Properties of a Novel Salmonella Phage L66 and Its Application Based on Electrochemical Sensor-Combined AuNPs to Detect Salmonella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical and Materials
2.2. Bacterial Strains, Bacteriophage, and Growth Conditions
2.3. Characteristics of Phage L66
2.3.1. Morphological Analysis
2.3.2. Host Lytic Range
2.3.3. Optimal Multiplicity of Infection
2.3.4. Adsorption Rate
2.3.5. One-Step Growth Curve
2.3.6. pH and Temperature Stability
2.4. Genome Sequencing, Annotation, and Comparison Analysis of Phage L66
2.5. Construction of Phage L66-Based Electrochemical Sensors
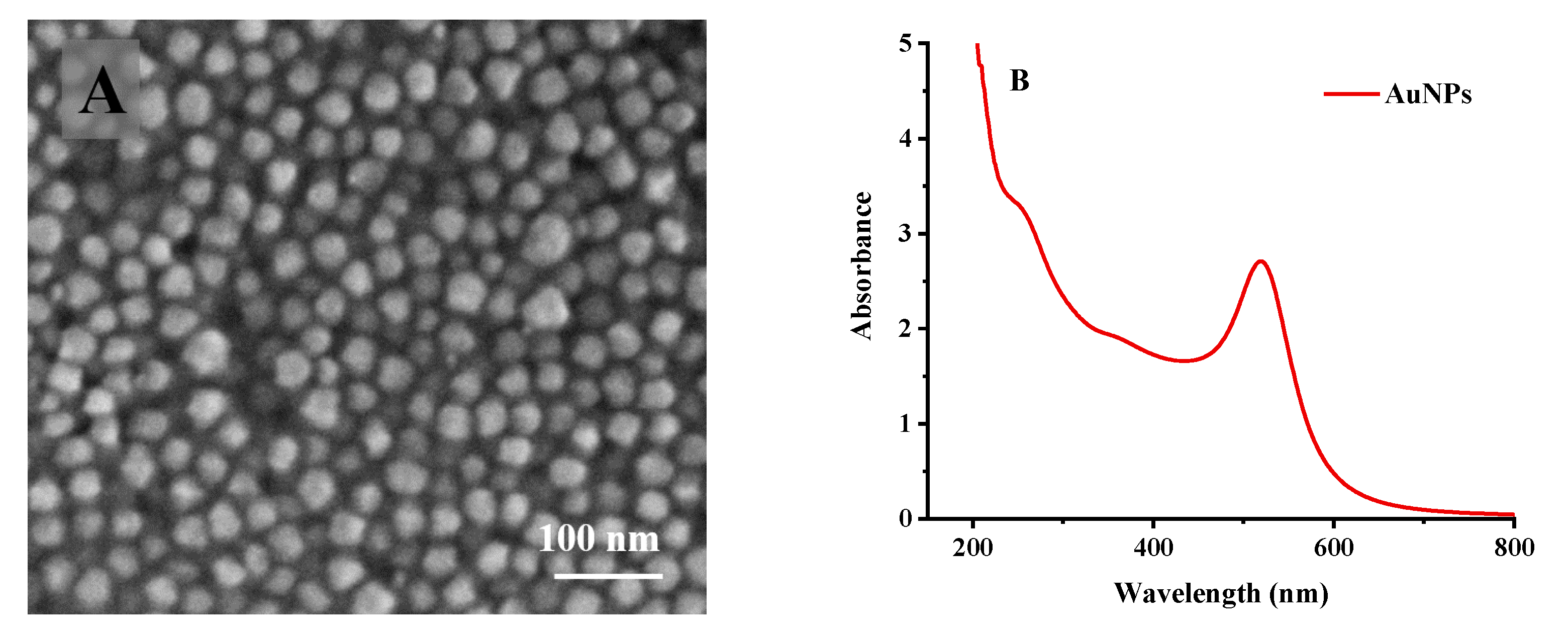
2.5.1. Activation of Electrode and Preparation of Gold Nanoparticles
2.5.2. Preparation of GDE-AuNPs-MPA-Phage L66 Composite Electrode
2.5.3. The Specificity and Stability Evaluation of the Sensor
2.6. Determination of Salmonella in Contaminated Food
2.7. Statistical Analysis
3. Results and Discussion
3.1. Characterization of Phage L66
3.1.1. Morphology of Phage L66
3.1.2. Host Range Properties of Phage L66
3.1.3. Basic Biological Characteristics of Phage L66
3.2. Bioinformatics Analysis of Phage L66 Genome
3.2.1. Analysis of the Phage L66 Genome
3.2.2. Evolutionary Analysis of Phage L66
3.3. Construction and Evaluation of Phage L66-Based Sensors
3.3.1. Optimization of Fabrication Conditions of Phage L66-Based Biosensor
3.3.2. Characterization of Phage L66-Based Biosensor
3.3.3. Standard Curve of Biosensor Measurement
3.3.4. Specificity of Phage L66-Based Biosensor
3.3.5. Stability of Phage L66-Based Biosensor
3.3.6. Detection of Salmonella in Different Substrates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pepe, T.; De Dominicis, R.; Esposito, G.; Ventrone, I.; Fratamico, P.M.; Cortesi, M.L. Detection of Campylobacter from poultry carcass skin samples at slaughter in Southern Italy. J. Food Prot. 2009, 72, 1718–1721. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, E.; Morales-Rueda, A.; Garcia-Gimeno, R.M. Cross-contamination and recontamination by Salmonella in foods: A review. Food Res. Int. 2012, 45, 545–556. [Google Scholar] [CrossRef]
- Humphrey, T.; Jorgensen, F. Pathogens on meat and infection in animals—Establishing a relationship using campylobacter and Salmonella as examples. Meat Sci. 2006, 74, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Tae-Hyun, L.; Myung-Seob, K.; Dong-Hun, L.; Yu-Na, L.; Jae-Keun, P.; Ha-Na, Y.; Hyun-Jeong, L.; Si-Yong, Y.; Young-Wook, C.; Joong-Bok, L. Use of bacteriophage for biological control of Salmonella Enteritidis infection in chicken. Res. Vet. Sci. 2012, 93, 1173–1178. [Google Scholar]
- Kosznik-Kwaśnicka, K.; Stasiłojć, M.; Grabowski, Ł.; Zdrojewska, K.; Węgrzyn, G.; Węgrzyn, A. Efficacy and safety of phage therapy against Salmonella enterica serovars Typhimurium and Enteritidis estimated by using a battery of in vitro tests and the Galleria mellonella animal model. Microbiol. Res. 2022, 261, 7052–7067. [Google Scholar] [CrossRef]
- Huang, C.; Shi, J.; Ma, W.; Li, Z.; Wang, J.; Li, J.; Wang, X. Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 2018, 111, 631–641. [Google Scholar] [CrossRef]
- Li, Z.W.; Ma, W.J.; Li, W.N.; Ding, Y.F.; Zhang, Y.; Yang, Q.L.; Wang, J.; Wang, X.H. A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 2020, 132, 9963–9969. [Google Scholar] [CrossRef] [PubMed]
- Moghimian, P.; Srot, V.; Pichon, B.P.; Facey, S.J.; Aken, P.A.V. Stability of M13 Phage in Organic Solvents. Biomater. Nanotechnol. 2016, 7, 72–77. [Google Scholar] [CrossRef]
- Mcleod, S.M.; Kimsey, H.H.; Davis, B.M.; Waldor, M.K. CTXphi and Vibrio cholerae: Exploring a newly recognized type of phage-host cell relationship. Mol. Microbiol. 2010, 57, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Lambrianou, A.; Demin, S.; Hall, E.A.H. Protein Engineering and Electrochemical Biosensors. Adv. Biochem. Eng. Biotechnol. 2008, 109, 65–96. [Google Scholar] [PubMed]
- Pérez-Ferńandez, B.; de la Escosura-Muniz, A. Electrochemical biosensors based on nanomaterials for aflatoxins detection: A review. Anal. Chim. Acta 2022, 1212, 339658. [Google Scholar] [CrossRef] [PubMed]
- Sedki, M.; Chen, C.; Ge, X.; Mulchandani, A. Non-lytic M13 phage-based highly sensitive impedimetric cytosensor for detection of coliforms. Biosens. Bioelectron. 2019, 148, 111794. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, Y.; Narita, F. Design and finite element simulation of metal-core piezoelectric fiber/epoxy matrix composites for virus detection. Sens. Actuators A Phys. 2021, 327, 112742. [Google Scholar] [CrossRef]
- Dong, J. Design and analysis of surface plasmon resonance sensor based on multi-core photonic crystal fiber. Optik 2022, 266, 169641. [Google Scholar] [CrossRef]
- Manera, M.G.; Pellegrini, G.; Lupo, P.; Bello, V.; Fernandez, C.; de Julian, C.; Casoli, F.; Rella, S.; Malitesta, C.; Albertini, F.; et al. Functional magneto-plasmonic biosensors transducers: Modelling and nanoscale analysis. Sens. Actuators B Chem. 2017, 239, 100–112. [Google Scholar] [CrossRef]
- Sargazi, S. Fluorescent-based nanosensors for selective detection of a wide range of biological macromolecules: A comprehensive review. Int. J. Biol. Macromol. 2022, 206, 115–147. [Google Scholar] [CrossRef]
- Wang, J.; Cui, X.; Zhu, J.; Tan, L.; Dong, L. A novel photosensitive dual-sensor for simultaneous detection of nucleic acids and small chemical molecules. Biosens. Bioelectron. 2019, 127, 108–117. [Google Scholar] [CrossRef]
- Yue, H.; He, Y.; Wang, L.; Lu, S.; Fu, Z. Label-free electrochemiluminescent biosensor for rapid and sensitive detection of pseudomonas aeruginosa using phage as highly specific recognition agent. Biosens. Bioelectron. 2017, 94, 429–432. [Google Scholar] [CrossRef]
- Farooq, U.; Yang, Q.L.; Ullah, M.W.; Wang, S.Q. Bacterial biosensing: Recent advances in phage-based bioassays and biosensors. Biosens. Bioelectron. 2018, 118, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Erturk, G.; Lood, R. Bacteriophages as biorecognition elements in capacitive biosensors: Phage and host bacteria detection. Sens. Actuators B Chem. 2018, 258, 535–543. [Google Scholar] [CrossRef]
- Horikawa, S.; Li, S.; Bedi, D.; Wen, S.; Chin, B.A. Effects of Surface Phage Coverage on the Performance of Wireless Phage-Immobilized Magnetoelastic Biosensors. ECS Trans. 2010, 33, 41–48. [Google Scholar] [CrossRef]
- Handa, H.; Gurczynski, S.; Jackson, M.; Auner, G.; Walker, J.; Mao, G. Recognition of Salmonella Typhimurium by immobilized phage P22 monolayers. Surf. Sci. 2008, 602, 1392–1400. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Ramesh, N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS ONE 2018, 13, e0206278. [Google Scholar] [CrossRef]
- Guo, Y.; Li, J.; Islam, M.S.; Yan, T.; Li, J. Application of a novel phage vB_SalS-LPSTLL for the biological control of Salmonella in foods. Food Res. Int. 2021, 147, 110492. [Google Scholar] [CrossRef] [PubMed]
- Green, S.; Ma, L.; Maresso, A. Phage Therapy. In Encyclopedia of Microbiology, 4th ed.; Academic Press: Oxford, UK, 2019; pp. 485–495. [Google Scholar]
- Younker, I.T.; Duffy, C. Jumbo Phages. In Encyclopedia of Virology, 4th ed.; Academic Press: Oxford, UK, 2021; pp. 229–241. [Google Scholar]
- Hungaro, H.M.; Mendon, R.C.S.; Gouvea, A.D.M.; Vanetti, M.C.D.; Pinto, C.L.D. Use of bacteriophages to reduce Salmonella in chicken skin in comparison with chemical agents. Food Res. Int. 2013, 52, 75–81. [Google Scholar] [CrossRef]
- Islam, M.S.; Yang, X.; Euler, C.W.; Han, X.; Li, J. Application of a novel phage ZPAH7 for controlling multidrug-resistant Aeromonas hydrophila on lettuce and reducing biofilms. Food Control 2021, 122, 107785. [Google Scholar] [CrossRef]
- Duc, H.M.; Son, H.M.; Yi, H.P.S.; Sato, J.; Miyamoto, T. Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157:H7 in different food matrices. Food Res. Int. 2020, 131, 108977. [Google Scholar] [CrossRef]
- Tian, Y.; Wu, L.; Lu, R.; Bao, H.; Zhou, Y.; Pang, M. Virulent phage vB_CpeP_HN02 inhibits Clostridium perfringens on the surface of the chicken meat. Int. J. Food Microbiol. 2022, 363, 109514. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.L.; Ding, Y.F.; Wang, X.H. Characterization of a novel T7-like Salmonella Typhimurium (ATCC13311) bacteriophage LPST144 and its endolysin. LWT 2020, 123, 109034. [Google Scholar] [CrossRef]
- Shoji, A.; Nakajima, M.; Morioka, K.; Fujimori, E.; Nakajima, H. Development of a surface plasmon resonance sensor using an optical fiber prepared by electroless displacement gold plating and its application to immunoassay. Talanta 2021, 240, 123–162. [Google Scholar] [CrossRef] [PubMed]
- Silveri, F.; Pelle, F.D.; Scroccarello, A.; Bukhari, Q.U.A.; Carlo, M.D.; Compagone, D. Modular graphene mediator film-based electrochemical pocket device for chlorpyrifos determination. Talanta 2022, 240, 13212. [Google Scholar] [CrossRef]
- Sari, A.K. The optimization of an electrochemical aptasensor to detect RBD protein S SARS-CoV-2 as a biomarker of COVID-19 using screen-printed carbon electrode/AuNP. J. Electrochem. Sci. Eng. 2022, 12, 219–235. [Google Scholar] [CrossRef]
- Hahn, J.; Kim, E.; You, Y.; Choi, Y.J. Colorimetric switchable linker-based bioassay for ultrasensitive detection of prostate-specific antigen as a cancer biomarker. Analyst 2019, 144, 4439–4446. [Google Scholar] [CrossRef] [PubMed]
- Tlili, C.; Sokullu, E.; Safavieh, M.; Tolba, M.; Ahmed, M.U.; Zourob, M. Bacteria screening, viability, and confirmation assays using bacteriophage-impedimetric/loop-mediated isothermal amplification dual-response biosensors. Anal. Chem. 2013, 85, 4893–4901. [Google Scholar] [CrossRef]
- Sangwan, S.; Seth, R. Synthesis, Characterization and Stability of Gold Nanoparticles (AuNPs) in Different Buffer Systems. J. Clust. Sci. 2021, 33, 749–764. [Google Scholar] [CrossRef]
- Yahaya, M.L.; Zakaria, N.D.; Noordin, R.; Razak, K.A. Synthesis of large and stable colloidal gold nanoparticles (AuNPs) by seeding-growth method. Mater. Today Proc. 2022. [Google Scholar] [CrossRef]
- Fernandes, E.; Deng, D.; Dias, J.; Azeredo, J.; Martins, V.C.; Cardoso, F.A.; Cardoso, S.; Kluskens, L.D.; Nobrega, C. A bacteriophage detection tool for viability assessment of Salmonella cells. Biosens. Bioelectron. 2014, 52, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Kavosi, B.; Salimi, A.; Hallaj, R.; Kamal, A. A highly sensitive prostate-specific antigen immunosensor based on gold nanoparticles/PAMAM dendrimer loaded on MWCNTS/chitosan/ionic liquid nanocomposite. Biosens. Bioelectron. 2014, 52, 20–28. [Google Scholar] [CrossRef]
- Wang, C.; Sauvageau, D.; Elias, A. Immobilization of Active Bacteriophages on Polyhydroxyalkanoate Surfaces. ACS Appl. Mater. Interfaces 2016, 8, 1128–1138. [Google Scholar] [CrossRef]
- Guan, G.M.; Sha, J.C.; Zhu, D.D. Heparin-MPA dual modified CdS quantum dots used as a simple and rapid label-free fluorescent sensor for protamine and hemin detection. Microchem. J. 2017, 133, 391–397. [Google Scholar] [CrossRef]
- Gervais, L.; Gel, M.; Allain, M.; Brovko, L.; Zourob, M.; Mandeville, R.; Griffiths, M.; Evoy, S. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens. Actuators B Chem. 2007, 125, 615–621. [Google Scholar] [CrossRef]
- Shabani, A.; Christophe, A.; Marquette, A.; Rosemonde, M.; Marcus, F.; Lawrence, F. Carbon microarrays for the direct impedimetric detection of Bacillus anthracis using Gamma phages as probes. Analyst 2013, 138, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Wang, X.L.; Li, Y.; Li, J.B. Cellulose nanocrystals composites with excellent thermal stability and high tensile strength for preparing flexible resistance strain sensors. Carbohydr. Polym. Technol. Appl. 2022, 3, 100214. [Google Scholar]
- Li, S.; Li, Y.; Chen, H.; Horikawa, S.; Wen, S.; Simonian, A.; Chin, B.A. Direct detection of Salmonella Typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens. Bioelectron. 2010, 26, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Maniloff, J.; Ackermann, H.W.; Jarvis, A. Phage Taxonomy and Classification. In Encyclopedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Elsevier: Oxford, UK, 1999; pp. 1221–1228. [Google Scholar]
- El-Dougdoug, N.K.; Cucic, S.; Abdelhamid, A.G.; Brovjo, L.; Kropinsik, A.M.; Griffiths, M.W.; Anany, H. Control of Salmonella Newport on cherry tomato using a cocktail of lytic bacteriophages. Int. J. Food Microbiol. 2019, 293, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Q.; Chong, Z.; Jie, Y.; Bie, X. Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res. 2017, 236, 14–23. [Google Scholar] [CrossRef]
- Cai, R.; Wang, Z.; Wang, G. Biological properties and genomics analysis of vB_KpnS_GH-K3, a Klebsiella phage with a putative depolymerase-like protein. Virus Genes 2019, 55, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.K.; Zhang, J.Y.; Liu, X.; Ning, H.Q.; Lin, H.; Wang, J.X. Biological characterization and complete genome analysis of a novel Stenotrophomonas maltophilia phage vB_SM_ytsc_ply2008005c. Virus Res. 2022, 318, 198856. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Ding, Y.; Huang, C.; Wang, X. Characterization of a novel Siphoviridae Salmonella bacteriophage T156 and its microencapsulation application in food matrix. Food Res. Int. 2021, 140, 110004. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Darius, P.; Summer, E.J.; Seto, D.; Mahadevan, P.; Milsson, A.S.; Ackermann, H.W.; Kropinski, A.M. Classification of myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009, 9, 224–240. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Johannes, W.; Jens, H.K.; Dann, T.; Rodney, J. Taxonomy of prokaryotic viruses: 2017 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 2018, 163, 1125–1129. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Yu, G.; Tian, K.; Xu, C. A highly sensitive and stable electrochemical sensor for simultaneous detection towards ascorbic acid, dopamine, and uric acid based on the hierarchical nanoporous PtTi alloy. Biosens. Bioelectron. 2016, 82, 119–126. [Google Scholar] [CrossRef]
- Tolba, M.; Ahmed, M.; Tlili, C.; Eichenseher, F.; Lossner, M.; Mohammed, Z. A bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of Listeria cells. Analyst 2012, 137, 5749–5756. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.; Aveyard, J.; Fernig, D. Determination of size and concentration of gold nanoparticles from UV-vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Qi, X.; Li, S.; Huang, C.; Su, W.; Huang, S. Novel N-Doped Carbon Dots/β-Cyclodextrin Nanocomposites for Enantioselective Recognition of Tryptophan Enantiomers. Sensors 2014, 16, 1874–1890. [Google Scholar]
- Zhang, L.; Li, D.; Song, W.; Shi, L.; Li, Y.; Long, Y. High Sensitive On-Site Cadmium Sensor Based on AuNPs Amalgam Modified Screen-Printed Carbon Electrodes. IEEE Sens. J. 2010, 10, 1583–1588. [Google Scholar] [CrossRef]
- Sakellari, G.I.; Hondow, N.; Gardine, P.H.E. Factors Influencing the Surface Functionalization of Citrate Stabilized Gold Nanoparticles with Cysteamine, 3-Mercaptopropionic Acid or l-Selenocystine for Sensor Applications. Chemosensors 2020, 8, 80. [Google Scholar] [CrossRef]
- Dudak, F.C.; Boyaci, I.H. Peptide-Based Surface Plasmon Resonance Biosensor for Detection of Staphylococcal Enterotoxin B. Food Anal. Methods 2014, 7, 506–511. [Google Scholar] [CrossRef]
- Gong, H.; Li, X. Y-type, C-rich DNA probe for electrochemical detection of silver ion and cysteine. Analyst 2011, 136, 2242–2246. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Marnix, V.; Patrick, A.; Jonge, D.; Lisa, L.; Dreesens, J.; Hubertus, J.E.; Beaumont, R.; Lavigne, R.; Dutilh, B.; et al. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [PubMed]
- Gómez, P.; Buckling, D. Bacteria-Phage Antagonistic Coevolution in Soil. Science 2011, 332, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Mohan, K.; Donavan, K.C.; Arter, J.A.; Penner, R.M.; Weeiss, G.A. Sub-nanomolar Detection of Prostate-Specific Membrane Antigen in Synthetic Urine by Synergistic, Dual-Ligand Phage. J. Am. Chem. Soc. 2013, 135, 7761–7772. [Google Scholar] [CrossRef] [Green Version]
- Mejri, M.B.; Baccar, H.; Campo, F.J.D.; Abdelghani, A. Impedance biosensing using phages for bacteria detection: Generation of dual signals as the clue for in-chip assay confirmation. Biosens. Bioelectron. 2010, 26, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Huss, P.; Meger, A.; Leander, M.; Nishikawa, K.; Raman, S. Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning. ELife Sci. 2021, 10, e63775. [Google Scholar] [CrossRef]
- Moghtader, F.; Congur, G.; Zareie, H.; Erdem, A.; Piskin, E. Impedimetric detection of pathogenic bacteria with bacteriophages using gold nanorod deposited graphite electrodes. RSC Adv. 2016, 6, 97832–97839. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Li, C.; Wang, Y.; Wang, X. EIS biosensor based on a novel Myoviridae bacteriophage SEP37 for rapid and specific detection of Salmonella in food matrixes. Food Res. Int. 2022, 158, 111479. [Google Scholar] [CrossRef]
- Guliy, O.I.; Velichko, N.S.; Fedonenko, Y.P.; Bunin, V.D. Use of an electro-optical sensor and phage antibodies for immunodetection of Herbaspirillum. Talanta 2019, 202, 362–368. [Google Scholar] [CrossRef]
- Saroh, N.; Warakorn, L.; Apon, N.; Proespichaya, K.; Ratthaphol, C.; Nitsara, K.; Panote, T. Phage-based capacitive biosensor for Salmonella detection. Talanta 2018, 188, 658–664. [Google Scholar]
- Chen, D.; Wang, X.; Zhang, K.; Cao, Y.; Tu, J.; Xiao, D.; Wu, Q. Glucose photoelectrochemical enzyme sensor based on competitive reaction of ascorbic acid. Biosens. Bioelectron. 2020, 166, 112466. [Google Scholar] [CrossRef]
- Chen, I.H.; Horikawa, S.; Bryant, K.; Riggs, R.; Chin, B.A.; Barbaree, J.M. Bacterial assessment of phage magnetoelastic sensors for Salmonella enterica Typhimurium detection in chicken meat. Food Control 2017, 71, 273–278. [Google Scholar] [CrossRef] [Green Version]









| Sample Name | Spiked (CFU/Ml) | Found (CFU/mL) | Recovery Rate (%) | RSD (%) |
|---|---|---|---|---|
| Spiked milk | 2.0 × 103 | 1885 | 94.3 | 3.2 |
| 2.0 × 102 | 218 | 109.0 | 4.6 | |
| 2.0 × 101 | 21 | 105 | 6.5 | |
| Eggs | 2.0 × 103 | 2070 | 103.5 | 4.1 |
| 2.0 × 102 | 199 | 99.5 | 5.6 | |
| 2.0 × 101 | 21 | 105.0 | 7.1 | |
| Chicken | 2.0 × 103 | 1975 | 98.8 | 4.2 |
| 2.0 × 102 | 203 | 105.5 | 5.3 | |
| 2.0 × 101 | 20 | 100 | 5.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Wang, Y.; Wang, J.; Wang, X. Properties of a Novel Salmonella Phage L66 and Its Application Based on Electrochemical Sensor-Combined AuNPs to Detect Salmonella. Foods 2022, 11, 2836. https://doi.org/10.3390/foods11182836
Li C, Wang Y, Wang J, Wang X. Properties of a Novel Salmonella Phage L66 and Its Application Based on Electrochemical Sensor-Combined AuNPs to Detect Salmonella. Foods. 2022; 11(18):2836. https://doi.org/10.3390/foods11182836
Chicago/Turabian StyleLi, Changbin, Yuanshang Wang, Jia Wang, and Xiaohong Wang. 2022. "Properties of a Novel Salmonella Phage L66 and Its Application Based on Electrochemical Sensor-Combined AuNPs to Detect Salmonella" Foods 11, no. 18: 2836. https://doi.org/10.3390/foods11182836
APA StyleLi, C., Wang, Y., Wang, J., & Wang, X. (2022). Properties of a Novel Salmonella Phage L66 and Its Application Based on Electrochemical Sensor-Combined AuNPs to Detect Salmonella. Foods, 11(18), 2836. https://doi.org/10.3390/foods11182836





