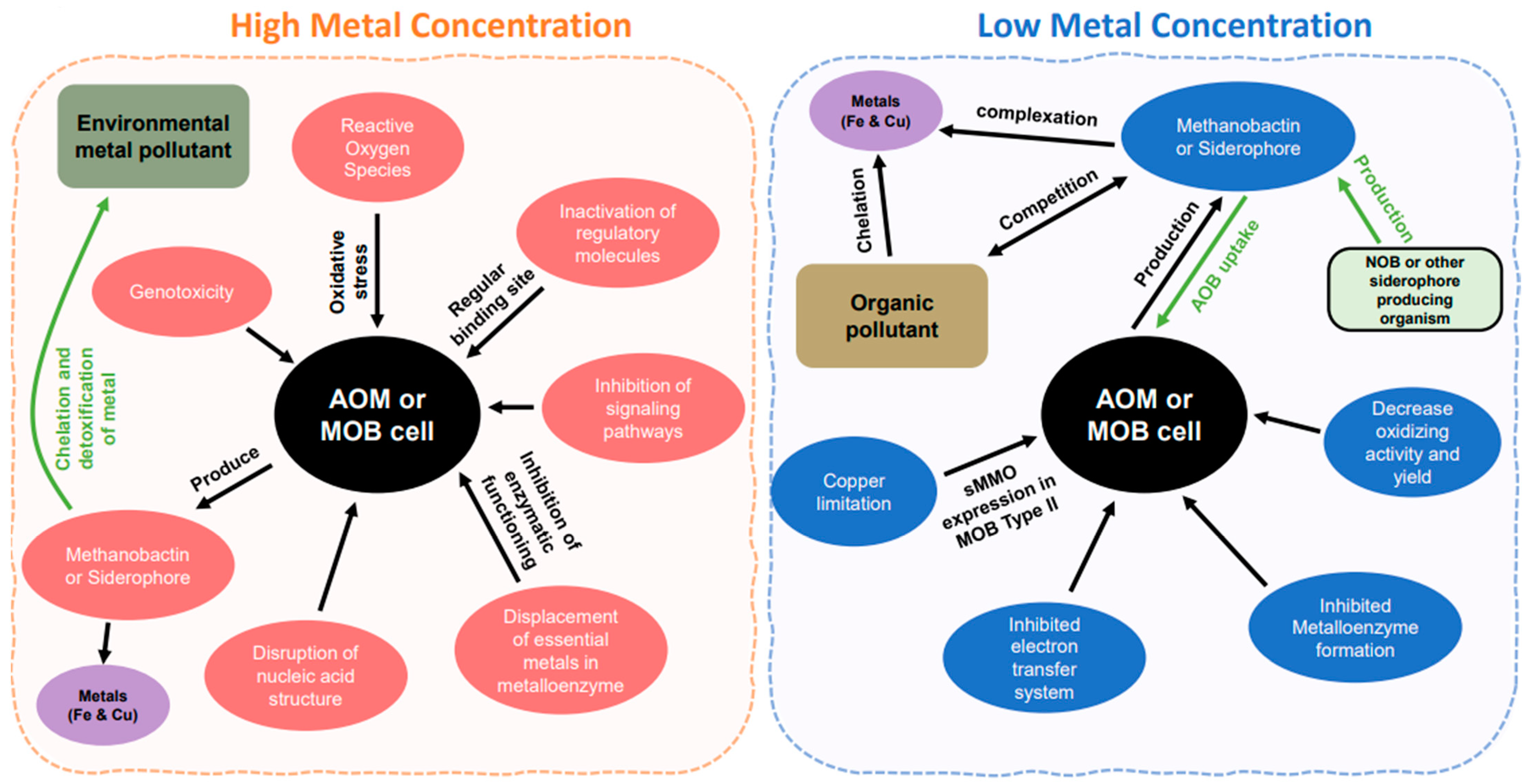
2.4. Environmental Pollutants with Ammonia and Methane Oxidizer
3. Ammonia and Methane Oxidizer Reactions with Metal in Elevated Metal Concentrations
3.1. High Concentrations of Metals
3.2. Low Concentrations of Metals
4. Chalkophores and Siderophore
5. Conclusions and Future Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| AOM | Ammonia-oxidizing microorganisms |
| MOB | Methane-oxidizing microorganism |
| Comammox | Complete ammonia-oxidizing bacteria |
| NOB | Nitrite-oxidizing bacteria |
| Fe | Iron |
| Cu | Copper |
| CH4 | Methane |
| NH4+ | Ammonium |
| NH3 | Ammonia |
| AMO | Ammonia monooxygenase |
| HAO | Hydroxylamine oxidoreductase |
| pMMO | Partial membrane monooxygenase |
| sMMO | soluble membrane monooxygenase |
References
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Montzka, S.A.; Dlugokencky, E.J.; Butler, J.H. Non-CO2 greenhouse gases and climate change. Nature 2011, 476, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Strous, M.; Kuenen, J.G.; Jetten, M.S. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 1999, 65, 3248–3250. [Google Scholar] [CrossRef] [PubMed]
- Martikainen, P.J. Heterotrophic nitrification–An eternal mystery in the nitrogen cycle. Soil Biol. Biochem. 2022, 168, 108611. [Google Scholar] [CrossRef]
- Smith, K. Changing views of nitrous oxide emissions from agricultural soil: Key controlling processes and assessment at different spatial scales. Eur. J. Soil Sci. 2017, 68, 137–155. [Google Scholar] [CrossRef]
- Thauer, R.K. Anaerobic oxidation of methane with sulfate: On the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr. Opin. Microbiol. 2011, 14, 292–299. [Google Scholar] [CrossRef]
- Stein, L.Y. Methane Oxidation. In Encyclopedia of Astrobiology; Gargaud, M., Irvine, W.M., Amils, R., Cleaves, H.J., Pinti, D., Cernicharo Quintanilla, J., Viso, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–4. [Google Scholar]
- Scheller, S.; Goenrich, M.; Boecher, R.; Thauer, R.K.; Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 2010, 465, 606–608. [Google Scholar] [CrossRef]
- Scheller, S.; Yu, H.; Chadwick, G.L.; McGlynn, S.E.; Orphan, V.J. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 2016, 351, 703–707. [Google Scholar] [CrossRef]
- Fashola, M.O.; Ngole-Jeme, V.M.; Babalola, O.O. Heavy Metal Pollution from Gold Mines: Environmental Effects and Bacterial Strategies for Resistance. Int. J. Environ. Res. Public Health 2016, 13, 1047. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Ehrlich, H. Microbes and metals. Appl. Microbiol. Biotechnol. 1997, 48, 687–692. [Google Scholar] [CrossRef]
- Miličević, A.; Branica, G.; Raos, N. Irving-Williams order in the framework of connectivity index 3 χv enables simultaneous prediction of stability constants of bivalent transition metal complexes. Molecules 2011, 16, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Morel, F.M.; Hering, J.G. Principles and Applications of Aquatic Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 1993. [Google Scholar]
- Buffle, J.; Wilkinson, K.J.; Van Leeuwen, H.P. Chemodynamics and bioavailability in natural waters. Environ. Sci. Technol. 2009, 43, 7170–7174. [Google Scholar] [CrossRef]
- Buffle, J.; Altmann, R.S.; Filella, M.; Tessier, A. Complexation by natural heterogeneous compounds: Site occupation distribution functions, a normalized description of metal complexation. Geochim. Cosmochim. Acta 1990, 54, 1535–1553. [Google Scholar] [CrossRef]
- Zhao, L.; Qiu, G.; Anderson, C.W.N.; Meng, B.; Wang, D.; Shang, L.; Yan, H.; Feng, X. Mercury methylation in rice paddies and its possible controlling factors in the Hg mining area, Guizhou province, Southwest China. Environ. Pollut. 2016, 215, 1–9. [Google Scholar] [CrossRef]
- Anderson, R.; Vinikour, W.; Brower, J. The distribution of Cd, Cu, Pb and Zn in the biota of two freshwater sites with different trace metal inputs. Ecography 1978, 1, 377–384. [Google Scholar] [CrossRef]
- Bjerregaard, P.; Andersen, C.B.; Andersen, O. Ecotoxicology of metals—sources, transport, and effects on the ecosystem. In Handbook on the Toxicology of Metals; Elsevier: Amsterdam, The Netherlands, 2022; pp. 593–627. [Google Scholar]
- Mohammadi, R.; Shahrokhian, S. In-situ fabrication of nanosheet arrays on copper foil as a new substrate for binder-free high-performance electrochemical supercapacitors. J. Electroanal. Chem. 2017, 802, 48–56. [Google Scholar] [CrossRef]
- Xia, X.; Gao, Y. Methane from microbial hydrogenolysis of sediment organic matter before the great oxidation event. Nat. Commun. 2021, 12, 5032. [Google Scholar] [CrossRef]
- Daebeler, A.; Bodelier, P.L.; Yan, Z.; Hefting, M.M.; Jia, Z.; Laanbroek, H.J. Interactions between Thaumarchaea, Nitrospira and methanotrophs modulate autotrophic nitrification in volcanic grassland soil. ISME J. 2014, 8, 2397–2410. [Google Scholar] [CrossRef]
- Stein, L.Y.; Roy, R.; Dunfield, P.F. Aerobic methanotrophy and nitrification: Processes and connections. eLS 2012. [Google Scholar] [CrossRef]
- Zheng, Y.; Huang, R.; Wang, B.; Bodelier, P.; Jia, Z. Competitive interactions between methane-and ammonia-oxidizing bacteria modulate carbon and nitrogen cycling in paddy soil. Biogeosciences 2014, 11, 3353–3368. [Google Scholar] [CrossRef]
- Srivastava, P.; Kowshik, M. Mechanisms of metal resistance and homeostasis in haloarchaea. Archaea 2013, 2013, 732864. [Google Scholar] [CrossRef]
- Shafiee, R.T.; Snow, J.T.; Zhang, Q.; Rickaby, R.E.M. Iron requirements and uptake strategies of the globally abundant marine ammonia-oxidising archaeon, Nitrosopumilus maritimus SCM1. ISME J. 2019, 13, 2295–2305. [Google Scholar] [CrossRef]
- Klotz, M.G.; Stein, L.Y. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett. 2008, 278, 146–156. [Google Scholar] [CrossRef]
- Kerou, M.; Offre, P.; Valledor, L.; Abby, S.S.; Melcher, M.; Nagler, M.; Weckwerth, W.; Schleper, C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. USA 2016, 113, E7937–E7946. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.B.; de la Torre, J.R.; Klotz, M.G.; Urakawa, H.; Pinel, N.; Arp, D.J.; Brochier-Armanet, C.; Chain, P.S.; Chan, P.P.; Gollabgir, A.; et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8818–8823. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.Y.; Kim, J.G.; Sinninghe Damste, J.S.; Rijpstra, W.I.; Madsen, E.L.; Kim, S.J.; Hong, H.; Si, O.J.; Kerou, M.; Schleper, C.; et al. A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environ. Microbiol. Rep. 2016, 8, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Amin, S.A.; Lundeen, R.A.; Heal, K.R.; Martens-Habbena, W.; Turkarslan, S.; Urakawa, H.; Costa, K.C.; Hendrickson, E.L.; Wang, T.; et al. Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 2018, 12, 508–519. [Google Scholar] [CrossRef]
- Santoro, A.E.; Dupont, C.L.; Richter, R.A.; Craig, M.T.; Carini, P.; McIlvin, M.R.; Yang, Y.; Orsi, W.D.; Moran, D.M.; Saito, M.A. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: An ammonia-oxidizing archaeon from the open ocean. Proc. Natl. Acad. Sci. USA 2015, 112, 1173–1178. [Google Scholar] [CrossRef]
- Gorman-Lewis, D.; Martens-Habbena, W.; Stahl, D.A. Cu (II) adsorption onto ammonia-oxidizing bacteria and archaea. Geochim. Cosmochim. Acta 2019, 255, 127–143. [Google Scholar] [CrossRef]
- Reyes, C.; Hodgskiss, L.H.; Kerou, M.; Pribasnig, T.; Abby, S.S.; Bayer, B.; Kraemer, S.M.; Schleper, C. Genome wide transcriptomic analysis of the soil ammonia oxidizing archaeon Nitrososphaera viennensis upon exposure to copper limitation. ISME J. 2020, 14, 2659–2674. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Vajrala, N.; Hauser, L.; Sayavedra-Soto, L.A.; Arp, D.J. Iron nutrition and physiological responses to iron stress in Nitrosomonas europaea. Arch. Microbiol. 2006, 186, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Morel, F.M.; Price, N.M. The biogeochemical cycles of trace metals in the oceans. Science 2003, 300, 944–947. [Google Scholar] [CrossRef] [PubMed]
- Whitby, H.; Posacka, A.M.; Maldonado, M.T.; Van Den Berg, C.M. Copper-binding ligands in the NE Pacific. Mar. Chem. 2018, 204, 36–48. [Google Scholar] [CrossRef]
- Hoffmann, S.R.; Shafer, M.M.; Armstrong, D.E. Strong colloidal and dissolved organic ligands binding copper and zinc in rivers. Environ. Sci. Technol. 2007, 41, 6996–7002. [Google Scholar] [CrossRef]
- Gwak, J.H.; Jung, M.Y.; Hong, H.; Kim, J.G.; Quan, Z.X.; Reinfelder, J.R.; Spasov, E.; Neufeld, J.D.; Wagner, M.; Rhee, S.K. Archaeal nitrification is constrained by copper complexation with organic matter in municipal wastewater treatment plants. ISME J. 2020, 14, 335–346. [Google Scholar] [CrossRef]
- Abendroth, J.; Buchko, G.W.; Liew, F.N.; Nguyen, J.N.; Kim, H.J. Structural Characterization of Cytochrome c’ β-Met from an Ammonia-Oxidizing Bacterium. Biochemistry 2022, 61, 563–574. [Google Scholar] [CrossRef]
- Kim, H.J.; Zatsman, A.; Upadhyay, A.K.; Whittaker, M.; Bergmann, D.; Hendrich, M.P.; Hooper, A.B. Membrane tetraheme cytochrome c(m552) of the ammonia-oxidizing Nitrosomonas europaea: A ubiquinone reductase. Biochemistry 2008, 47, 6539–6551. [Google Scholar] [CrossRef][Green Version]
- Shafiee, R.T.; Diver, P.J.; Snow, J.T.; Zhang, Q.; Rickaby, R.E. Marine ammonia-oxidising archaea and bacteria occupy distinct iron and copper niches. ISME Commun. 2021, 1, 1. [Google Scholar] [CrossRef]
- Palomo, A.; Pedersen, A.G.; Fowler, S.J.; Dechesne, A.; Sicheritz-Ponten, T.; Smets, B.F. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 2018, 12, 1779–1793. [Google Scholar] [CrossRef]
- Feissner, R.E.; Richard-Fogal, C.L.; Frawley, E.R.; Loughman, J.A.; Earley, K.W.; Kranz, R.G. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol. Microbiol. 2006, 60, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Glass, J.B.; Orphan, V.J. Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol. 2012, 3, 61. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lücker, S.; Wagner, M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Koike, K.; Smith, G.J.; Yamamoto-Ikemoto, R.; Lucker, S.; Matsuura, N. Distinct comammox Nitrospira catalyze ammonia oxidation in a full-scale groundwater treatment bioreactor under copper limited conditions. Water Res. 2022, 210, 117986. [Google Scholar] [CrossRef] [PubMed]
- Strous, M.; Fuerst, J.A.; Kramer, E.H.; Logemann, S.; Muyzer, G.; van de Pas-Schoonen, K.T.; Webb, R.; Kuenen, J.G.; Jetten, M.S. Missing lithotroph identified as new planctomycete. Nature 1999, 400, 446–449. [Google Scholar] [CrossRef]
- Van De Graaf, A.A.; De Bruijn, P.; Robertson, L.A.; Jetten, M.S.; Kuenen, J.G. Metabolic pathway of anaerobic ammonium oxidation on the basis of 15N studies in a fluidized bed reactor. Microbiology 1997, 143, 2415–2421. [Google Scholar] [CrossRef]
- Ni, S.-Q.; Zhang, J. Anaerobic ammonium oxidation: From laboratory to full-scale application. Biomed Res. Int. 2013, 2013, 469360. [Google Scholar] [CrossRef]
- Wang, H.; Yu, G.; He, W.; Du, C.; Deng, Z.; Wang, D.; Yang, M.; Yang, E.; Zhou, Y.; Sanjaya, E.H. Enhancing autotrophic nitrogen removal with a novel dissolved oxygen-differentiated airlift internal circulation reactor: Long-term operational performance and microbial characteristics. J. Environ. Manag. 2021, 296, 113271. [Google Scholar] [CrossRef]
- Chen, H.; Tu, Z.; Wu, S.; Yu, G.; Du, C.; Wang, H.; Yang, E.; Zhou, L.; Deng, B.; Wang, D. Recent advances in partial denitrification-anaerobic ammonium oxidation process for mainstream municipal wastewater treatment. Chemosphere 2021, 278, 130436. [Google Scholar] [CrossRef]
- Zekker, I.; Raudkivi, M.; Artemchuk, O.; Rikmann, E.; Priks, H.; Jaagura, M.; Tenno, T. Mainstream-sidestream wastewater switching promotes anammox nitrogen removal rate in organic-rich, low-temperature streams. Environ. Technol. 2021, 42, 3073–3082. [Google Scholar] [CrossRef]
- Cui, M.; Ma, A.; Qi, H.; Zhuang, X.; Zhuang, G. Anaerobic oxidation of methane: An “active” microbial process. MicrobiologyOpen 2015, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.-D.; Wu, H.-S.; Gao, Z.-Q. Distribution and environmental significance of nitrite-dependent anaerobic methane-oxidising bacteria in natural ecosystems. Appl. Microbiol. Biotechnol. 2015, 99, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Versantvoort, W.; Guerrero-Cruz, S.; Speth, D.R.; Frank, J.; Gambelli, L.; Cremers, G.; van Alen, T.; Jetten, M.S.M.; Kartal, B.; Op den Camp, H.J.M.; et al. Comparative Genomics of Candidatus Methylomirabilis Species and Description of Ca. Methylomirabilis Lanthanidiphila. Front. Microbiol. 2018, 9, 1672. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, K.K.; Goswami, G.; Das, D. Biotransformation of Methane and Carbon Dioxide Into High-Value Products by Methanotrophs: Current State of Art and Future Prospects. Front. Microbiol. 2021, 12, 636486. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, O.P.; Smith, T.J.; Dandare, S.U.; Parwin, K.S.; Singh, H.; Loh, H.X.; Cunningham, M.R.; Williams, P.N.; Nichol, T.; Subramanian, A.; et al. Metal(loid) speciation and transformation by aerobic methanotrophs. Microbiome 2021, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Zahn, J.A.; DiSpirito, A.A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. Res. 1996, 178, 1018–1029. [Google Scholar] [CrossRef]
- Ross, M.O.; MacMillan, F.; Wang, J.; Nisthal, A.; Lawton, T.J.; Olafson, B.D.; Mayo, S.L.; Rosenzweig, A.C.; Hoffman, B.M. Particulate methane monooxygenase contains only mononuclear copper centers. Science 2019, 364, 566–570. [Google Scholar] [CrossRef]
- Takeguchi, M.; Ohashi, M.; Okura*, I. Role of iron in particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biometals 1999, 12, 123–129. [Google Scholar] [CrossRef]
- Leak, D.J.; Dalton, H. Growth yields of methanotrophs. Appl. Microbiol. Biotechnol. 1986, 23, 470–476. [Google Scholar] [CrossRef]
- Semrau, J.D.; DiSpirito, A.A.; Gu, W.; Yoon, S. Metals and methanotrophy. Appl. Environ. Microbiol. 2018, 84, e02289-17. [Google Scholar] [CrossRef] [PubMed]
- Merkx, M.; Kopp, D.A.; Sazinsky, M.H.; Blazyk, J.L.; Muller, J.; Lippard, S.J. Dioxygen Activation and Methane Hydroxylation by Soluble Methane Monooxygenase: A Tale of Two Irons and Three Proteins. Angew. Chem. Int. Ed. 2001, 40, 2782–2807. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Zahn, J.A.; Bergmann, D.J.; Boyd, J.M.; Kunz, R.C.; DiSpirito, A.A. Membrane-associated quinoprotein formaldehyde dehydrogenase from Methylococcus capsulatus Bath. J. Bacteriol. Res. 2001, 183, 6832–6840. [Google Scholar] [CrossRef]
- Stein, L.Y.; Yoon, S.; Semrau, J.D.; Dispirito, A.A.; Crombie, A.; Murrell, J.C.; Vuilleumier, S.; Kalyuzhnaya, M.G.; Op den Camp, H.J.; Bringel, F.; et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J. Bacteriol. 2010, 192, 6497–6498. [Google Scholar] [CrossRef]
- Arp, D.J.; Stein, L.Y. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 471–495. [Google Scholar] [CrossRef]
- Jung, M.Y.; Sedlacek, C.J.; Kits, K.D.; Mueller, A.J.; Rhee, S.K.; Hink, L.; Nicol, G.W.; Bayer, B.; Lehtovirta-Morley, L.; Wright, C.; et al. Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 2022, 16, 272–283. [Google Scholar] [CrossRef]
- Mohammadi, S.S.; Pol, A.; van Alen, T.; Jetten, M.S.; Op den Camp, H.J. Ammonia oxidation and nitrite reduction in the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. Front. Microbiol. 2017, 8, 1901. [Google Scholar] [CrossRef]
- Duine, J.A.; Frank, J., Jr. Studies on methanol dehydrogenase from Hyphomicrobium X. Isolation of an oxidized form of the enzyme. Biochem. J. 1980, 187, 213–219. [Google Scholar] [CrossRef]
- Stein, L.Y.; Klotz, M.G. Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem. Soc. Trans. 2011, 39, 1826–1831. [Google Scholar] [CrossRef]
- Poret-Peterson, A.T.; Graham, J.E.; Gulledge, J.; Klotz, M.G. Transcription of nitrification genes by the methane-oxidizing bacterium, Methylococcus capsulatus strain Bath. ISME J. 2008, 2, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Nyerges, G.; Kozlowski, J.A.; Poret-Peterson, A.T.; Stein, L.Y.; Klotz, M.G. Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol. Lett. 2011, 322, 82–89. [Google Scholar] [CrossRef]
- Versantvoort, W.; Pol, A.; Jetten, M.S.M.; van Niftrik, L.; Reimann, J.; Kartal, B.; Op den Camp, H.J.M. Multiheme hydroxylamine oxidoreductases produce NO during ammonia oxidation in methanotrophs. Proc. Natl. Acad. Sci. USA 2020, 117, 24459–24463. [Google Scholar] [CrossRef]
- Jones, R.D.; Morita, R.Y. Methane Oxidation by Nitrosococcus oceanus and Nitrosomonas europaea. Appl. Environ. Microbiol. 1983, 45, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Hyman, M.R.; Wood, P.M. Methane oxidation by Nitrosomonas europaea. Biochem. J. 1983, 212, 31–37. [Google Scholar] [CrossRef]
- Ward, B.B. Kinetic studies on ammonia and methane oxidation by Nitrosococcus oceanus. Arch. Microbiol. 1987, 147, 126–133. [Google Scholar] [CrossRef]
- Su, Q.; Schittich, A.R.; Jensen, M.M.; Ng, H.; Smets, B.F. Role of Ammonia Oxidation in Organic Micropollutant Transformation during Wastewater Treatment: Insights from Molecular, Cellular, and Community Level Observations. Environ. Sci. Technol. 2021, 55, 2173–2188. [Google Scholar] [CrossRef]
- Helbling, D.E.; Johnson, D.R.; Honti, M.; Fenner, K. Micropollutant biotransformation kinetics associate with WWTP process parameters and microbial community characteristics. Environ. Sci. Technol. 2012, 46, 10579–10588. [Google Scholar] [CrossRef]
- Men, Y.; Han, P.; Helbling, D.E.; Jehmlich, N.; Herbold, C.; Gulde, R.; Onnis-Hayden, A.; Gu, A.Z.; Johnson, D.R.; Wagner, M. Biotransformation of two pharmaceuticals by the ammonia-oxidizing archaeon Nitrososphaera gargensis. Environ. Sci. Technol. 2016, 50, 4682–4692. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.M.; Cui, G.J.; Nunoura, T.; Takaki, Y.; Li, W.L.; Li, J.; Gao, Z.M.; Takai, K.; Zhang, A.Q. Genomics insights into ecotype formation of ammonia-oxidizing archaea in the deep ocean. Environ. Microbiol. 2019, 21, 716–729. [Google Scholar] [CrossRef]
- Burrows, K.J.; Cornish, A.; Scott, D.; Higgins, I.J. Substrate specificities of the soluble and particulate methane mono-oxygenases of Methylosinus trichosporium OB3b. Microbiology 1984, 130, 3327–3333. [Google Scholar] [CrossRef]
- Colby, J.; Stirling, D.I.; Dalton, H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem. J. 1977, 165, 395–402. [Google Scholar] [CrossRef]
- Green, J.; Dalton, H. Substrate specificity of soluble methane monooxygenase: Mechanistic implications. Biol. Chem. 1989, 264, 17698–17703. [Google Scholar] [CrossRef]
- Sirajuddin, S.; Rosenzweig, A.C. Enzymatic oxidation of methane. Biochemistry 2015, 54, 2283–2294. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Vu, D.Q.; Bui, T.P.L.; Mai, T.L.A.; Jensen, L.S.; de Neergaard, A. Organic matter and water management strategies to reduce methane and nitrous oxide emissions from rice paddies in Vietnam. Agric. Ecosyst. Environ. 2014, 196, 137–146. [Google Scholar] [CrossRef]
- Ehrlich, H.L.; Newman, D.K.; Kappler, A. Ehrlich’s Geomicrobiology; Dekker, D., Ed.; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Vázquez, M.N.; Guerrero, Y.R.; González, L.M.; de la Noval, W.T. Brassinosteroids and plant responses to heavy metal stress. An overview. Open J. Metal. 2013, 3, 34–41. [Google Scholar] [CrossRef]
- Banfalvi, G. (Ed.) Cellular Effects of Heavy Metals; Springer: Dordrecht, The Netherlands, 2011; pp. XIV, 348. [Google Scholar]
- Principi, P.; Villa, F.; Bernasconi, M.; Zanardini, E. Metal toxicity in municipal wastewater activated sludge investigated by multivariate analysis and in situ hybridization. Water Res. 2006, 40, 99–106. [Google Scholar] [CrossRef]
- Subrahmanyam, G.; Hu, H.-W.; Zheng, Y.-M.; Gattupalli, A.; He, J.-Z.; Liu, Y.-R. Response of ammonia oxidizing microbes to the stresses of arsenic and copper in two acidic alfisols. Appl. Soil Ecol. 2014, 77, 59–67. [Google Scholar] [CrossRef]
- Mertens, J.; Broos, K.; Wakelin, S.A.; Kowalchuk, G.A.; Springael, D.; Smolders, E. Bacteria, not archaea, restore nitrification in a zinc-contaminated soil. ISME J. 2009, 3, 916–923. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Smith, S.M.; Rawat, S.; Yatsunyk, L.A.; Stemmler, T.L.; Rosenzweig, A.C. Oxidation of methane by a biological dicopper centre. Nature 2010, 465, 115–119. [Google Scholar] [CrossRef]
- Yu, S.S.-F.; Chen, K.H.-C.; Tseng, M.Y.-H.; Wang, Y.-S.; Tseng, C.-F.; Chen, Y.-J.; Huang, D.-S.; Chan, S.I. Production of high-quality particulate methane monooxygenase in high yields from Methylococcus capsulatus (Bath) with a hollow-fiber membrane bioreactor. J. Bacteriol. Res. 2003, 185, 5915–5924. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Kenney, G.E.; Rosenzweig, A.C. Dual pathways for copper uptake by methanotrophic bacteria. Biol. Chem. 2011, 286, 37313–37319. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Rosenzweig, A.C. Copper methanobactin: A molecule whose time has come. Curr. Opin. Chem. Biol. 2008, 12, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I.; Nicol, G.W. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M. Principles of Bioinorganic Chemistry; University Science Books: Sausalito, CA, USA, 1994. [Google Scholar]
- Yang, R.; Van den Berg, C.M. Metal complexation by humic substances in seawater. Environ. Sci. Technol. 2009, 43, 7192–7197. [Google Scholar] [CrossRef]
- Shank, G.C.; Skrabal, S.A.; Whitehead, R.F.; Kieber, R.J. Strong copper complexation in an organic-rich estuary: The importance of allochthonous dissolved organic matter. Mar. Chem. 2004, 88, 21–39. [Google Scholar] [CrossRef]
- Voelker, B.M.; Kogut, M.B. Interpretation of metal speciation data in coastal waters: The effects of humic substances on copper binding as a test case. Mar. Chem. 2001, 74, 303–318. [Google Scholar] [CrossRef]
- Sanders, J. The effect of pH on the total and free ionic concentrations of mnganese, zinc and cobalt in soil solutions. Open J. Soil Sci. 1983, 34, 315–323. [Google Scholar] [CrossRef]
- Coale, K.H.; Bruland, K.W. Copper complexation in the Northeast Pacific. Limnol. Oceanogr. 1988, 33, 1084–1101. [Google Scholar] [CrossRef]
- Jacquot, J.E.; Horak, R.E.; Amin, S.A.; Devol, A.H.; Ingalls, A.E.; Armbrust, E.V.; Stahl, D.A.; Moffett, J.W. Assessment of the potential for copper limitation of ammonia oxidation by Archaea in a dynamic estuary. Mar. Chem. 2014, 162, 37–49. [Google Scholar] [CrossRef]
- Yang, Y.; Herbold, C.W.; Jung, M.-Y.; Qin, W.; Cai, M.; Du, H.; Lin, J.-G.; Li, X.; Li, M.; Gu, J.-D. Survival strategies of ammonia-oxidizing archaea (AOA) in a full-scale WWTP treating mixed landfill leachate containing copper ions and operating at low-intensity of aeration. Water Res. 2021, 191, 116798. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, M.; Liu, K.; Yang, E.; Chen, J.; Wu, S.; Xie, M.; Wang, D.; Deng, H.; Chen, H. Insights into the synergy between functional microbes and dissolved oxygen partition in the single-stage partial nitritation-anammox granules system. Bioresour. Technol. 2022, 347, 126364. [Google Scholar] [CrossRef]
- Chen, H.; Wu, J.; Liu, B.; Li, Y.Y.; Yasui, H. Competitive dynamics of anaerobes during long-term biological sulfate reduction process in a UASB reactor. Bioresour. Technol. 2019, 280, 173–182. [Google Scholar] [CrossRef]
- Villaverde, P.; Gondar, D.; Antelo, J.; Lopez, R.; Fiol, S.; Arce, F. Influence of pH on copper, lead and cadmium binding by an ombrotrophic peat. Eur. J. Soil Sci. 2009, 60, 377–385. [Google Scholar] [CrossRef]
- Reyes, C.; Hodgskiss, L.H.; Baars, O.; Kerou, M.; Bayer, B.; Schleper, C.; Kraemer, S.M. Copper limiting threshold in the terrestrial ammonia oxidizing archaeon Nitrososphaera viennensis. Res. Microbiol. 2020, 171, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef]
- Zhang, L.-M.; Hu, H.-W.; Shen, J.-P.; He, J.-Z. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 2012, 6, 1032–1045. [Google Scholar] [CrossRef]
- Kenney, G.E.; Rosenzweig, A.C. Chalkophores. Annu. Rev. Biochem. 2018, 87, 645. [Google Scholar] [CrossRef]
- El Ghazouani, A.; Baslé, A.; Gray, J.; Graham, D.W.; Firbank, S.J.; Dennison, C. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8400–8404. [Google Scholar] [CrossRef]
- Klotz, M.G.; Arp, D.J.; Chain, P.S.; El-Sheikh, A.F.; Hauser, L.J.; Hommes, N.G.; Larimer, F.W.; Malfatti, S.A.; Norton, J.M.; Poret-Peterson, A.T.; et al. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl. Environ. Microbiol. 2006, 72, 6299–6315. [Google Scholar] [CrossRef]
- Franza, T.; Mahe, B.; Expert, D. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 2005, 55, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Chain, P.; Lamerdin, J.; Larimer, F.; Regala, W.; Lao, V.; Land, M.; Hauser, L.; Hooper, A.; Klotz, M.; Norton, J.; et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 2003, 185, 2759–2773. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Lücker, S.; Albertsen, M.; Kitzinger, K.; Herbold, C.; Spieck, E.; Nielsen, P.H.; Wagner, M.; Daims, H. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. USA 2015, 112, 11371–11376. [Google Scholar] [CrossRef]
- Peng, P.; Gu, W.; DiSpirito, A.A.; Semrau, J.D. Multiple Mechanisms for Copper Uptake by Methylosinus trichosporium OB3b in the Presence of Heterologous Methanobactin. mBio 2022, 13, e0223922. [Google Scholar] [CrossRef]
- Choi, D.W.; Do, Y.S.; Zea, C.J.; McEllistrem, M.T.; Lee, S.W.; Semrau, J.D.; Pohl, N.L.; Kisting, C.J.; Scardino, L.L.; Hartsel, S.C.; et al. Spectral and thermodynamic properties of Ag(I), Au(III), Cd(II), Co(II), Fe(III), Hg(II), Mn(II), Ni(II), Pb(II), U(IV), and Zn(II) binding by methanobactin from Methylosinus trichosporium OB3b. J. Inorg. Biochem. 2006, 100, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Matsen, J.B.; Yang, S.; Stein, L.Y.; Beck, D.; Kalyuzhnaya, M.G. Global Molecular Analyses of Methane Metabolism in Methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part I: Transcriptomic Study. Front. Microbiol. 2013, 4, 40. [Google Scholar] [CrossRef] [PubMed]
- Hakemian, A.S.; Tinberg, C.E.; Kondapalli, K.C.; Telser, J.; Hoffman, B.M.; Stemmler, T.L.; Rosenzweig, A.C. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds copper(I). J. Am. Chem. Soc. 2005, 127, 17142–17143. [Google Scholar] [CrossRef]
- Gu, W.; Haque, M.F.U.; Baral, B.S.; Turpin, E.A.; Bandow, N.L.; Kremmer, E.; Flatley, A.; Zischka, H.; DiSpirito, A.A.; Semrau, J.D. A TonB-Dependent Transporter Is Responsible for Methanobactin Uptake by Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 2016, 82, 1917–1923. [Google Scholar] [CrossRef]
- Knapp, C.W.; Fowle, D.A.; Kulczycki, E.; Roberts, J.A.; Graham, D.W. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc. Natl. Acad. Sci. USA 2007, 104, 12040–12045. [Google Scholar] [CrossRef]
| Scheme | Siderophore Biosynthesis | Siderophore Transporter | Chalkophore Biosynthesis | Chalkophore Transporter | Note |
|---|---|---|---|---|---|
| Nitrosococcus oceani ATCC19707 | + | + | N.D. | N.D. | Nitrosococcus oceani has hydroxamate-type siderophore aerobactin synthesis and aerobactin receptor genes, and the iron transporter contains an ABC-type Fe3+/cobalamin siderophore transport system [116]. |
| Nitrosomonas europaea ATCC19718 | − | + | − | − | Nitrosomonas europaea has many iron-related genes to obtain iron from the environment; one of them is siderophore. However, no evidence was found for siderophore production [118]. |
| Nitrosopumilus maritimus SCM1 a | − | − | − | − | Most ammonia-oxidizing archaea do not have siderophore and chalkophore-related functions, but only N. maritimus SCM1 has a siderophore-related gene. |
| Nitrosocosmicus oleophilus MY3 | − | − | − | − | |
| Nitrososphaera viennensis EN76 | − | − | − | − | |
| Nitrospira inopinata | ± b | + | − | − | Nitrospira inopinata has putative TonB-dependent receptors (NITINOP_0357, NITINOP_1937, NITINOP_2355). |
| Methylosinus trichosporium OB3b | − c | ± | + | + | After binding copper, methanobactin is reinternalized through a specific outer membrane TonB-dependent transporter [114,120,121]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayub, H.; Kang, M.-J.; Farooq, A.; Jung, M.-Y. Ecological Aerobic Ammonia and Methane Oxidation Involved Key Metal Compounds, Fe and Cu. Life 2022, 12, 1806. https://doi.org/10.3390/life12111806
Ayub H, Kang M-J, Farooq A, Jung M-Y. Ecological Aerobic Ammonia and Methane Oxidation Involved Key Metal Compounds, Fe and Cu. Life. 2022; 12(11):1806. https://doi.org/10.3390/life12111806
Chicago/Turabian StyleAyub, Hina, Min-Ju Kang, Adeel Farooq, and Man-Young Jung. 2022. "Ecological Aerobic Ammonia and Methane Oxidation Involved Key Metal Compounds, Fe and Cu" Life 12, no. 11: 1806. https://doi.org/10.3390/life12111806
APA StyleAyub, H., Kang, M.-J., Farooq, A., & Jung, M.-Y. (2022). Ecological Aerobic Ammonia and Methane Oxidation Involved Key Metal Compounds, Fe and Cu. Life, 12(11), 1806. https://doi.org/10.3390/life12111806







