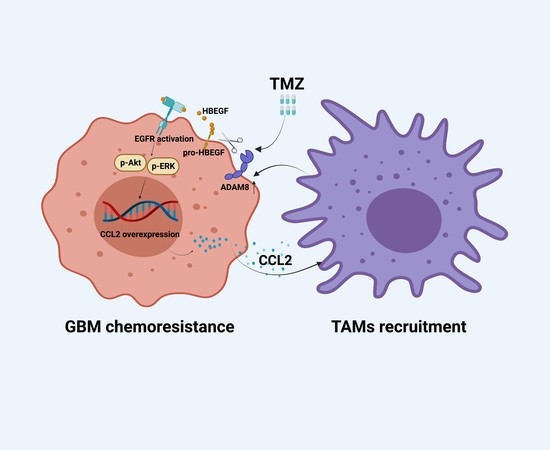
The GBM Tumor Microenvironment as a Modulator of Therapy Response: ADAM8 Causes Tumor Infiltration of Tams through HB-EGF/EGFR-Mediated CCL2 Expression and Overcomes TMZ Chemosensitization in Glioblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Specimens
2.2. Cell Lines, Cell Culture, and Construction of Stable Cell Lines
2.3. Antibodies and Agents
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. ELISA
2.6. Cell Proliferation Assay
2.7. Cell Migration and Invasion Assays
2.8. Immunofluorescence Staining
2.9. Immunohistochemistry Staining
2.10. Xenograft Studies in Nude Mice
2.11. Western Blot
2.12. RNA—Seq Data Analysis
2.13. Statistical Analysis
3. Results
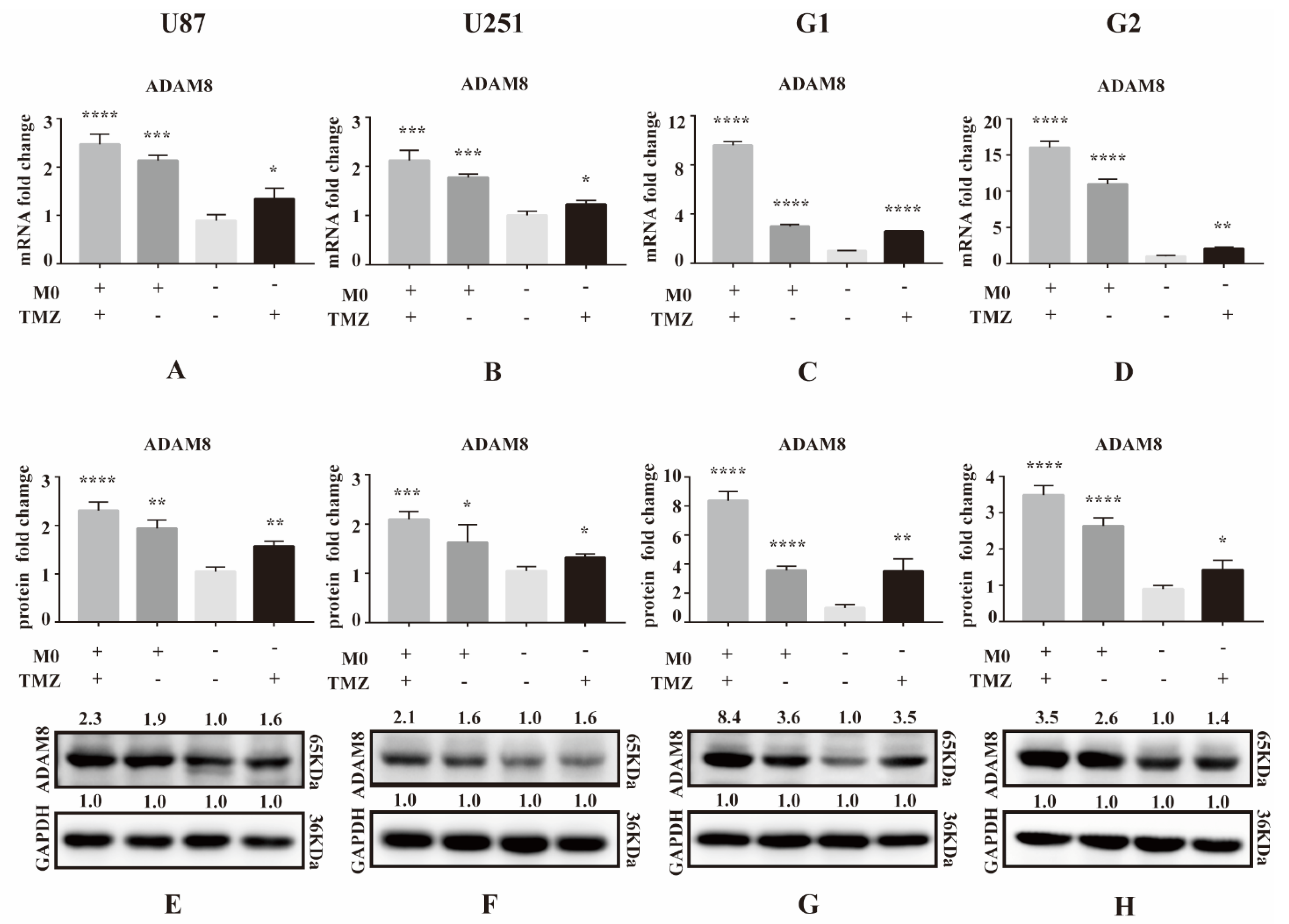
3.1. ADAM8 Expression Induced by TMZ and Macrophage Co-Culture
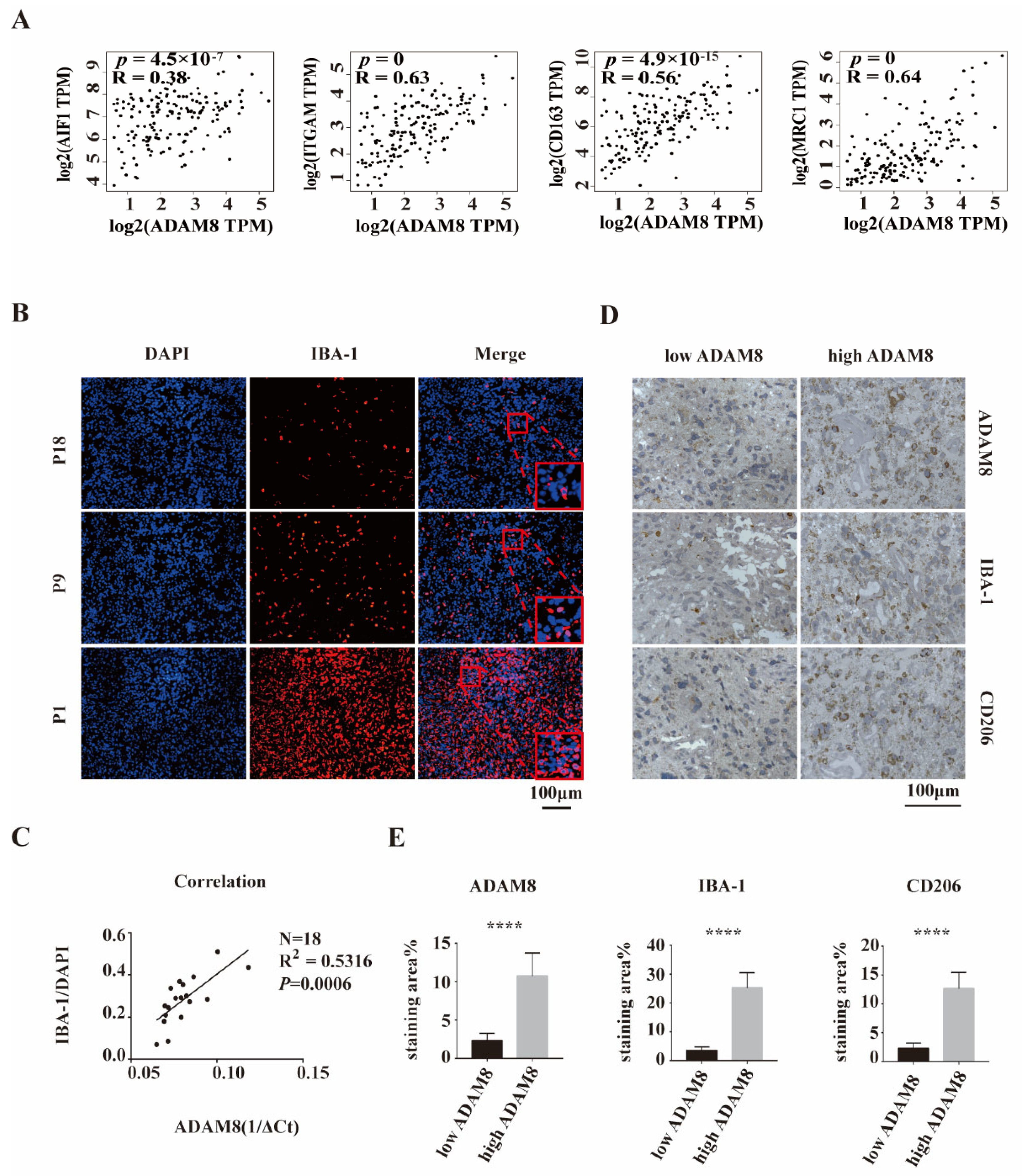
3.2. Positive Correlation of ADAM8 Expression and Macrophages Infiltration in GBM Tissue
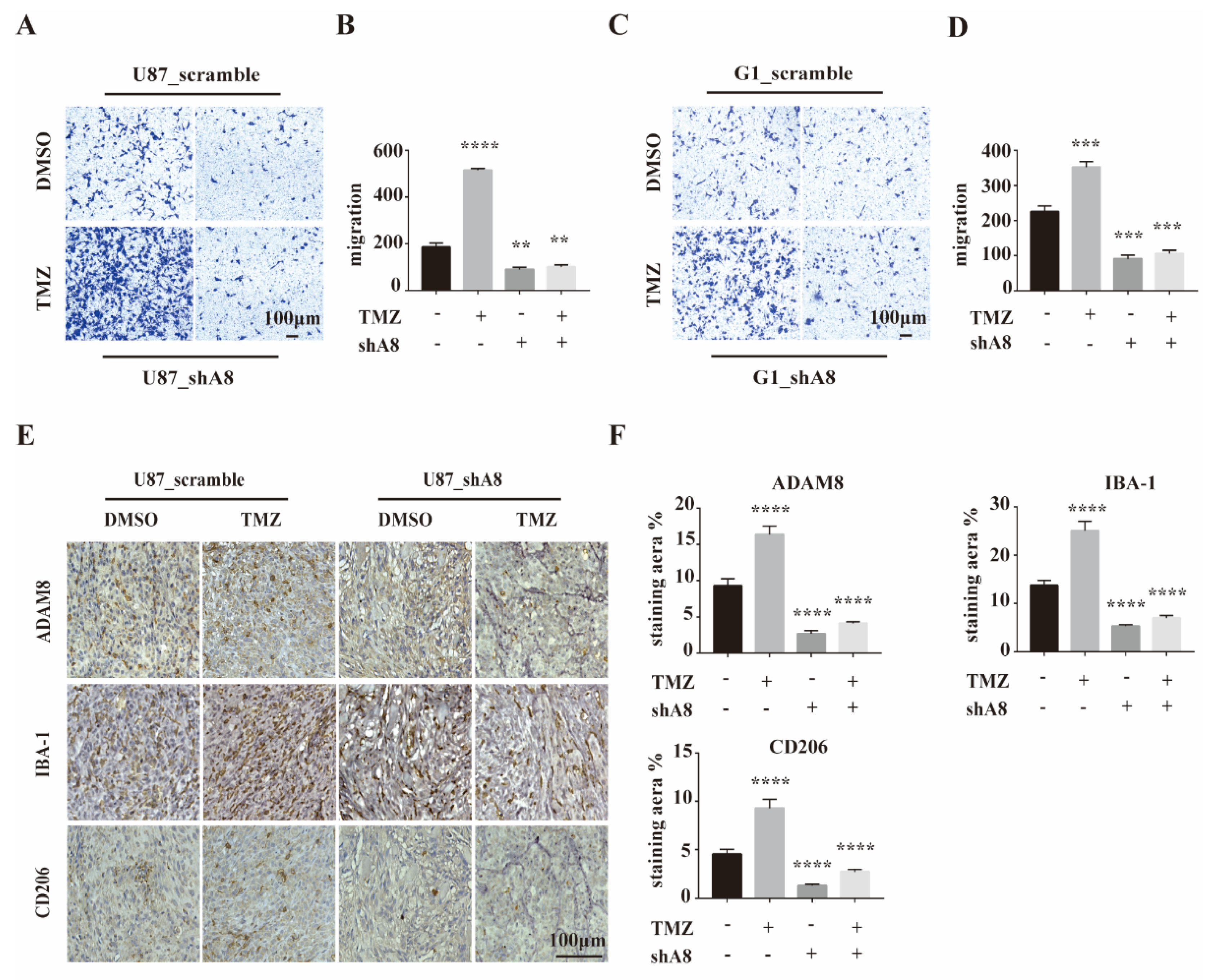
3.3. ADAM8 Induces Macrophage Recruitment In Vitro and In Vivo
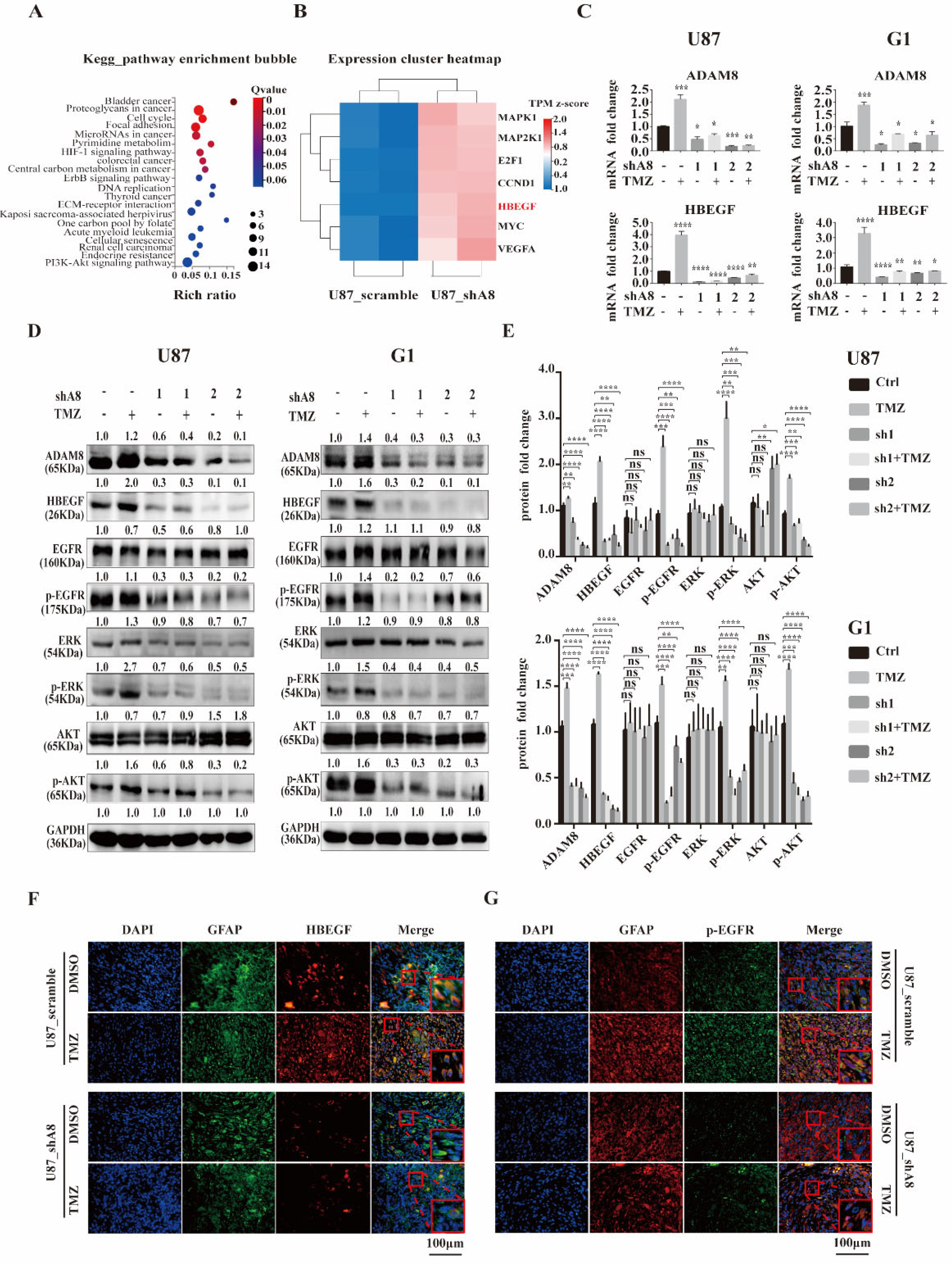
3.4. ADAM8 Regulates the HB-EGF/EGFR Signal Pathway
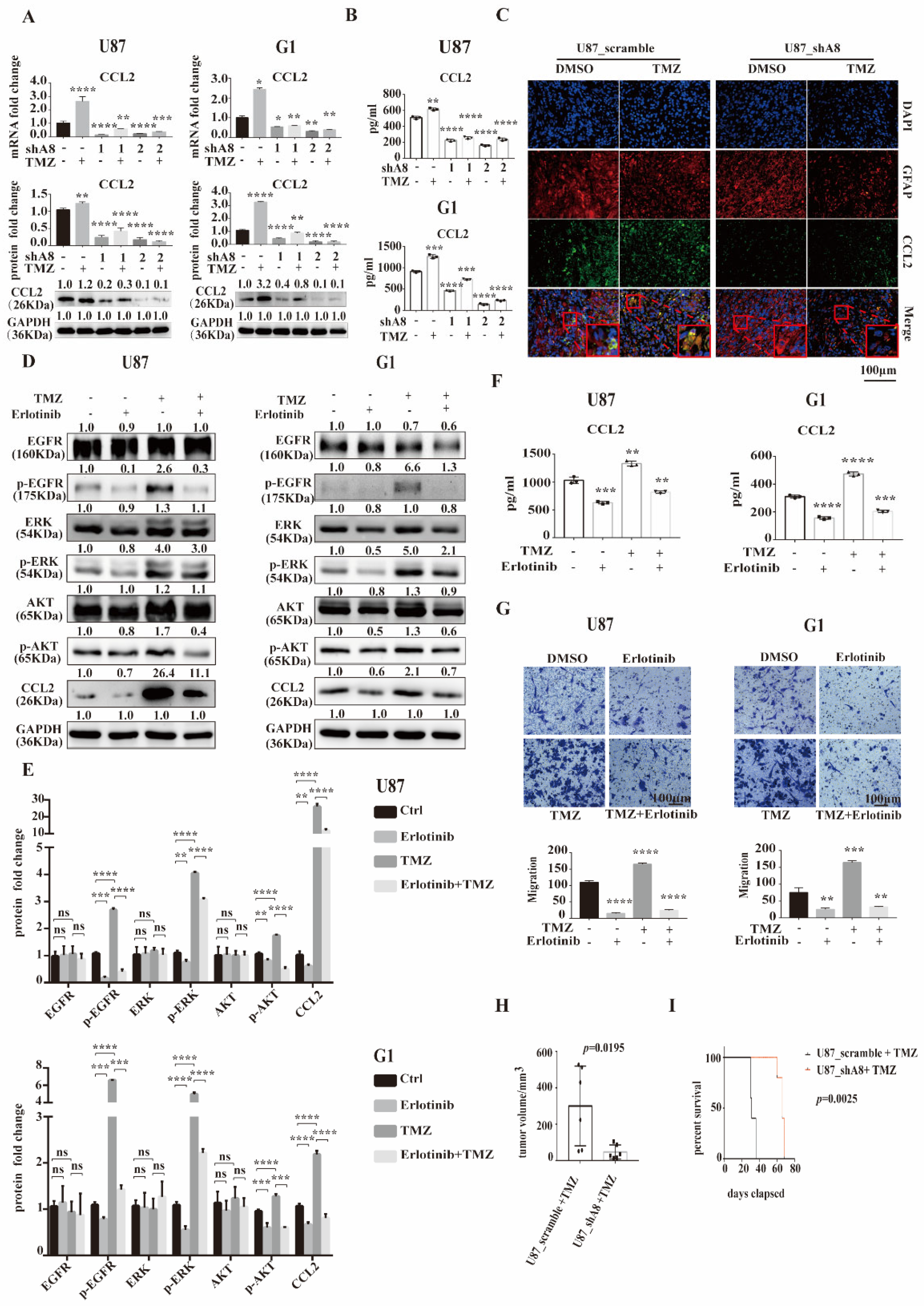
3.5. ADAM8 Induce HB-EGF/EGFR Mediated CCL2 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; de Blank, P.M.; Finlay, J.L.; Gurney, J.G.; McKean-Cowdin, R.; Stearns, D.S.; Wolff, J.E.; Liu, M.; Wolinsky, Y.; et al. American Brain Tumor Association Adolescent and Young Adult Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol. 2016, 18 (Suppl. S1), i1–i50. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol. 2015, 17 (Suppl. S4), iv1–iv62. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.P.; Adamson, D.C. Current FDA-Approved Therapies for High-Grade Malignant Gliomas. Biomedicines 2021, 9, 324. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Maugeri-Sacca, M.; Bartucci, M.; De Maria, R. DNA damage repair pathways in cancer stem cells. Mol. Cancer Ther. 2012, 11, 1627–1636. [Google Scholar] [CrossRef]
- Agnihotri, S.; Gajadhar, A.S.; Ternamian, C.; Gorlia, T.; Diefes, K.L.; Mischel, P.S.; Kelly, J.; McGown, G.; Thorncroft, M.; Carlson, B.L.; et al. Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J. Clin. Investig. 2012, 122, 253–266. [Google Scholar] [CrossRef]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef]
- Geraldo, L.H.M.; Garcia, C.; da Fonseca, A.C.C.; Dubois, L.G.F.; de Sampaio, E.S.T.C.L.; Matias, D.; de Camargo Magalhaes, E.S.; do Amaral, R.F.; da Rosa, B.G.; Grimaldi, I.; et al. Glioblastoma Therapy in the Age of Molecular Medicine. Trends Cancer 2019, 5, 46–65. [Google Scholar] [CrossRef]
- Hou, J.; Karin, M.; Sun, B. Targeting cancer-promoting inflammation—Have anti-inflammatory therapies come of age? Nat. Rev. Clin. Oncol. 2021, 18, 261–279. [Google Scholar] [CrossRef]
- Buonfiglioli, A.; Hambardzumyan, D. Macrophages and microglia: The cerberus of glioblastoma. Acta Neuropathol. Commun. 2021, 9, 54. [Google Scholar] [CrossRef]
- Mitchem, J.B.; Brennan, D.J.; Knolhoff, B.L.; Belt, B.A.; Zhu, Y.; Sanford, D.E.; Belaygorod, L.; Carpenter, D.; Collins, L.; Piwnica-Worms, D.; et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013, 73, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Zhang, I.Y.; Chen, X.; Da Fonseca, A.; Wu, S.; Ren, H.; Badie, S.; Sadeghi, S.; Ouyang, M.; et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin. Cancer Res. 2013, 19, 3764–3775. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Dzaye, O.; Hahn, A.; Yu, Y.; Scavetta, R.J.; Dittmar, G.; Kaczmarek, A.K.; Dunning, K.R.; Ricciardelli, C.; Rinnenthal, J.L.; et al. Glioma-derived versican promotes tumor expansion via glioma-associated microglial/macrophages Toll-like receptor 2 signaling. Neuro Oncol. 2015, 17, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ke, S.Q.; Huang, Z.; Flavahan, W.; Fang, X.; Paul, J.; Wu, L.; Sloan, A.E.; McLendon, R.E.; Li, X.; et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015, 17, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.; Benzel, J.; Dorzweiler, K.; Cook, L.; Schlomann, U.; Zarbock, A.; Slater, E.P.; Nimsky, C.; Bartsch, J.W. ADAM8 in invasive cancers: Links to tumor progression, metastasis, and chemoresistance. Clin. Sci. 2019, 133, 83–99. [Google Scholar] [CrossRef]
- Schlomann, U.; Rathke-Hartlieb, S.; Yamamoto, S.; Jockusch, H.; Bartsch, J.W. Tumor necrosis factor alpha induces a metalloprotease-disintegrin, ADAM8 (CD 156): Implications for neuron-glia interactions during neurodegeneration. J. Neurosci. 2000, 20, 7964–7971. [Google Scholar] [CrossRef]
- Koller, G.; Schlomann, U.; Golfi, P.; Ferdous, T.; Naus, S.; Bartsch, J.W. ADAM8/MS2/CD156, an emerging drug target in the treatment of inflammatory and invasive pathologies. Curr. Pharm. Des. 2009, 15, 2272–2281. [Google Scholar] [CrossRef]
- Hsia, H.E.; Tushaus, J.; Brummer, T.; Zheng, Y.; Scilabra, S.D.; Lichtenthaler, S.F. Functions of ‘A disintegrin and metalloproteases (ADAMs)’ in the mammalian nervous system. Cell. Mol. Life Sci. 2019, 76, 3055–3081. [Google Scholar] [CrossRef]
- Wildeboer, D.; Naus, S.; Amy Sang, Q.X.; Bartsch, J.W.; Pagenstecher, A. Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. J. Neuropathol. Exp. Neurol. 2006, 65, 516–527. [Google Scholar] [CrossRef]
- He, S.; Ding, L.; Cao, Y.; Li, G.; Deng, J.; Tu, Y.; Wang, B. Overexpression of a disintegrin and metalloprotease 8 in human gliomas is implicated in tumor progression and prognosis. Med. Oncol. 2012, 29, 2032–2037. [Google Scholar] [CrossRef]
- Li, Y.; Guo, S.; Zhao, K.; Conrad, C.; Driescher, C.; Rothbart, V.; Schlomann, U.; Guerreiro, H.; Bopp, M.H.; Konig, A.; et al. ADAM8 affects glioblastoma progression by regulating osteopontin-mediated angiogenesis. Biol. Chem. 2021, 402, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.R.; Markesbery, M.G.; Harding, P.A. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) and proteolytic processing by a disintegrin and metalloproteinases (ADAM): A regulator of several pathways. Semin. Cell Dev. Biol. 2014, 28, 22–30. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Knobbe-Thomsen, C.B.; Wan, X.; Fan, Q.W.; Reifenberger, G.; Weiss, W.A. EGFR Cooperates with EGFRvIII to Recruit Macrophages in Glioblastoma. Cancer Res. 2018, 78, 6785–6794. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Eibach, M.; Bartsch, J.W.; Dolga, A.M.; Schlomann, U.; Conrad, C.; Schieber, S.; Schilling, O.; Biniossek, M.L.; Culmsee, C.; et al. The metalloprotease-disintegrin ADAM8 contributes to temozolomide chemoresistance and enhanced invasiveness of human glioblastoma cells. Neuro Oncol. 2015, 17, 1474–1485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, J.; Yao, X.; Wu, S.; Tian, W.; Gan, C.; Wan, X.; You, C.; Hu, F.; Zhang, S.; et al. Programmed Cell Death 10 Mediated CXCL2-CXCR2 Signaling in Regulating Tumor-Associated Microglia/Macrophages Recruitment in Glioblastoma. Front. Immunol. 2021, 12, 637053. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef]
- Andersen, R.S.; Anand, A.; Harwood, D.S.L.; Kristensen, B.W. Tumor-Associated Microglia and Macrophages in the Glioblastoma Microenvironment and Their Implications for Therapy. Cancers 2021, 13, 4255. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Dang, W.Q.; Cao, M.F.; Xiao, J.F.; Lv, S.Q.; Jiang, W.J.; Yao, X.H.; Lu, H.M.; Miao, J.Y.; et al. CCL8 secreted by tumor-associated macrophages promotes invasion and stemness of glioblastoma cells via ERK1/2 signaling. Lab. Investig. 2020, 100, 619–629. [Google Scholar] [CrossRef]
- Liu, X.J.; Liu, Y.; Qi, Y.W.; Huang, Y.M.; Hu, F.; Dong, F.Y.; Shu, K.; Lei, T. Signal Pathways Involved in the Interaction between Tumor-Associated Macrophages/TAMs and Glioblastoma Cells. Front. Oncol. 2022, 12. [Google Scholar] [CrossRef]
- Puolakkainen, P.; Koski, A.; Vainionpaa, S.; Shen, Z.; Repo, H.; Kemppainen, E.; Mustonen, H.; Seppanen, H. Anti-inflammatory macrophages activate invasion in pancreatic adenocarcinoma by increasing the MMP9 and ADAM8 expression. Med. Oncol. 2014, 31, 884. [Google Scholar] [CrossRef] [PubMed]
- Schafer, A.; Evers, L.; Meier, L.; Schlomann, U.; Bopp, M.H.A.; Dreizner, G.L.; Lassmann, O.; Ben Bacha, A.; Benescu, A.C.; Pojskic, M.; et al. The Metalloprotease-Disintegrin ADAM8 Alters the Tumor Suppressor miR-181a-5p Expression Profile in Glioblastoma Thereby Contributing to Its Aggressiveness. Front. Oncol. 2022, 12, 826273. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Marisetty, A.; Schrand, B.; Gabrusiewicz, K.; Hashimoto, Y.; Ott, M.; Grami, Z.; Kong, L.Y.; Ling, X.; Caruso, H.; et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2019, 129, 137–149. [Google Scholar] [CrossRef]
- Tao, W.; Chu, C.; Zhou, W.; Huang, Z.; Zhai, K.; Fang, X.; Huang, Q.; Zhang, A.; Wang, X.; Yu, X.; et al. Dual Role of WISP1 in maintaining glioma stem cells and tumor-supportive macrophages in glioblastoma. Nat. Commun. 2020, 11, 3015. [Google Scholar] [CrossRef]
- Han, S.; Zhen, W.; Guo, T.; Zou, J.; Li, F. SETDB1 promotes glioblastoma growth via CSF-1-dependent macrophage recruitment by activating the AKT/mTOR signaling pathway. J. Exp. Clin. Cancer Res. 2020, 39, 218. [Google Scholar] [CrossRef] [PubMed]
- Larionova, I.; Cherdyntseva, N.; Liu, T.; Patysheva, M.; Rakina, M.; Kzhyshkowska, J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology 2019, 8, 1596004. [Google Scholar] [CrossRef] [PubMed]
- Dreymueller, D.; Pruessmeyer, J.; Schumacher, J.; Fellendorf, S.; Hess, F.M.; Seifert, A.; Babendreyer, A.; Bartsch, J.W.; Ludwig, A. The metalloproteinase ADAM8 promotes leukocyte recruitment in vitro and in acute lung inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 313, L602–L614. [Google Scholar] [CrossRef]
- Munoz, J.L.; Rodriguez-Cruz, V.; Greco, S.J.; Nagula, V.; Scotto, K.W.; Rameshwar, P. Temozolomide induces the production of epidermal growth factor to regulate MDR1 expression in glioblastoma cells. Mol. Cancer Ther. 2014, 13, 2399–2411. [Google Scholar] [CrossRef]
- Meng, X.; Zhao, Y.; Han, B.; Zha, C.; Zhang, Y.; Li, Z.; Wu, P.; Qi, T.; Jiang, C.; Liu, Y.; et al. Dual functionalized brain-targeting nanoinhibitors restrain temozolomide-resistant glioma via attenuating EGFR and MET signaling pathways. Nat. Commun. 2020, 11, 594. [Google Scholar] [CrossRef]
- Huang, K.; Liu, X.; Li, Y.; Wang, Q.; Zhou, J.; Wang, Y.; Dong, F.; Yang, C.; Sun, Z.; Fang, C.; et al. Genome-Wide CRISPR-Cas9 Screening Identifies NF-kappaB/E2F6 Responsible for EGFRvIII-Associated Temozolomide Resistance in Glioblastoma. Adv. Sci. 2019, 6, 1900782. [Google Scholar] [CrossRef]
- Higashiyama, S.; Nanba, D. ADAM-mediated ectodomain shedding of HB-EGF in receptor cross-talk. Biochim. Biophys. Acta 2005, 1751, 110–117. [Google Scholar] [CrossRef]
- Felsenstein, M.; Blank, A.; Bungert, A.D.; Mueller, A.; Ghori, A.; Kremenetskaia, I.; Rung, O.; Broggini, T.; Turkowski, K.; Scherschinski, L.; et al. CCR2 of Tumor Microenvironmental Cells Is a Relevant Modulator of Glioma Biology. Cancers 2020, 12, 1882. [Google Scholar] [CrossRef] [PubMed]
- Vakilian, A.; Khorramdelazad, H.; Heidari, P.; Sheikh Rezaei, Z.; Hassanshahi, G. CCL2/CCR2 signaling pathway in glioblastoma multiforme. Neurochem. Int. 2017, 103, 1–7. [Google Scholar] [CrossRef]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Liu, Q.; Dou, R.; Xiong, B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer 2019, 18, 64. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Kretz, A.; Naumann, U.; Aulwurm, S.; Egashira, K.; Isenmann, S.; Weller, M. Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann. Neurol. 2003, 54, 388–392. [Google Scholar] [CrossRef]
- Chen, Z.; Feng, X.; Herting, C.J.; Garcia, V.A.; Nie, K.; Pong, W.W.; Rasmussen, R.; Dwivedi, B.; Seby, S.; Wolf, S.A.; et al. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res. 2017, 77, 2266–2278. [Google Scholar] [CrossRef]
- Hutter, G.; Theruvath, J.; Graef, C.M.; Zhang, M.; Schoen, M.K.; Manz, E.M.; Bennett, M.L.; Olson, A.; Azad, T.D.; Sinha, R.; et al. Microglia are effector cells of CD47-SIRPalpha antiphagocytic axis disruption against glioblastoma. Proc. Natl. Acad. Sci. USA 2019, 116, 997–1006. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Huang, Y.; Qi, Y.; Wu, S.; Hu, F.; Wang, J.; Shu, K.; Zhang, H.; Bartsch, J.W.; Nimsky, C.; et al. The GBM Tumor Microenvironment as a Modulator of Therapy Response: ADAM8 Causes Tumor Infiltration of Tams through HB-EGF/EGFR-Mediated CCL2 Expression and Overcomes TMZ Chemosensitization in Glioblastoma. Cancers 2022, 14, 4910. https://doi.org/10.3390/cancers14194910
Liu X, Huang Y, Qi Y, Wu S, Hu F, Wang J, Shu K, Zhang H, Bartsch JW, Nimsky C, et al. The GBM Tumor Microenvironment as a Modulator of Therapy Response: ADAM8 Causes Tumor Infiltration of Tams through HB-EGF/EGFR-Mediated CCL2 Expression and Overcomes TMZ Chemosensitization in Glioblastoma. Cancers. 2022; 14(19):4910. https://doi.org/10.3390/cancers14194910
Chicago/Turabian StyleLiu, Xiaojin, Yimin Huang, Yiwei Qi, Shiqiang Wu, Feng Hu, Junwen Wang, Kai Shu, Huaqiu Zhang, Jörg W. Bartsch, Christopher Nimsky, and et al. 2022. "The GBM Tumor Microenvironment as a Modulator of Therapy Response: ADAM8 Causes Tumor Infiltration of Tams through HB-EGF/EGFR-Mediated CCL2 Expression and Overcomes TMZ Chemosensitization in Glioblastoma" Cancers 14, no. 19: 4910. https://doi.org/10.3390/cancers14194910
APA StyleLiu, X., Huang, Y., Qi, Y., Wu, S., Hu, F., Wang, J., Shu, K., Zhang, H., Bartsch, J. W., Nimsky, C., Dong, F., & Lei, T. (2022). The GBM Tumor Microenvironment as a Modulator of Therapy Response: ADAM8 Causes Tumor Infiltration of Tams through HB-EGF/EGFR-Mediated CCL2 Expression and Overcomes TMZ Chemosensitization in Glioblastoma. Cancers, 14(19), 4910. https://doi.org/10.3390/cancers14194910