Establishment of a Protoplasts-Based Transient Expression System in Banana (Musa spp.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Protoplast Isolation
2.3. Counting of Protoplasts
2.4. Protoplast Transfection
- (1)
- Add 15 μg plasmids into 100 μL of freshly isolated protoplasts in the MMG solution and mix well.
- (2)
- Add 110 μL of 45% (w/v) PEG solution (PEG 4000, 0.2 mmol/L Mannitol and 0.1 mol/L CaCl2) into the tube and mix well by gently inverting the tubes.
- (3)
- After a 15 min incubation in the dark, the protoplasts are gently washed by adding a 1 mL W5 buffer and harvested by being centrifuged for 3 min at 300 G and then resuspended in a 1 mL W5 buffer.
- (4)
- Repeat the above wash step two times.
- (5)
- The transfected protoplasts in the W5 solution are incubated under dim light at 25 °C for 12–16 h.
2.5. Bimolecular Fluorescent Complimentary (BiFC) Assay
2.6. Microscopy
3. Result
3.1. The Banana Sucker Is the Best Source Material for Isolation of Protoplasts
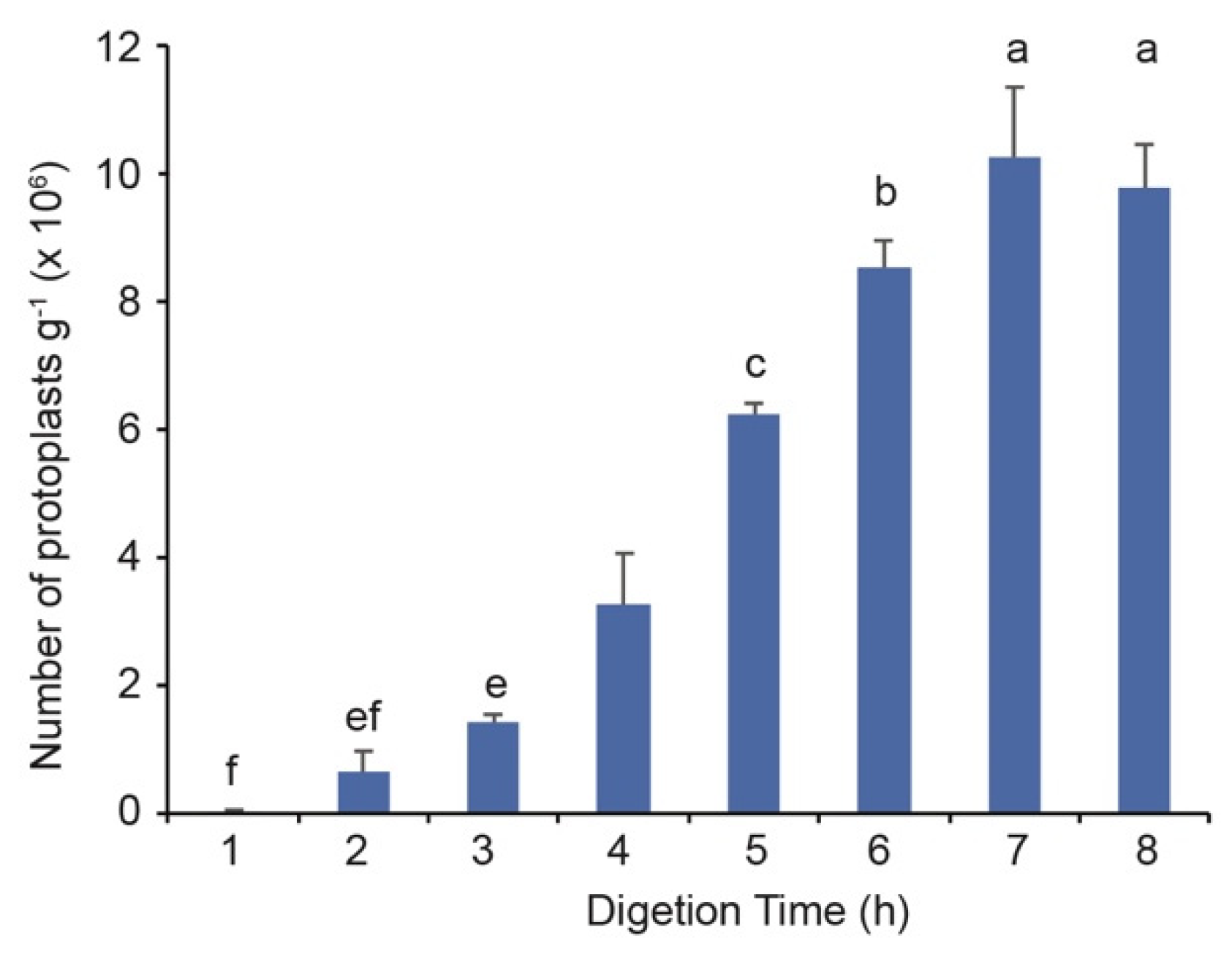
3.2. Five Hours of Enzymatic Hydrolysis Is Sufficient for the Protoplast Isolation from Banana Suckers
3.3. Transient Expression of GFP in Banana Protoplasts
3.4. Effects of DNA Amount on the Protoplast Transformation Efficiency
3.5. Effects of PEG Concentration and Incubation Time on the Protoplast Transformation Efficiency
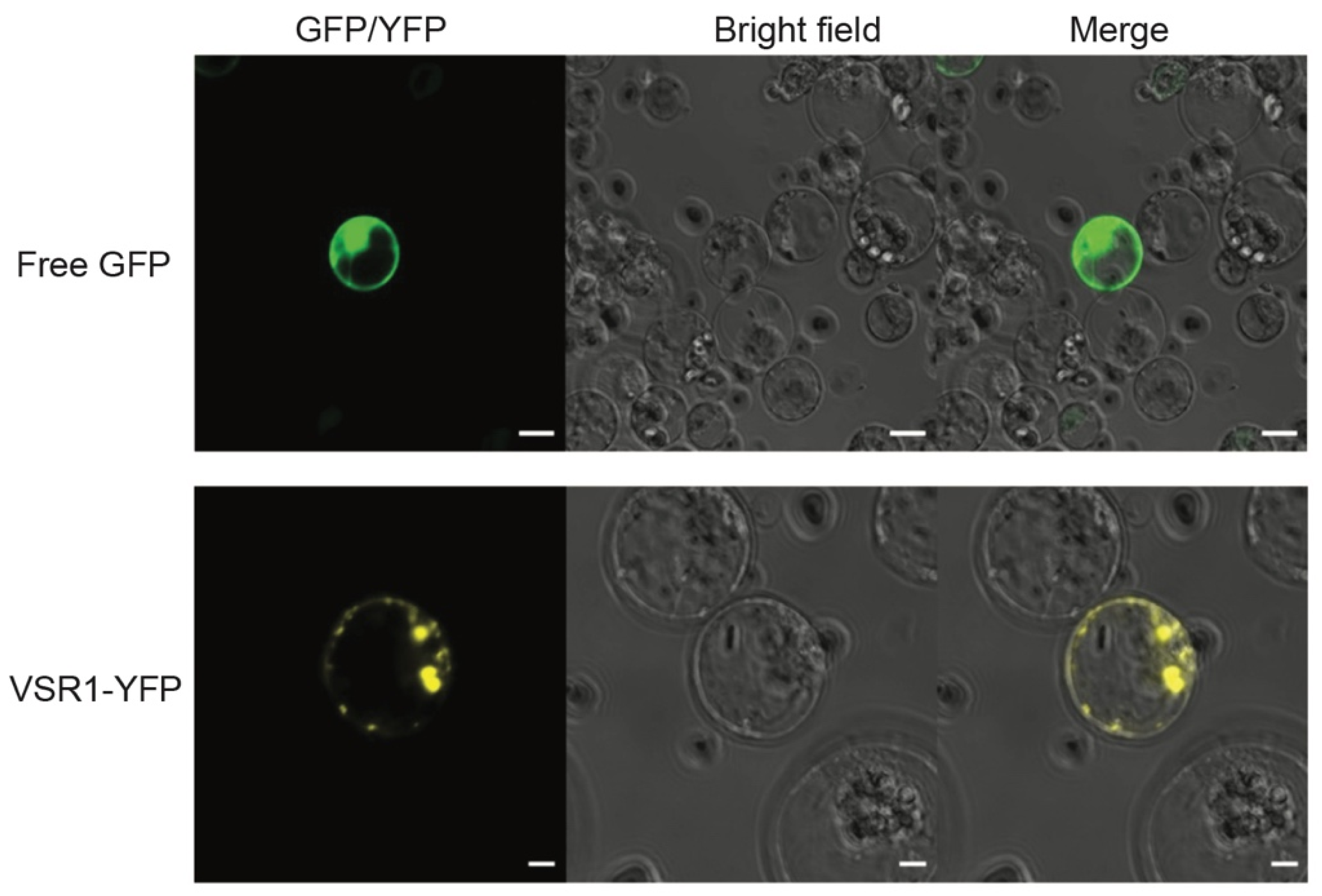
3.6. Protein Localization Assay Using Transient Expression System in Banana Protoplasts
3.7. BiFC Assay Validates Protein-Protein Interactions in Banana Protoplasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.P.; Jenny, C.; et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef]
- Butler, D. Fungus threatens top banana. Nature 2013, 504, 195–196. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef]
- Pegg, K.G.; Coates, L.M.; O’Neill, W.T.; Turner, D.W. The Epidemiology of Fusarium Wilt of Banana. Front. Plant Sci. 2019, 10, 1395. [Google Scholar] [CrossRef] [PubMed]
- Ghag, S.B.; Ganapathi, T.R. Genetically modified bananas: To mitigate food security concerns. Sci. Hortic. 2017, 214, 91–98. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Assani, A.; Haïcour, R.; Wenzel, G.; Foroughi-Wehr, B.; Bakry, F.; Côte, F.-X.; Ducreux, G.; Ambroise, A.; Grapin, A. Influence of donor material and genotype on protoplast regeneration in banana and plantain cultivars (Musa spp.). Plant Sci. 2002, 162, 355–362. [Google Scholar] [CrossRef]
- Shivani; Kaur, N.; Awasthi, P.; Tiwari, S. Identification and expression analysis of genes involved in somatic embryogenesis of banana. Acta Physiol. Plant. 2018, 40, 139. [Google Scholar] [CrossRef]
- Sheen, J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001, 127, 1466–1475. [Google Scholar] [CrossRef]
- He, P.; Shan, L.; Sheen, J. The Use of Protoplasts to Study Innate Immune Responses. Methods Mol. Biol. 2007, 354, 1–9. [Google Scholar]
- Confraria, A.; Baena-González, E. Using Arabidopsis Protoplasts to Study Cellular Responses to Environmental Stress. Methods Mol. Biol. 2016, 1398, 247–269. [Google Scholar]
- Jia, X.; Zhang, X.; Qu, J.; Han, R. Optimization Conditions of Wheat Mesophyll Protoplast Isolation. Agric. Sci. 2016, 7, 850–858. [Google Scholar] [CrossRef]
- Priyadarshani, S.V.G.N.; Hu, B.; Li, W.; Ali, H.; Jia, H.; Zhao, L.; Ojolo, S.P.; Azam, S.M.; Xiong, J.; Yan, M.; et al. Simple protoplast isolation system for gene expression and protein interaction studies in pineapple (Ananas comosus L.). Plant Methods 2018, 14, 95. [Google Scholar] [CrossRef]
- Wu, F.; Hanzawa, Y. A Simple Method for Isolation of Soybean Protoplasts and Application to Transient Gene Expression Analyses. J. Vis. Exp. 2018, 57258. [Google Scholar] [CrossRef]
- Ye, S.; Chen, G.; Kohnen, M.V.; Wang, W.; Cai, C.; Ding, W.; Wu, C.; Gu, L.; Zheng, Y.; Ma, X.; et al. Robust CRISPR/Cas9 mediated genome editing and its application in manipulating plant height in the first generation of hexaploid Ma bamboo (Dendrocalamus latiflorus Munro). Plant Biotechnol. J. 2019, 18, 1501–1503. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, G.; Chen, Z.; Han, J.; Hu, Y.; Wang, K. Optimization of protoplast isolation, transformation and its application in sugarcane (Saccharum spontaneum L.). Crop J. 2020, 9, 133–142. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, H.; Liu, J.; Yang, Q.; Shao, X.; Bi, F.; Hu, C.; Huo, H.; Chen, K.; Yi, G. Establishment of a PEG-mediated protoplast transformation system based on DNA and CRISPR/Cas9 ribonucleoprotein complexes for banana. BMC Plant Biol. 2020, 20, 425. [Google Scholar] [CrossRef]
- Khatri, A.; Dahot, M.; Khan, I.; Nizamani, G.S. An efficient method of protoplast isolation in banana (Musa spp.). Pak. J. Bot. 2010, 42, 1267–1271. [Google Scholar]
- Miao, Y.; Yan, P.K.; Kim, H.; Hwang, I.; Jiang, L. Localization of Green Fluorescent Protein Fusions with the Seven Arabidopsis Vacuolar Sorting Receptors to Prevacuolar Compartments in Tobacco BY-2 Cells. Plant Physiol. 2006, 142, 945–962. [Google Scholar] [CrossRef]
- Zhao, C.; Tang, Y.; Wang, J.; Zeng, Y.; Sun, H.; Zheng, Z.; Su, R.; Schneeberger, K.; Parker, J.E.; Cui, H. A mis-regulated cyclic nucleotide-gated channel mediates cytosolic calcium elevation and activates immunity in Arabidopsis. New Phytol. 2021, 230, 1078–1094. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- He, F.; Chen, S.; Ning, Y.; Wang, G.L. Rice (Oryza sativa) Protoplast Isolation and Its Application for Transient Expression Analysis: Rice Protoplast Isolation and Transient Expression Analysis. Curr. Protoc. Plant Biol. 2016, 1, 373–383. [Google Scholar] [CrossRef]
- Shen, Y.; Meng, D.; McGrouther, K.; Zhang, J.; Cheng, L. Efficient isolation of Magnolia protoplasts and the application to subcellular localization of MdeHSF1. Plant Methods 2017, 13, 44. [Google Scholar] [CrossRef]
- Megia, R.; Haïcour, R.; Rossignol, L.; Sihachakr, D. Callus formation from cultured protoplasts of banana (Musa sp.). Plant Sci. 1992, 85, 91–98. [Google Scholar] [CrossRef]
- Shabanian, N.; Rahmani, M.S.; Zarrinjoei, F. The effect of enzyme concentrations and digestion time on protoplast isolation from in vitro grown leaves of Oak (Quercus brantii). J. Biotechnol. 2010, 150, 565. [Google Scholar] [CrossRef]
- Armstrong, C.; Petersen, W.; Buchholz, W.; Bowen, B.; Sulc, S. Factors affecting PEG-mediated stable transformation of maize protoplasts. Plant Cell Rep. 1990, 9, 335–339. [Google Scholar] [CrossRef]
- Ge, C.-Y.; Duan, Y.-B.; Zhou, M.-G.; Chen, C.-J. A Protoplast Transformation System for Gene Deletion and Complementation in Sclerotinia sclerotiorum. J. Phytopathol. 2013, 161, 800–806. [Google Scholar] [CrossRef]
- Chang, S.; Cohen, S.N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol. Gen. Genet. 1979, 168, 111–115. [Google Scholar] [CrossRef]
- Reed, K.M.; Bargmann, B.O.R. Protoplast Regeneration and Its Use in New Plant Breeding Technologies. Front. Genome Ed. 2021, 3, 734951. [Google Scholar] [CrossRef]
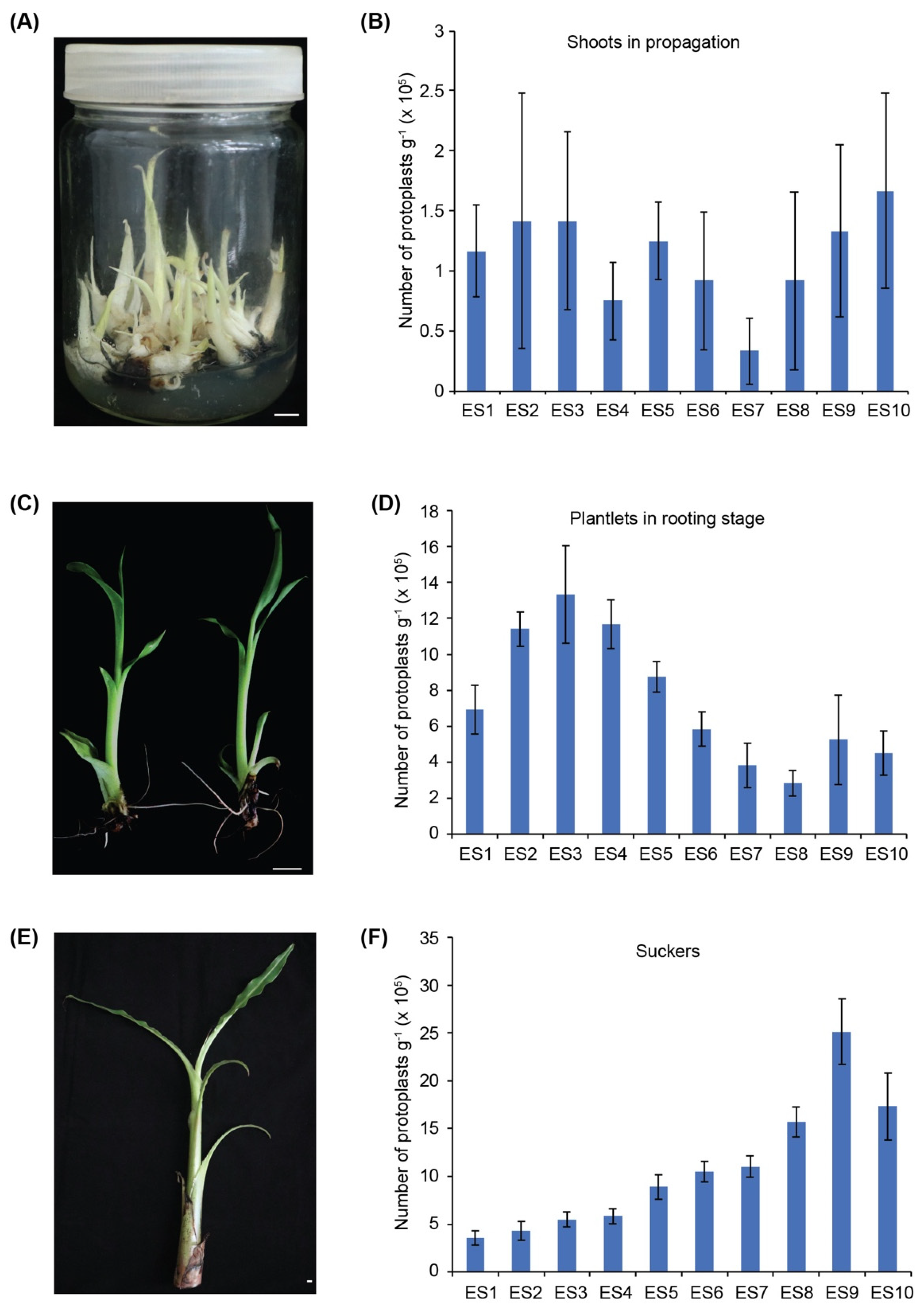
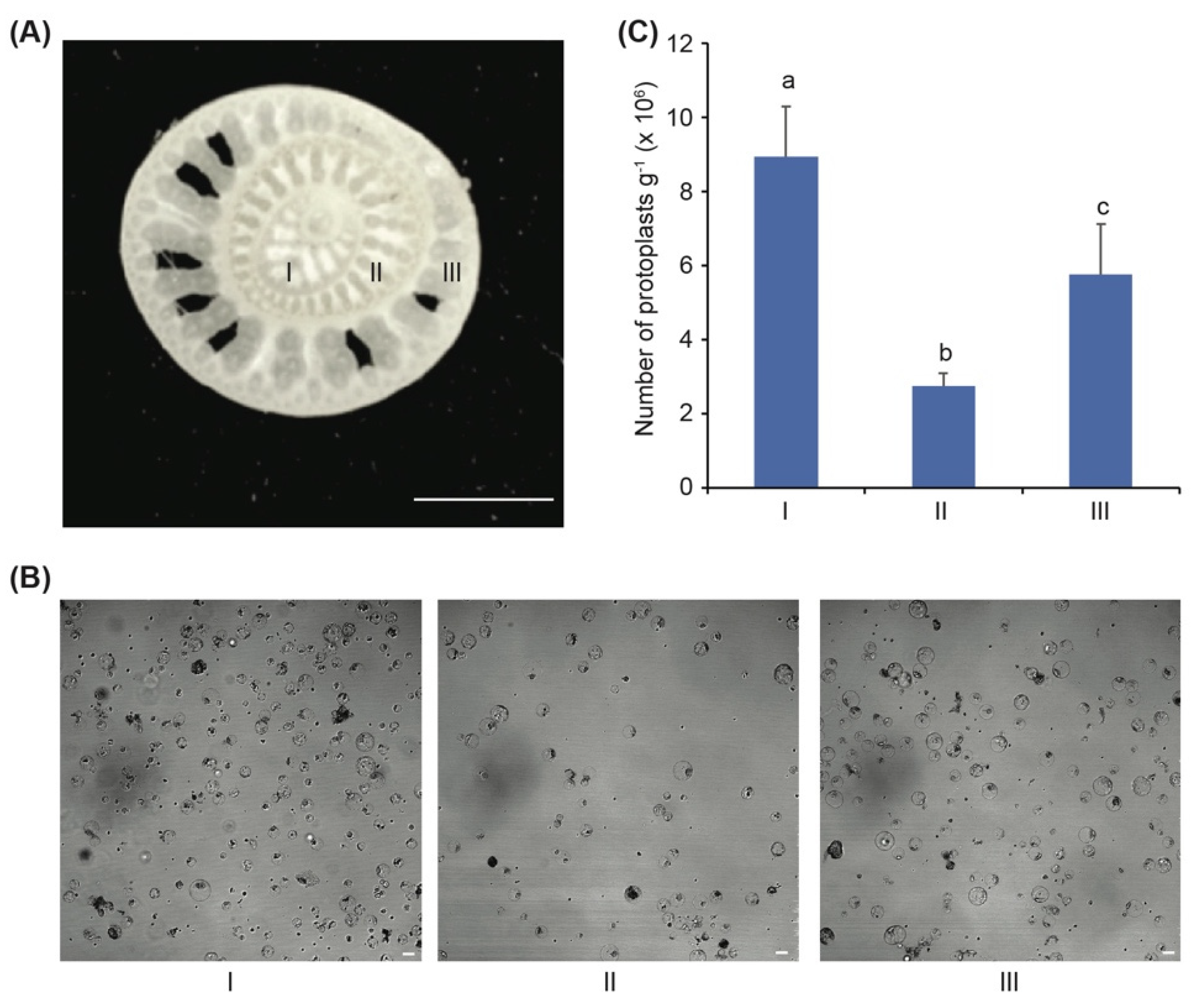


| Enzyme Solutions | Cellulase R-10 (%) | Macerozyme R-10 (%) | Pectinase Y-23 (%) |
|---|---|---|---|
| ES1 | 1.5 | 0.3 | 0 |
| ES2 | 3 | 1 | 0.2 |
| ES3 | 3 | 1 | 0.5 |
| ES4 | 3 | 1 | 1 |
| ES5 | 3 | 2 | 1 |
| ES6 | 1.5 | 0.75 | 0 |
| ES7 | 2 | 0.75 | 0 |
| ES8 | 2.5 | 0.75 | 0 |
| ES9 | 2.5 | 1 | 0 |
| ES10 | 2 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Li, S.; Du, C.; Gao, H.; Yang, D.; Fu, G.; Cui, H. Establishment of a Protoplasts-Based Transient Expression System in Banana (Musa spp.). Agronomy 2022, 12, 2648. https://doi.org/10.3390/agronomy12112648
Zhao C, Li S, Du C, Gao H, Yang D, Fu G, Cui H. Establishment of a Protoplasts-Based Transient Expression System in Banana (Musa spp.). Agronomy. 2022; 12(11):2648. https://doi.org/10.3390/agronomy12112648
Chicago/Turabian StyleZhao, Chunhui, Shuyu Li, Chanjuan Du, Hui Gao, Di Yang, Gang Fu, and Haitao Cui. 2022. "Establishment of a Protoplasts-Based Transient Expression System in Banana (Musa spp.)" Agronomy 12, no. 11: 2648. https://doi.org/10.3390/agronomy12112648
APA StyleZhao, C., Li, S., Du, C., Gao, H., Yang, D., Fu, G., & Cui, H. (2022). Establishment of a Protoplasts-Based Transient Expression System in Banana (Musa spp.). Agronomy, 12(11), 2648. https://doi.org/10.3390/agronomy12112648








