Metabolome Profiling in Aging Studies
Abstract
Simple Summary
Abstract
1. Introduction
2. Theories of Aging
3. Approaches for Metabolomic Profiling
4. Untargeted Metabolomic Profiling in the Study of Aging in Animal Models
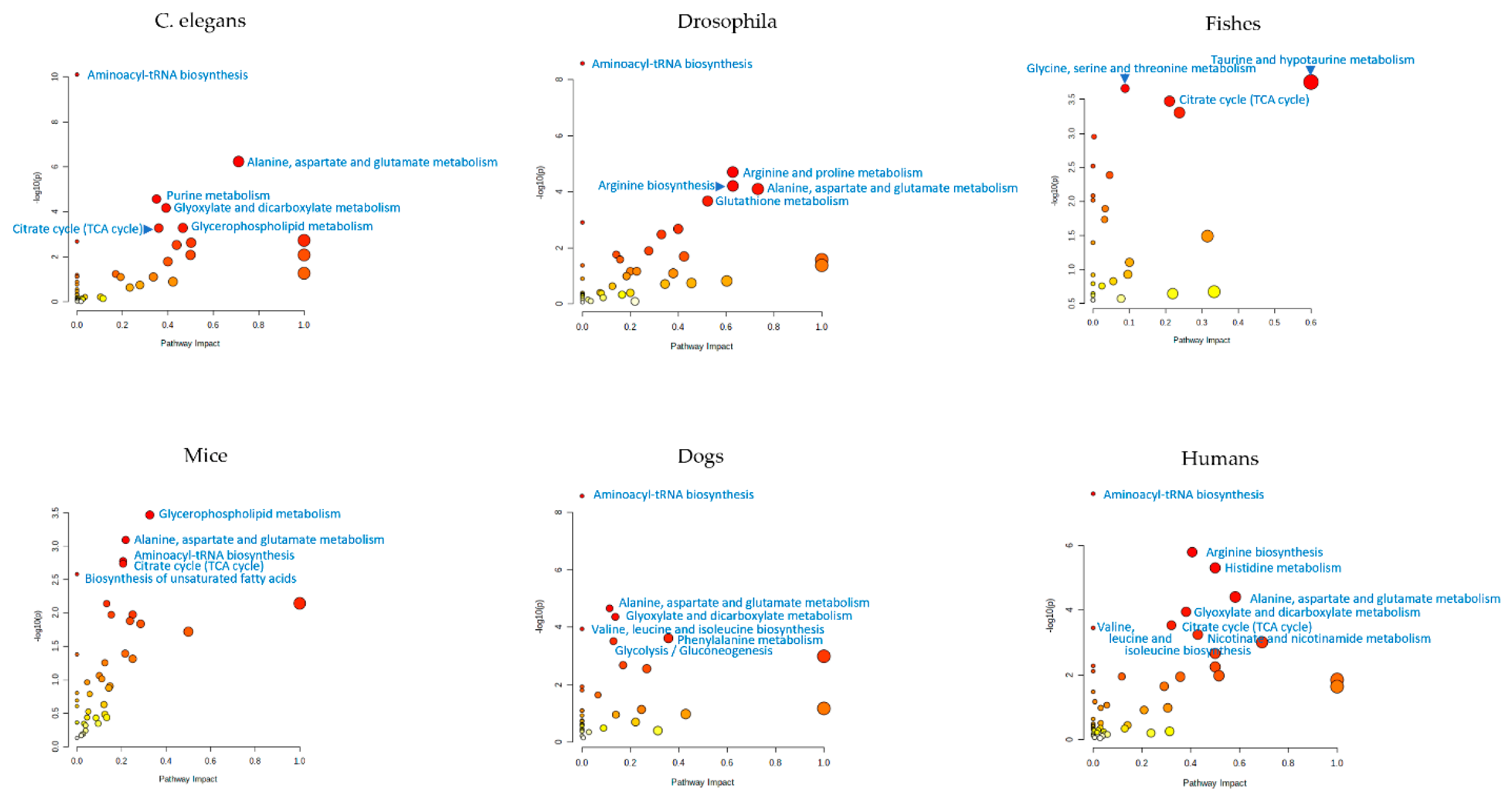
5. Metabolomic Profiling of Caenorhabditis elegans
6. Metabolomic Profiling of Drosophila
7. Metabolomic Profiling of Fishes
8. Metabolomic Profiling of Rodents
9. Metabolomic Profiling of Dogs
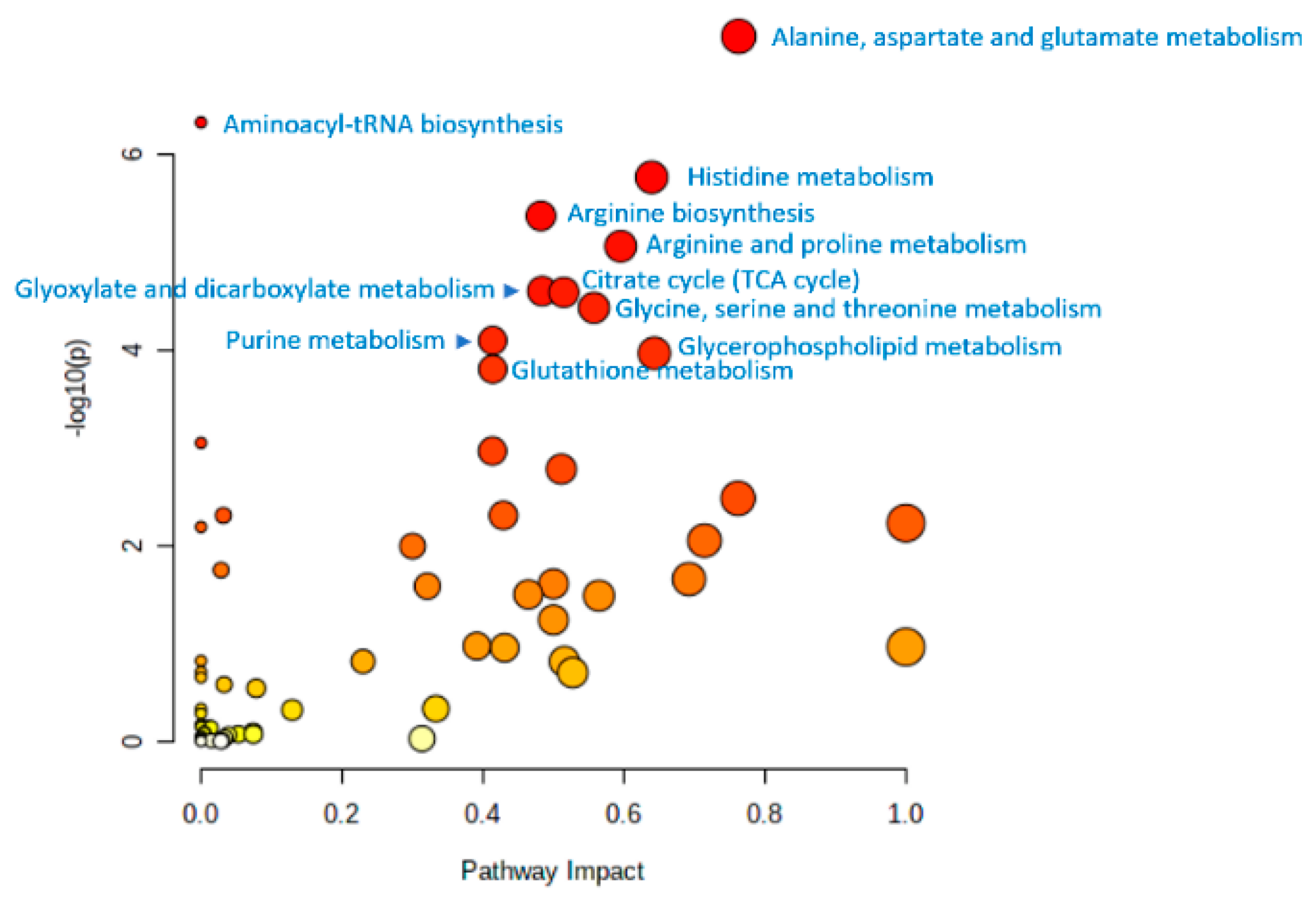
10. Untargeted Metabolomic Profiling in the Study of Human Aging
11. Summary of Metabolome Profiling Data from Aging Studies
12. Final Remarks
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferrucci, L.; Giallauria, F.; Guralnik, J.M. Epidemiology of Aging. Radiol. Clin. N. Am. 2008, 46, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Butler, R.N.; Miller, R.A.; Perry, D.; Carnes, B.A.; Williams, T.F.; Cassel, C.; Brody, J.; Bernard, M.A.; Partridge, L.; Kirkwood, T.; et al. New model of health promotion and disease prevention for the 21st century. BMJ 2008, 337, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Wijsman, C.A.; Rozing, M.P.; Streefland, T.C.M.; Le Cessie, S.; Mooijaart, S.P.; Slagboom, P.E.; Westendorp, R.G.J.; Pijl, H.; Van Heemst, D. Familial longevity is marked by enhanced insulin sensitivity. Aging Cell 2011, 10, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, H.; Kameda, M.; Yanagida, M. Whole blood metabolomics in aging research. Int. J. Mol. Sci. 2021, 22, 175. [Google Scholar] [CrossRef] [PubMed]
- Balliu, B.; Durrant, M.; De Goede, O.; Abell, N.; Li, X.; Liu, B.; Gloudemans, M.J.; Cook, N.L.; Smith, K.S.; Knowles, D.A.; et al. Genetic regulation of gene expression and splicing during a 10-year period of human aging. Genome Biol. 2019, 20, 230. [Google Scholar] [CrossRef]
- Melzer, D.; Pilling, L.C.; Ferrucci, L. The genetics of human ageing. Nat. Rev. Genet. 2020, 21, 88–101. [Google Scholar] [CrossRef]
- Karasik, D.; Demissie, S.; Cupples, L.A.; Kiel, D.P. Disentangling the genetic determinants of human aging: Biological age as an alternative to the use of survival measures. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2005, 60, 574–587. [Google Scholar] [CrossRef]
- Piper, M.D.W.; Bartke, A. Diet and Aging. Cell Metab. 2008, 8, 99–104. [Google Scholar] [CrossRef]
- Kerber, R.A.; O’Brien, E.; Cawthon, R.M. Gene expression profiles associated with aging and mortality in humans. Aging Cell 2009, 8, 239–250. [Google Scholar] [CrossRef]
- Deelen, J.; Beekman, M.; Uh, H.W.; Helmer, Q.; Kuningas, M.; Christiansen, L.; Kremer, D.; van der Breggen, R.; Suchiman, H.E.D.; Lakenberg, N.; et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 2011, 10, 686–698. [Google Scholar] [CrossRef]
- Phillip, J.M.; Aifuwa, I.; Walston, J.; Wirtz, D. The Mechanobiology of Aging. Annu. Rev. Biomed. Eng. 2015, 17, 113–141. [Google Scholar] [CrossRef]
- Amaro, A.; Petretto, A.; Angelini, G.; Pfeffer, U. Advancements in Omics Sciences. In Translational Medicine: Tools and Techniques; Shahzad, A., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 67–108. ISBN 9780128034941. [Google Scholar]
- Sun, N.; Youle, R.J.; Finkel, T. The Mitochondrial Basis of Aging. Mol. Cell 2016, 61, 654–666. [Google Scholar] [CrossRef]
- Franceschi, C.; Capri, M.; Monti, D.; Giunta, S.; Olivieri, F.; Sevini, F.; Panourgia, M.P.; Invidia, L.; Celani, L.; Scurti, M.; et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007, 128, 92–105. [Google Scholar] [CrossRef]
- Harman, D. The Free Radical Theory of Aging. Antioxid. Redox Signal. 2003, 5, 557–561. [Google Scholar] [CrossRef]
- Lehmann, A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 2003, 85, 1101–1111. [Google Scholar] [CrossRef]
- Harman, D. The Biologic Clock: The Mitochondria? J. Am. Geriatr. Soc. 1972, 20, 145–147. [Google Scholar] [CrossRef]
- Schriner, S.E.; Linford, N.J.; Martin, G.M.; Treuting, P.; Ogburn, C.E.; Emond, M.; Coskun, P.E.; Ladiges, W.; Wolf, N.; Van Remmen, H.; et al. Medecine: Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 2005, 308, 1909–1911. [Google Scholar] [CrossRef]
- Sun, J.; Folk, D.; Bradley, T.J.; Tower, J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics 2002, 161, 661–672. [Google Scholar] [CrossRef]
- Matheu, A.; Maraver, A.; Klatt, P.; Flores, I.; Garcia-Cao, I.; Borras, C.; Flores, J.M.; Viña, J.; Blasco, M.A.; Serrano, M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 2007, 448, 375–379. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Mouchiroud, L.; Ryu, D.; Moullan, N.; Katsyuba, E.; Knott, G.; Williams, R.W.; Auwerx, J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013, 497, 451–457. [Google Scholar] [CrossRef]
- Yang, W.; Hekimi, S. A mitochondrial superoxide signal triggers increased longevity in caenorhabditis elegans. PLoS Biol. 2010, 8, e1000556. [Google Scholar] [CrossRef] [PubMed]
- Timblin, G.A.; Tharp, K.M.; Ford, B.; Winchester, J.M.; Wang, J.; Zhu, S.; Khan, R.I.; Louie, S.K.; Iavarone, A.T.; ten Hoeve, J.; et al. Mitohormesis reprogrammes macrophage metabolism to enforce tolerance. Nat. Metab. 2021, 3, 618–635. [Google Scholar] [CrossRef] [PubMed]
- McCay, C.M.; Crowell, M.F.; Maynard, L.A. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition 1989, 5, 155–171, discussion 172. [Google Scholar] [CrossRef] [PubMed]
- Sohal, R.S.; Ku, H.H.; Agarwal, S.; Forster, M.J.; Lal, H. Oxidative damage, mitochondrial oxidant generation and antioxidant defenses during aging and in response to food restriction in the mouse. Mech. Ageing Dev. 1994, 74, 121–133. [Google Scholar] [CrossRef]
- Haigis, M.C.; Guarente, L.P. Mammalian sirtuins—Emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006, 20, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Cantó, C.; Jiang, L.Q.; Deshmukh, A.S.; Mataki, C.; Coste, A.; Lagouge, M.; Zierath, J.R.; Auwerx, J. Interdependence of AMPK and SIRT1 for Metabolic Adaptation to Fasting and Exercise in Skeletal Muscle. Cell Metab. 2010, 11, 213–219. [Google Scholar] [CrossRef]
- Stanfel, M.N.; Shamieh, L.S.; Kaeberlein, M.; Kennedy, B.K. The TOR pathway comes of age. Biochim. Biophys. Acta Gen. Subj. 2009, 1790, 1067–1074. [Google Scholar] [CrossRef]
- Greer, E.L.; Dowlatshahi, D.; Banko, M.R.; Villen, J.; Hoang, K.; Blanchard, D.; Gygi, S.P.; Brunet, A. An AMPK-FOXO Pathway Mediates Longevity Induced by a Novel Method of Dietary Restriction in C. elegans. Curr. Biol. 2007, 17, 1646–1656. [Google Scholar] [CrossRef]
- Morshneva, A.V. FoxO transcription factors as multifunctional cell regulators. Tsitologiya 2020, 62, 687–698. [Google Scholar] [CrossRef]
- Baar, M.P.; Brandt, R.M.; Putavet, D.A.; Klein, J.D.; Derks, K.W.; Bourgeois, B.R.; Stryeck, S.; Rijksen, Y.; van Willigenburg, H.; Feijtel, D.A.; et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 2017, 169, 132–147. [Google Scholar] [CrossRef]
- Onken, B.; Driscoll, M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans healthspan via AMPK, LKB1, and SKN-1. PLoS ONE 2010, 5, e8758. [Google Scholar] [CrossRef]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Mouchiroud, L.; Molin, L.; Dallière, N.; Solari, F. Life span extension by resveratrol, rapamycin, and metformin: The promise of dietary restriction mimetics for an healthy aging. BioFactors 2010, 36, 377–382. [Google Scholar] [CrossRef]
- Imai, S.I.; Guarente, L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014, 24, 464–471. [Google Scholar] [CrossRef]
- Verdin, E. NAD+ in aging, metabolism, and neurodegeneration. Science 2015, 350, 1208–1213. [Google Scholar] [CrossRef]
- Yoshino, J.; Baur, J.A.; Imai, S. ichiro NAD + Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018, 27, 513–528. [Google Scholar] [CrossRef]
- Corrada, M.M.; Kawas, C.H.; Mozaffar, F.; Paganini-Hill, A. Association of body mass index and weight change with all-cause mortality in the elderly. Am. J. Epidemiol. 2006, 163, 938–949. [Google Scholar] [CrossRef]
- Irie, J.; Inagaki, E.; Fujita, M.; Nakaya, H.; Mitsuishi, M.; Yamaguchi, S.; Yamashita, K.; Shigaki, S.; Ono, T.; Yukioka, H.; et al. Effect of oral administration of nicotinamide mononucleotide on clinical parameters and nicotinamide metabolite levels in healthy Japanese men. Endocr. J. 2020, 67, 153–160. [Google Scholar] [CrossRef]
- Ikeda, T.; Aizawa, J.; Nagasawa, H.; Gomi, I.; Kugota, H.; Nanjo, K.; Jinno, T.; Masuda, T.; Morita, S. Effects and feasibility of exercise therapy combined with branched-chain amino acid supplementation on muscle strengthening in frail and pre-frail elderly people requiring long-term care: A crossover trial. Appl. Physiol. Nutr. Metab. 2016, 41, 438–445. [Google Scholar] [CrossRef]
- Bjelakovic, G.; Nikolova, D.; Gluud, C. Antioxidant supplements and mortality. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Parkhitko, A.A.; Filine, E.; Mohr, S.E.; Moskalev, A.; Perrimon, N. Targeting metabolic pathways for extension of lifespan and healthspan across multiple species. Ageing Res. Rev. 2020, 64, 101188. [Google Scholar] [CrossRef] [PubMed]
- Nanda, T.; Das, M. Metabolomics: The Future of Systems Biology. J. Comput. Sci. Syst. Biol. 2011, 4, S13. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhai, G.; Singmann, P.; He, Y.; Xu, T.; Prehn, C.; Römisch-Margl, W.; Lattka, E.; Gieger, C.; Soranzo, N.; et al. Human serum metabolic profiles are age dependent. Aging Cell 2012, 11, 960–967. [Google Scholar] [CrossRef]
- Srivastava, S. Emerging insights into the metabolic alterations in aging using metabolomics. Metabolites 2019, 9, 301. [Google Scholar] [CrossRef]
- Gao, A.W.; Smith, R.L.; van Weeghel, M.; Kamble, R.; Janssens, G.E.; Houtkooper, R.H. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp. Gerontol. 2018, 113, 128–140. [Google Scholar] [CrossRef]
- Cox, J.E.; Thummel, C.S.; Tennessen, J.M. Metabolomic studies in Drosophila. Genetics 2017, 206, 1169–1185. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Soltow, Q.A.; Li, S.; Sidik, A.; Jones, D.P.; Promislow, D.E.L. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell 2014, 13, 596–604. [Google Scholar] [CrossRef]
- Avanesov, A.S.; Ma, S.; Pierce, K.A.; Yim, S.H.; Lee, B.C.; Clish, C.B.; Gladyshev, V.N. Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. eLife 2014, 3, e02077. [Google Scholar] [CrossRef]
- Sarup, P.; Pedersen, S.M.M.; Nielsen, N.C.; Malmendal, A.; Loeschcke, V. The Metabolic Profile of Long-Lived Drosophila melanogaster. PLoS ONE 2012, 7, e47461. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Lyu, Y.; Pletcher, S.D.; Promislow, D.E.L. Proteomics and metabolomics in ageing research: From biomarkers to systems biology. Essays Biochem. 2017, 61, 379–388. [Google Scholar] [CrossRef]
- Kristal, B.S.; Shurubor, Y.I. Metabolomics: Opening another window into aging. Sci. Aging Knowl. Environ. 2005, 2005, pe19. [Google Scholar] [CrossRef]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef]
- Kotze, H.L.; Armitage, E.G.; Sharkey, K.J.; Allwood, J.W.; Dunn, W.B.; Williams, K.J.; Goodacre, R. A novel untargeted metabolomics correlation-based network analysis incorporating human metabolic reconstructions. BMC Syst. Biol. 2013, 7, 107. [Google Scholar] [CrossRef]
- Adav, S.S.; Wang, Y. Metabolomics signatures of aging: Recent advances. Aging Dis. 2021, 12, 646–661. [Google Scholar] [CrossRef]
- Bunning, B.J.; Contrepois, K.; Lee-McMullen, B.; Dhondalay, G.K.R.; Zhang, W.; Tupa, D.; Raeber, O.; Desai, M.; Nadeau, K.C.; Snyder, M.P.; et al. Global metabolic profiling to model biological processes of aging in twins. Aging Cell 2020, 19, e13073. [Google Scholar] [CrossRef]
- Van Den Akker, E.B.; Trompet, S.; Barkey Wolf, J.J.H.; Beekman, M.; Suchiman, H.E.D.; Deelen, J.; Asselbergs, F.W.; Boersma, E.; Cats, D.; Elders, P.M.; et al. Metabolic age based on the BBMRI-NL 1H-NMR metabolomics repository as biomarker of age-related disease. Circ. Genom. Precis. Med. 2020, 13, 541–547. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Nagana Gowda, G.A.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. Nmr spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Wang, L.; Naser, F.J.; Spalding, J.L.; Patti, G.J. A protocol to compare methods for untargeted metabolomics. Methods Mol. Biol. 2019, 1862, 1–15. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Cantó, C.; Jeninga, E.H.; Andreux, Ṕ.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The metabolic footprint of aging in mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Huhman, D.V.; Sumner, L.W. Mass spectrometry strategies in metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef] [PubMed]
- Haijes, H.A.; Willemsen, M.; van der Ham, M.; Gerrits, J.; Pras-Raves, M.L.; Prinsen, H.C.M.T.; van Hasselt, P.M.; de Sain-Van der Velden, M.G.M.; Verhoeven-Duif, N.M.; Jans, J.J.M. Direct infusion based metabolomics identifies metabolic disease in patients’ dried blood spots and plasma. Metabolites 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.; Ruttkies, C.; Treutler, H.; Neumann, S. Bioinformatics can boost metabolomics research. J. Biotechnol. 2017, 261, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Wulff, J.E.; Mitchell, M.W. A Comparison of Various Normalization Methods for LC/MS Metabolomics Data. Adv. Biosci. Biotechnol. 2018, 9, 339–351. [Google Scholar] [CrossRef]
- Nam, S.L.; de la Mata, A.P.; Dias, R.P.; Harynuk, J.J. Towards standardization of data normalization strategies to improve urinary metabolomics studies by gc×gc-tofms. Metabolites 2020, 10, 376. [Google Scholar] [CrossRef]
- Sugimoto, M.; Kawakami, M.; Robert, M.; Soga, T.; Tomita, M. Bioinformatics Tools for Mass Spectroscopy-Based Metabolomic Data Processing and Analysis. Curr. Bioinform. 2012, 7, 96–108. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef]
- Jollife, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. A. Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Scholz, M.; Gatzek, S.; Sterling, A.; Fiehn, O.; Selbig, J. Metabolite fingerprinting: Detecting biological features by independent component analysis. Bioinformatics 2004, 20, 2447–2454. [Google Scholar] [CrossRef]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.J.A.N.; van der Greef, J.; Timmerman, M.E. ANOVA-simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef]
- Vis, D.J.; Westerhuis, J.A.; Smilde, A.K.; van der Greef, J. Statistical validation of megavariate effects in ASCA. BMC Bioinform. 2007, 8, 322. [Google Scholar] [CrossRef]
- Jonsson, P.; Bruce, S.J.; Moritz, T.; Trygg, J.; Sjöström, M.; Plumb, R.; Granger, J.; Maibaum, E.; Nicholson, J.K.; Holmes, E.; et al. Extraction, interpretation and validation of information for comparing samples in metabolic LC/MS data sets. Analyst 2005, 130, 701–707. [Google Scholar] [CrossRef]
- Linden, A. Measuring diagnostic and predictive accuracy in disease management: An introduction to receiver operating characteristic (ROC) analysis. J. Eval. Clin. Pract. 2006, 12, 132–139. [Google Scholar] [CrossRef]
- Broeckling, C.D.; Reddy, I.R.; Duran, A.L.; Zhao, X.; Sumner, L.W. MET-IDEA: Data extraction tool for mass spectrometry-based metabolomics. Anal. Chem. 2006, 78, 4334–4341. [Google Scholar] [CrossRef]
- Baran, R.; Kochi, H.; Saito, N.; Suematsu, M.; Soga, T.; Nishioka, T.; Robert, M.; Tomita, M. MathDAMP: A package for differential analysis of metabolite profiles. BMC Bioinform. 2006, 7, 530. [Google Scholar] [CrossRef]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. TagFinder for the quantitative analysis of gas chromatography—Mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef]
- Bucaciuc Mracica, T.; Anghel, A.; Ion, C.F.; Moraru, C.V.; Tacutu, R.; Lazar, G.A. MetaboAge DB: A repository of known ageing-related changes in the human metabolome. Biogerontology 2020, 21, 763–771. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38 (Suppl. S2), W71–W77. [Google Scholar] [CrossRef]
- Nevedomskaya, E.; Meissner, A.; Goraler, S.; De Waard, M.; Ridwan, Y.; Zondag, G.; Van Der Pluijm, I.; Deelder, A.M.; Mayboroda, O.A. Metabolic profiling of accelerated aging ERCC1d/-mice. J. Proteome Res. 2010, 9, 3680–3687. [Google Scholar] [CrossRef] [PubMed]
- Taormina, G.; Ferrante, F.; Vieni, S.; Grassi, N.; Russo, A.; Mirisola, M.G. Longevity: Lesson from model organisms. Genes 2019, 10, 518. [Google Scholar] [CrossRef] [PubMed]
- Toth, M.J.; Tchernof, A. Lipid metabolism in the elderly. Eur. J. Clin. Nutr. 2000, 54, S121–S125. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.W.; Nabi, I.R.; Demetriou, M. Metabolism, Cell Surface Organization, and Disease. Cell 2009, 139, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Feltes, B.C.; De Faria Poloni, J.; Bonatto, D. The developmental aging and origins of health and disease hypotheses explained by different protein networks. Biogerontology 2011, 12, 293–308. [Google Scholar] [CrossRef]
- Partridge, L.; Thornton, J.; Bates, G. The new science of ageing. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 6–8. [Google Scholar] [CrossRef]
- Piper, M.D.W.; Partridge, L. Drosophila as a model for ageing. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2707–2717. [Google Scholar] [CrossRef]
- Allard, J.B.; Duan, C. Comparative endocrinology of aging and longevity regulation. Front. Endocrinol. 2011, 2, 75. [Google Scholar] [CrossRef]
- Strange, K. Drug discovery in fish, flies, and worms. ILAR J. 2016, 57, 133–143. [Google Scholar] [CrossRef]
- Barré-Sinoussi, F.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Future Sci. OA 2015, 1, FSO63. [Google Scholar] [CrossRef]
- Ball, H.C.; Levari-Shariati, S.; Cooper, L.N.; Aliani, M. Comparative metabolomics of aging in a long-lived bat: Insights into the physiology of extreme longevity. PLoS ONE 2018, 13, e0196154. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Poonawalla, A.; Icyuz, M.; Swindell, W.R.; Wilson, L.; Barnes, S.; Sun, L.Y. Transcriptomic and metabolomic profiling of long-lived growth hormone releasing hormone knock-out mice: Evidence for altered mitochondrial function and amino acid metabolism. Aging 2020, 12, 3473–3485. [Google Scholar] [CrossRef]
- Maslov, D.L.; Zemskaya, N.V.; Trifonova, O.P.; Lichtenberg, S.; Balashova, E.E.; Lisitsa, A.V.; Moskalev, A.A.; Lokhov, P.G. Comparative metabolomic study of drosophila species with different lifespans. Int. J. Mol. Sci. 2021, 22, 12873. [Google Scholar] [CrossRef]
- Chaleckis, R.; Murakami, I.; Takada, J.; Kondoh, H.; Yanagida, M. Individual variability in human blood Metabolites identifies age-related differences. Proc. Natl. Acad. Sci. USA 2016, 113, 4252–4259. [Google Scholar] [CrossRef]
- Holtze, S.; Gorshkova, E.; Braude, S.; Cellerino, A.; Dammann, P.; Hildebrandt, T.B.; Hoeflich, A.; Hoffmann, S.; Koch, P.; Terzibasi Tozzini, E.; et al. Alternative Animal Models of Aging Research. Front. Mol. Biosci. 2021, 8, 660959. [Google Scholar] [CrossRef]
- Williams, R.E.; Lenz, E.M.; Lowden, J.S.; Rantalainen, M.; Wilson, I.D. The metabonomics of aging and development in the rat: An investigation into the effect of age on the profile of endogenous metabolites in the urine of male rats using 1H NMR and HPLC-TOF MS. Mol. Biosyst. 2005, 1, 166–175. [Google Scholar] [CrossRef]
- Wang, Y.; Lawler, D.; Larson, B.; Ramadan, Z.; Kochhar, S.; Holmes, E.; Nicholson, J.K. Metabonomic investigations of aging and caloric restriction in a life-long dog study. J. Proteome Res. 2007, 6, 1846–1854. [Google Scholar] [CrossRef]
- Copes, N.; Edwards, C.; Chaput, D.; Saifee, M.; Barjuca, I.; Nelson, D.; Paraggio, A.; Saad, P.; Lipps, D.; Stevens, S.M.; et al. Metabolome and proteome changes with aging in Caenorhabditis elegans. Exp. Gerontol. 2015, 72, 67–84. [Google Scholar] [CrossRef]
- Wan, Q.L.; Shi, X.; Liu, J.; Ding, A.J.; Pu, Y.Z.; Li, Z.; Wu, G.S.; Luo, H.R. Metabolomic signature associated with reproduction-regulated aging in Caenorhabditis elegans. Aging 2017, 9, 447–474. [Google Scholar] [CrossRef]
- Gao, A.W.; Chatzispyrou, I.A.; Kamble, R.; Liu, Y.J.; Herzog, K.; Smith, R.L.; Van Lenthe, H.; Vervaart, M.A.T.; Van Cruchten, A.; Luyf, A.C.; et al. A sensitive mass spectrometry platform identifies metabolic changes of life history traits in C. elegans. Sci. Rep. 2017, 7, 2407. [Google Scholar] [CrossRef]
- Trifonova, O.P.; Maslov, D.L.; Mikhailov, A.N.; Zolotarev, K.V.; Nakhod, K.V.; Nakhod, V.I.; Belyaeva, N.F.; Mikhailova, M.V.; Lokhov, P.G.; Archakov, A.I. Comparative analysis of the blood plasma metabolome of negligible, gradual and rapidly ageing fishes. Fishes 2018, 3, 46. [Google Scholar] [CrossRef]
- Maslov, D.L.; Trifonova, O.P.; Mikhailov, A.N.; Zolotarev, K.V.; Nakhod, K.V.; Nakhod, V.I.; Belyaeva, N.F.; Mikhailova, M.V.; Lokhov, P.G.; Archakov, A.I. Comparative analysis of skeletal muscle metabolites of fish with various rates of aging. Fishes 2019, 4, 25. [Google Scholar] [CrossRef]
- Zhao, X.; Golic, F.T.; Harrison, B.R.; Manoj, M.; Hoffman, E.V.; Simon, N.; Johnson, R.; MacCoss, M.J.; McIntyre, L.M.; Promislow, D.E.L. The metabolome as a biomarker of aging in Drosophila melanogaster. Aging Cell 2022, 21, e13548. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yin, Y.; Li, J.; Wang, H.; Lv, W.; Gao, Y.; Wang, T.; Zhong, Y.; Zhou, Z.; Cai, Y.; et al. Global stable-isotope tracing metabolomics reveals system-wide metabolic alternations in aging Drosophila. Nat. Commun. 2022, 13, 3518. [Google Scholar] [CrossRef] [PubMed]
- Laye, M.J.; Tran, V.; Jones, D.P.; Kapahi, P.; Promislow, D.E.L. The effects of age and dietary restriction on the tissue-specific metabolome of Drosophila. Aging Cell 2015, 14, 797–808. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, H.; Cai, Y.; Wang, H.; Niu, K.; Wu, X.; Ma, H.; Yang, Y.; Tong, W.; Liu, F.; et al. Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. eLife 2018, 7, e35368. [Google Scholar] [CrossRef]
- Tomás-Loba, A.; Bernardes de Jesus, B.; Mato, J.M.; Blasco, M.A. A metabolic signature predicts biological age in mice. Aging Cell 2013, 12, 93–101. [Google Scholar] [CrossRef]
- Han, Q.; Li, H.; Jia, M.; Wang, L.; Zhao, Y.; Zhang, M.; Zhang, Q.; Meng, Z.; Shao, J.; Yang, Y.; et al. Age-related changes in metabolites in young donor livers and old recipient sera after liver transplantation from young to old rats. Aging Cell 2021, 20, e13425. [Google Scholar] [CrossRef]
- Puurunen, J.; Ottka, C.; Salonen, M.; Niskanen, J.E.; Lohi, H. Age, breed, sex and diet influence serum metabolite profiles of 2000 pet dogs. R. Soc. Open Sci. 2022, 9, 211642. [Google Scholar] [CrossRef]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the adult human plasma metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef]
- Darst, B.F.; Koscik, R.L.; Hogan, K.J.; Johnson, S.C.; Engelman, C.D. Longitudinal plasma metabolomics of aging and sex. Aging 2019, 11, 1262–1282. [Google Scholar] [CrossRef]
- Menni, C.; Kastenmüller, G.; Petersen, A.K.; Bell, J.T.; Psatha, M.; Tsai, P.C.; Gieger, C.; Schulz, H.; Erte, I.; John, S.; et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013, 42, 1111–1119. [Google Scholar] [CrossRef]
- Krumsiek, J.; Mittelstrass, K.; Do, K.T.; Stückler, F.; Ried, J.; Adamski, J.; Peters, A.; Illig, T.; Kronenberg, F.; Friedrich, N.; et al. Gender-specific pathway differences in the human serum metabolome. Metabolomics 2015, 11, 1815–1833. [Google Scholar] [CrossRef]
- Tissenbaum, H.A. Using C. elegans for aging research. Invertebr. Reprod. Dev. 2015, 59, 59–63. [Google Scholar] [CrossRef]
- Clark, A.G.; Eisen, M.B.; Smith, D.R.; Bergman, C.M.; Oliver, B.; Markow, T.A.; Kaufman, T.C.; Kellis, M.; Gelbart, W.; Iyer, V.N.; et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef]
- Ma, S.; Avanesov, A.S.; Porter, E.; Lee, B.C.; Mariotti, M.; Zemskaya, N.; Guigo, R.; Moskalev, A.A.; Gladyshev, V.N. Comparative transcriptomics across 14 Drosophila species reveals signatures of longevity. Aging Cell 2018, 17, e12740. [Google Scholar] [CrossRef]
- Patnaik, B.K.; Mahapatro, N.; Jena, B.S. Ageing in fishes. Gerontology 1994, 40, 113–132. [Google Scholar] [CrossRef]
- Anderson, R.M.; Shanmuganayagam, D.; Weindruch, R. Caloric restriction and aging: Studies in mice and monkeys. Toxicol. Pathol. 2009, 37, 47–51. [Google Scholar] [CrossRef]
- Schumacher, B.; Van Der Pluijm, I.; Moorhouse, M.J.; Kosteas, T.; Robinson, A.R.; Suh, Y.; Breit, T.M.; Van Steeg, H.; Niedernhofer, L.J.; Van Ijcken, W.; et al. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008, 4, e1000161. [Google Scholar] [CrossRef]
- Radakovich, L.B.; Pannone, S.C.; Truelove, M.P.; Olver, C.S.; Santangelo, K.S. Hematology and biochemistry of aging—Evidence of “anemia of the elderly” in old dogs. Vet. Clin. Pathol. 2017, 46, 34–45. [Google Scholar] [CrossRef]
- Xenoulis, P.G.; Steiner, J.M. Lipid metabolism and hyperlipidemia in dogs. Vet. J. 2010, 183, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Contrepois, K.; Jiang, L.; Snyder, M. Optimized analytical procedures for the untargeted metabolomic profiling of human urine and plasma by combining hydrophilic interaction (HILIC) and reverse-phase liquid chromatography (RPLC)-mass spectrometry. Mol. Cell Proteom. 2015, 14, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Smith, M.; Buffenstein, R. Naked mole-rat mortality rates defy gompertzian laws by not increasing with age. eLife 2018, 7, 31157. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, J.U.M.; Bennett, N.C. Eusociality has evolved independently in two genera of bathyergid mole-rats—But occurs in no other subterranean mammal. Behav. Ecol. Sociobiol. 1993, 33, 253–260. [Google Scholar] [CrossRef]
- Pryor, S.H.; Hill, R.; Dixson, D.L.; Fraser, N.J.; Kelaher, B.P.; Scott, A. Anemonefish facilitate bleaching recovery in a host sea anemone. Sci. Rep. 2020, 10, 18586. [Google Scholar] [CrossRef]
- Nielsen, J.; Hedeholm, R.B.; Heinemeier, J.; Bushnell, P.G.; Christiansen, J.S.; Olsen, J.; Ramsey, C.B.; Brill, R.W.; Simon, M.; Steffensen, K.F.; et al. Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 2016, 353, 702–704. [Google Scholar] [CrossRef]
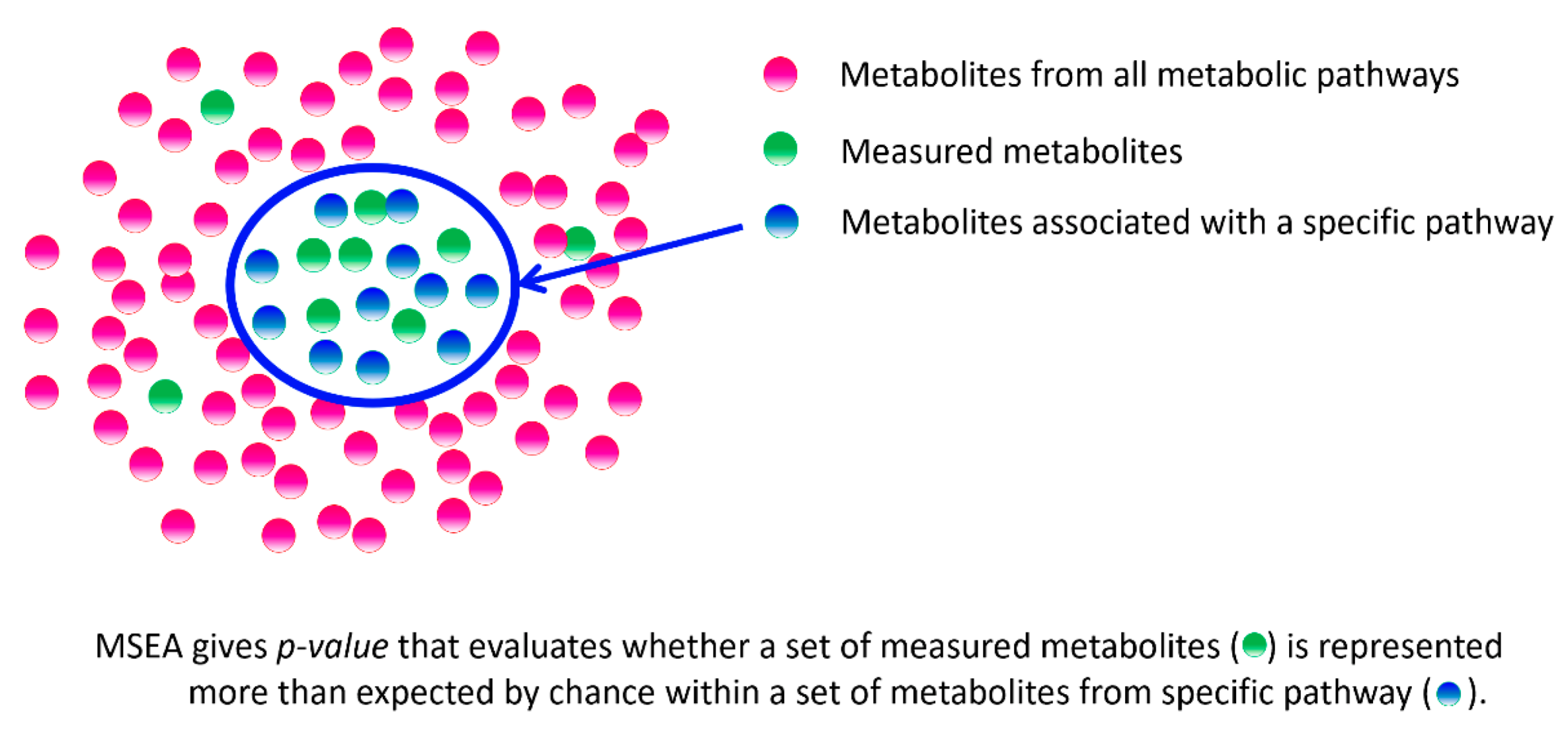
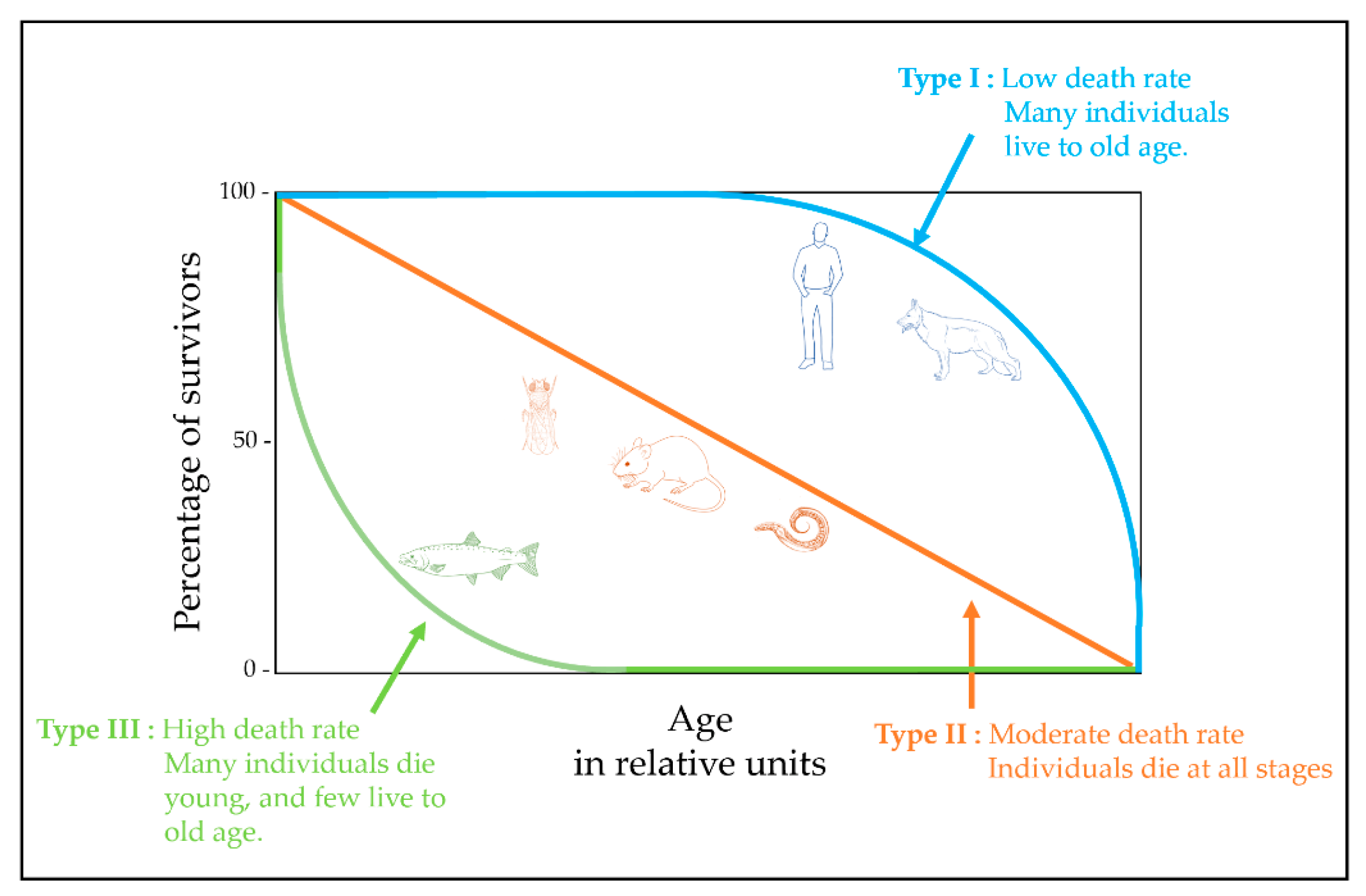
| Characteristics | C. elegans | Drosophila | Fishes | Rodents | Dogs | Humans | |||
|---|---|---|---|---|---|---|---|---|---|
| Pacific Salmon | Pike | Carp | Mice | Rats | |||||
| Similarity to the human genome | ~25% | ~50% | >70% | >70% | >70% | ~83% | ~90% | ~85% | 100% |
| Genome size | 11/12 chromosomes 21,305 genes | 8 chromosomes 14,065 genes | 52–74 chromosomes Up to 40,000 genes | 18 chromosomes ~22,000 genes | 104 chromosomes | 40 chromosomes 29,083 genes | 42 chromosomes ~25,000 genes | 78 chromosomes 36,322 genes | 46 chromosomes 63,494 genes |
| Lifespan 1 | 2–3 weeks | 4–6 weeks | Several years | 7–10 years | 20 years | 1–3 years | 2–3 years | 6–16 years | ∼80 years |
| Age of puberty 1 | 50 h | 10 days | 2–5 years | 3–5 years | 2–5 years | 9–12 weeks | 1.5–3 months | 14–18 months | 10–16 years |
| Number of offspring 1 | 300–1400 offspring | ∼120 eggs | no more than 20,000 eggs | from 18 000 up to 220 000 eggs | up to 1.5 million eggs | 6–12 cubs | 8–10 cubs | 3–8 cubs | 1–2 children |
| Advantages | - low cost of animals and maintenance - no ethical requirements - short LS - easy to work with - genetically tractable - strains can be archived by cryopreservation | - low cost of animals and maintenance - no ethical requirements - the breadth of LS variation - rapid onset of puberty and high fertility - a wide range of phenotypes - some intracellular processes are similar or homologous to human cells | - vertebrates - no need for ethical requirements - the breadth of LS variation - most intracellular processes and many physiological processes are similar to mammals - high fertility | - mammals - high similarity with the human genome - most cellular processes and physiological processes are similar to humans - available for many genetic manipulations - a wide range of phenotypes - strains archived by cryopreservation of embryos and sperm | - mammals - high similarity with the human genome - most cellular processes and physiological processes are similar to humans | - research is most relevant for improving health | |||
| Disadvantages | - relatively simple anatomy - lacks distinct endocrine tissues and various other tissue types - evolutionarily very distant from humans | - strains needed maintain constantly - evolutionarily distant from humans | - evolutionarily distant from humans - long LS | - the expensive cost of animals and maintenance - the need for ethical requirements - relatively long LS - over-reliance on pre-clinical models: many drugs that are effective in mice and rats do not work in humans | - the very expensive cost of animals and maintenance - the need for ethical requirements - long LS | - limited ability to do experiments - the need for ethical requirements - long LS - “diversity of aging” | |||
| Object of Study | Research Materials | Age of Objects | Profiling Methods | Metabolites and Metabolic Pathways that Change with Aging | References |
|---|---|---|---|---|---|
| C. elegans | Whole worms | day 4 (young adult), day 10 (the mean length of LS) | GC–MS | Purine and pyrimidine metabolism, free hydrophobic amino acids, S-adenosylmethionine metabolism, sorbitol, free fatty acids, cellular redox balance, amino acid biosynthesis | [99] |
| Whole worms | young adult and day 10 | NMR spectroscopy, LC–MS | Glutathione metabolism, glutamate metabolism, purine and pyrimidine metabolism, taurine and hypotaurine metabolism, tricarboxylic acid cycle | [100] | |
| Whole worms | days 1, 3, 5, 7, 9 and 10 | LC–MS | Metabolism of fatty acids, amino acids, and phospholipids | [101] | |
| Drosophila | Whole flies | every 2–6 days throughout the life | DIMS | Carbohydrates, amino acids, carnitines, biogenic amines, lipids | [94] |
| Whole flies | days 1–80 | LC–MS | Lifetime dynamics of many metabolites | [51] | |
| Whole flies | days 3, 10, 24, 36, 51, 66, 81 | LC–MS | Metabolism of carbohydrates, glycerophospholipids, neurotransmitters, amino acids, and the carnitine shuttle | [50] | |
| Heads, thoraces, abdomens, whole flies | days 10, 25, and 40 | LC–MS | Metabolism of amino acids and NAD+ | [106] | |
| Whole flies | days 4, 10, 24, 45, 69, 80 | LC–MS | Arginine-ornithine metabolism, tryptophan metabolism | [104] | |
| Whole flies | day 3, day 30 | LC–MS | Glycolysis | [107] | |
| Heads, muscle tissue | day 3, day 30 | LC–MS | Carbohydrate metabolism (galactose, starch, sucrose metabolism), amino acids metabolism (alanine, asparagine, glutamine, serine metabolism), purine metabolism | [105] | |
| Fishes | Blood plasma | 2.4 ± 0.5 1 years 3.4 ± 0.5 1 years 6.7 ± 2.4 1 years 4.3 ± 1.9 1 years 6.1 ± 1.9 1 years 4.0 ± 0.4 1 years (from groups of short-lived to long-lived fish species) | DIMS | Dipeptides, di- and triglycerides, fatty acids, phosphoethanolamines, and phosphatidylcholines | [102] |
| Skeletal muscles | Amino acids, lipids, biogenic amines, intermediates of glycolysis, glycogenolysis, and the citric acid cycle | [103] | |||
| Mice | Blood plasma, muscle tissue (quadriceps), liver | 13 weeks (“young”), 93 weeks (“old”) | GC–MS, LC–MS | Metabolism of fatty acids and glucose | [62] |
| Serum | 8–129 weeks | LC–MS | Phospholipids, fatty acids, organic acids, creatine, methionine, uric acid | [108] | |
| Serum, urine | 8, 12, 16, and 20 weeks (mutants with accelerated aging) | NMR spectroscopy | Changes in lipid and energy metabolism, transition to ketosis | [82] | |
| Rats | Liver, serum | 3–5 months (young), 15–17 months (old) | LC–MS | Organic acids and their derivatives, lipids and lipid-like molecules, glycerophospholipids, arachidonic acid, histidine, linoleate | [109] |
| Dogs | Urine | 13, 18, 32 weeks, 1, 1.5, and 2 years, annually after 5 years until the death | NMR spectroscopy | Metabolites associated with energy metabolism | [98] |
| Serum | 1 month-16 years | NMR spectroscopy | Lipids, cholesterols, triglycerides, lipoproteins, protein glycosylation marker GlycA | [110] | |
| Humans | Blood plasma | 20–65 years | LC–MS, GC–MS | Tricarboxylic acid intermediates, creatine, essential and non-essential amino acids, urea, ornithine, polyamines, markers of oxidative stress, lipid metabolism products (including fatty acids, carnitine, β-hydroxybutyrate, and cholesterol), dehydroepiandrosterone sulfate (putative antiaging androgen), xenobiotics (e.g., caffeine) | [111] |
| Whole blood, blood plasma, and erythrocytes | 29 ± 4 1 years (young), 81 ± 7 1 years (elder) | LC–MS | 1,5-anhydroglucitol, dimethylguanosine, acetylcarnosine, carnosine, ophthalmic acid, UDP-acetylglucosamine, N-acetylarginine, N6-acetyllysine, pantothenate, citrulline, leucine, isoleucine, NAD+, and NADP+ | [95] | |
| Blood plasma | 6 months–82 years | LC–MS | Metabolism of progestin steroids, xanthine, and long-chain fatty acids | [58] | |
| Blood plasma | every two years from middle-aged adults for 10 years | LC–MS | Sphingolipids, lipid steroids (including androgens, progestins, and pregnenolones), amino acids | [112] | |
| Blood plasma, serum | 17–85 years | LC–MS | Lipids (long-chain fatty acids, polyunsaturated fatty acids, and other fatty acids), amino acids (including glutamine, tyrosine, histidine) | [113] | |
| Serum | 60.51 ± 8.77 1 for females, 61.17 ± 8.79 1 for males | LC–MS, GC–MS | Amino acids, lipids (fatty acids, androgenic steroids) | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balashova, E.E.; Maslov, D.L.; Trifonova, O.P.; Lokhov, P.G.; Archakov, A.I. Metabolome Profiling in Aging Studies. Biology 2022, 11, 1570. https://doi.org/10.3390/biology11111570
Balashova EE, Maslov DL, Trifonova OP, Lokhov PG, Archakov AI. Metabolome Profiling in Aging Studies. Biology. 2022; 11(11):1570. https://doi.org/10.3390/biology11111570
Chicago/Turabian StyleBalashova, Elena E., Dmitry L. Maslov, Oxana P. Trifonova, Petr G. Lokhov, and Alexander I. Archakov. 2022. "Metabolome Profiling in Aging Studies" Biology 11, no. 11: 1570. https://doi.org/10.3390/biology11111570
APA StyleBalashova, E. E., Maslov, D. L., Trifonova, O. P., Lokhov, P. G., & Archakov, A. I. (2022). Metabolome Profiling in Aging Studies. Biology, 11(11), 1570. https://doi.org/10.3390/biology11111570










