The Ability of a Novel Trypsin-like Peptidase Activity Assay Kit to Detect Red-Complex Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples
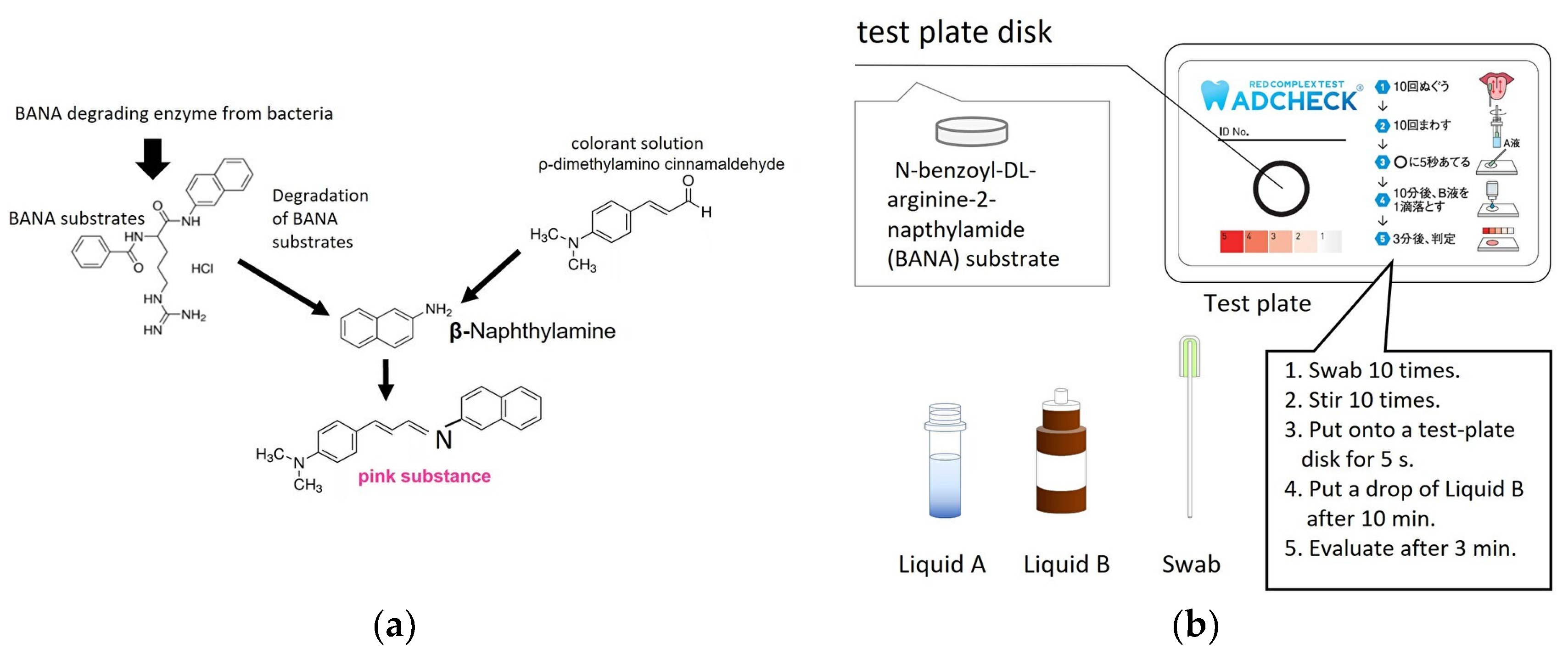
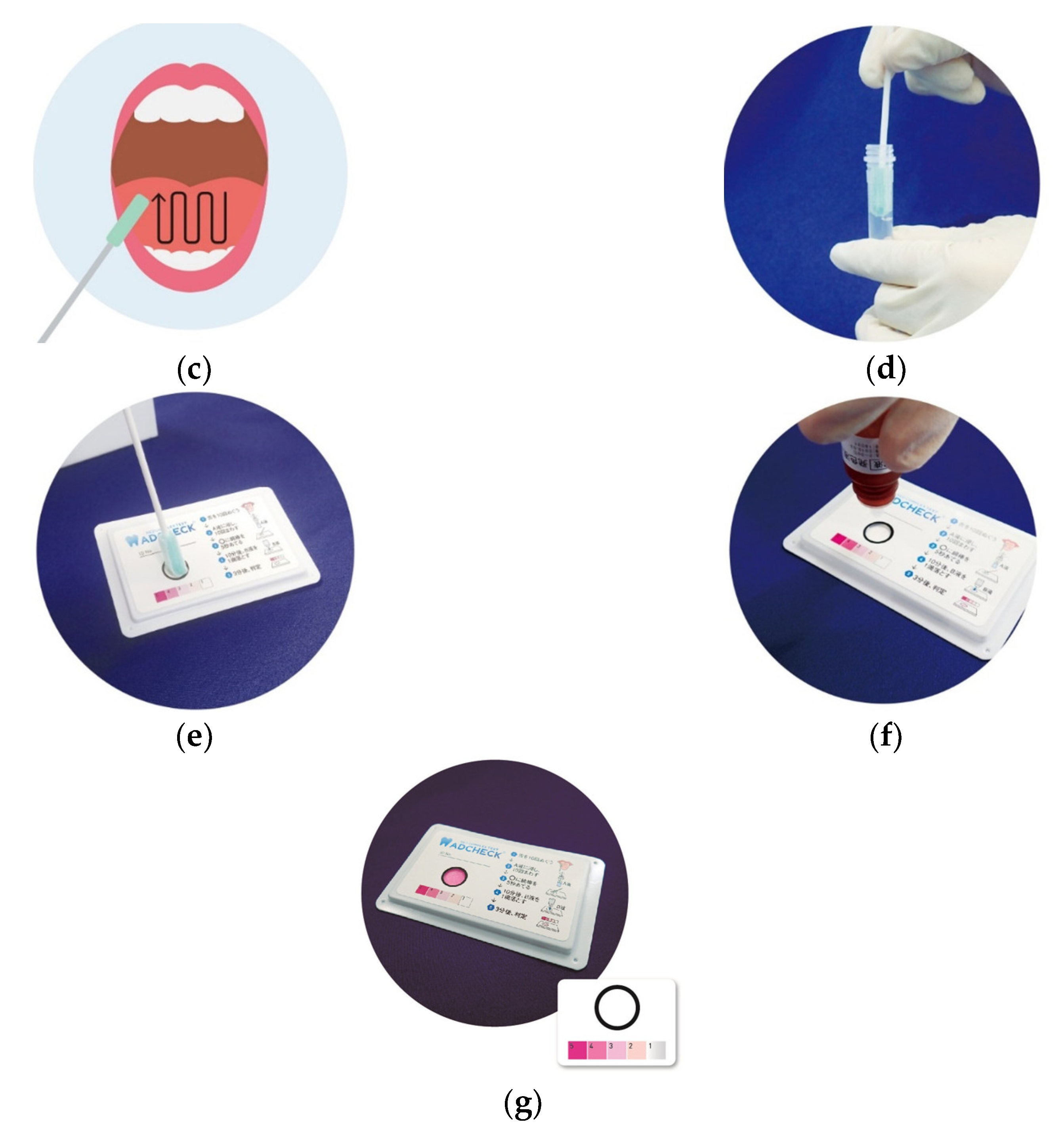
2.2. TLP-AA Kit
2.3. Bacterial Cultures
2.4. Real-Time PCR
2.5. Statistical Analysis
3. Results
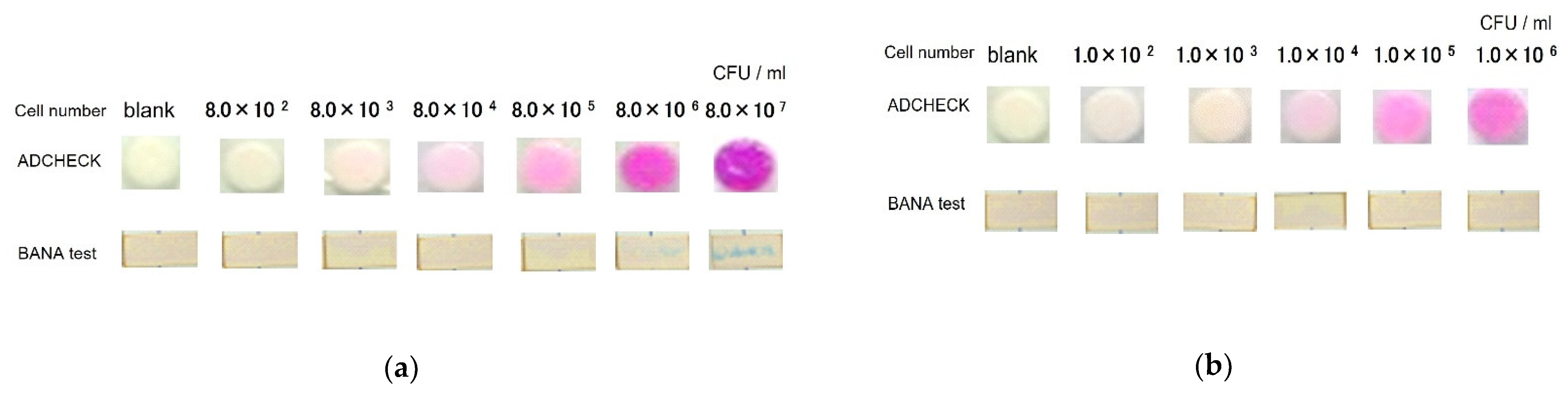
3.1. Detection of TLPs from P. gingivalis Bacterial Strains by a Novel TLP-AA Kit
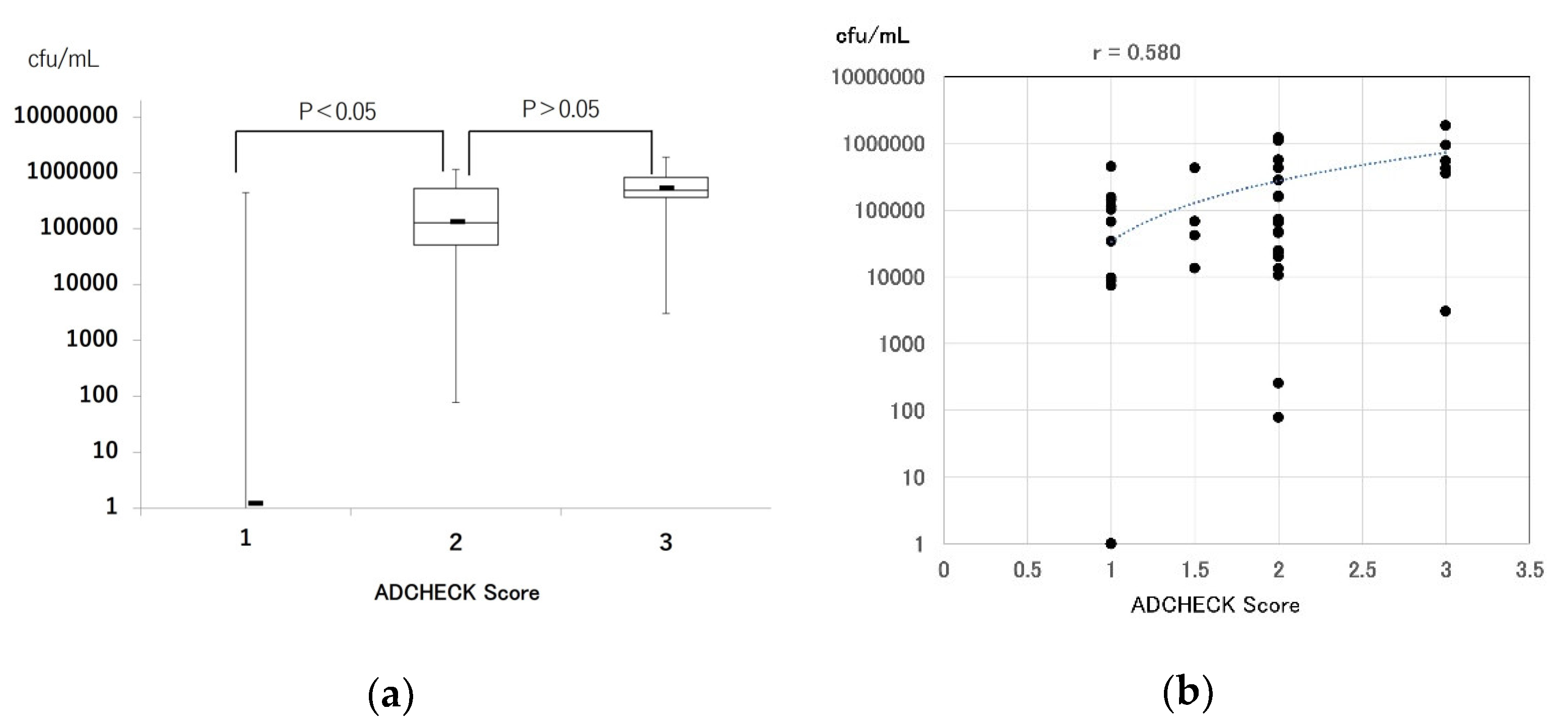
3.2. Comparison of Real-Time PCR and a Novel TLP-AA Kit in Clinical Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Usui, M.; Onizuka, S.; Sato, T.; Kokabu, S.; Ariyoshi, W.; Nakashima, K. Mechanism of Alveolar Bone Destruction in Periodontitis—Periodontal Bacteria and Inflammation. Jpn. Dent. Sci. Rev. 2021, 57, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial Complexes in Subgingival Plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Bretz, W.A.; Kerschensteiner, D.; Stoll, J.; Socransky, S.S.; Hujoel, P.; Lopatin, D.E. Development of a Diagnostic Test for Anaerobic Periodontal Infections Based on Plaque Hydrolysis of Benzoyl-DL-Arginine-Naphthylamide. J. Clin. Microbiol. 1990, 28, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Bretz, W.A.; Lopatin, D.; Stoll, J.; Rau, C.F.; Hillenburg, K.L.; Killoy, W.J.; Drisko, C.L.; Williams, R.; Weber, H.P. Multi-center Clinical Evaluation of a Chairside Method for Detecting Certain Periodontopathic Bacteria in Periodontal Disease. J. Periodontol. 1990, 61, 189–196. [Google Scholar] [CrossRef]
- Wilson, M.; Lopatin, D.; Osborne, G.; Kieser, J.B. Prevalence of Treponema denticola and Porphyromonas gingivalis in Plaque from Periodontally-Healthy and Periodontally-Diseased Sites. J. Med. Microbiol. 1993, 38, 406–410. [Google Scholar] [CrossRef]
- Loesche, W.J.; Lopatin, D.E.; Giordano, J.; Alcoforado, G.; Hujoel, P. Comparison of the Benzoyl-DL-Arginine-Naphthylamide NA) test, DNA Probes, and Immunological Reagents for Ability to Detect Anaerobic Periodontal Infections due to Porphyromonas Gingivalis, Treponema Denticola, and Bacteroides Forsythus. J. Clin. Microbiol. 1992, 30, 427–433. [Google Scholar] [CrossRef]
- Iwasaki, M.; Usui, M.; Ariyoshi, W.; Nakashima, K.; Nagai-Yoshioka, Y.; Inoue, M.; Nishihara, T. A Preliminary Study on the Ability of the Trypsin-Like Peptidase Activity Assay Kit to Detect Periodontitis. Dent. J. 2020, 8, 98. [Google Scholar] [CrossRef]
- Iwasaki, M.; Usui, M.; Ariyoshi, W.; Nakashima, K.; Nagai-Yoshioka, Y.; Inoue, M.; Kobayashi, K.; Nishihara, T. Evaluation of the Ability of the Trypsin-Like Peptidase Activity Assay to Detect Severe Periodontitis. PLoS ONE 2021, 16, e0256538. [Google Scholar] [CrossRef]
- Ishikawa, I.; Nakashima, K.; Koseki, T.; Nagasawa, T.; Watanabe, H.; Arakawa, S.; Nitta, H.; Nishihara, T. Induction of the Immune Response to Periodontopathic Bacteria and Its Role in the Pathogenesis of Periodontitis. Periodontology 1997, 14, 79–111. [Google Scholar] [CrossRef]
- Lyons, S.R.; Griffen, A.L.; Leys, E.J. Quantitative Real-Time PCR for Porphyromonas gingivalis and Total Bacteria. J. Clin. Microbiol. 2000, 38, 2362–2365. [Google Scholar] [CrossRef]
- Eke, P.I.; Page, R.C.; Wei, L.; Thornton-Evans, G.; Genco, R.J. Update of the case definitions for population-based surveillance of periodontitis. J. Periodontol. 2012, 83, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Takeuchi, Y.; Umeda, M.; Ishikawa, I.; Benno, Y. Rapid Detection and Quantification of Five Periodontopathic Bacteria by Real-Time PCR. Microbiol. Immunol. 2001, 45, 39–44. [Google Scholar] [CrossRef]
- Ishihara, K.; Naito, Y.; Kato, T.; Takazoe, I.; Okuda, K.; Eguchi, T.; Nakashima, K.; Matsuda, N.; Yamasaki, K.; Hasegawa, K. A sensitive enzymatic method (SK-013) for detection and quantification of specific periodontopathogens. J. Periodontal Res. 1992, 27, 81–85. [Google Scholar] [CrossRef]
- Bujang, M.A.; Adnan, T.H. Requirements for Minimum Sample Size for Sensitivity and Specificity Analysis. J. Clin. Diagn. Res. 2016, 10, YE01–YE06. [Google Scholar] [CrossRef] [PubMed]
- Andrade, J.A.; Feres, M.; Figueiredo, L.C.; Salvador, S.L.; Cortelli, S.C. The Ability of the BANA Test to Detect Different Levels of P. gingivalis, T. denticola and T. Forsythia. Braz. Oral Res. 2010, 24, 224–230. [Google Scholar] [CrossRef]
- Bostanci, N.; Belibasakis, G.N. Porphyromonas gingivalis: An Invasive and Evasive Opportunistic Oral Pathogen. FEMS Microbiol. Lett. 2012, 333, 1–9. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Liang, S.; Payne, M.A.; Hashim, A.; Jotwani, R.; Eskan, M.A.; McIntosh, M.L.; Alsam, A.; Kirkwood, K.L.; Lambris, J.D.; et al. Low-Abundance Biofilm Species Orchestrates Inflammatory Periodontal Disease through the Commensal Microbiota and Complement. Cell Host Microbe 2011, 10, 497–506. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R.J. Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 2016, 24, 477–489. [Google Scholar] [CrossRef]
- Lang, N.P.; Joss, A.; Orsanic, T.; Gusberti, F.A.; Siegrist, B.E. Bleeding on Probing. A Predictor for the Progression of Periodontal Disease? J. Clin. Periodontol. 1986, 13, 590–596. [Google Scholar] [CrossRef]
- Lang, N.P.; Adler, R.; Joss, A.; Nyman, S. Absence of Bleeding on Probing. An Indicator of Periodontal Stability. J. Clin. Periodontol. 1990, 17, 714–721. [Google Scholar] [CrossRef]
- Arias-Bujanda, N.; Regueira-Iglesias, A.; Balsa-Castro, C.; Nibali, L.; Donos, N.; Tomás, I. Accuracy of Single Molecular Biomarkers in Gingival Crevicular Fluid for the Diagnosis of Periodontitis: A Systematic Review and Meta-analysis. J. Clin. Periodontol. 2019, 46, 1166–1182. [Google Scholar] [CrossRef] [PubMed]
- Hieda, Y.; Usui, M.; Nakamura, T.; Morishita, M.; Hanatani, T.; Moritani, Y.; Koga, Y.; Kasai, S.; Inoue, M.; Nakashima, K. Decreased Ratios of Interleukin-1β and Intercellular Adhesion Molecule 1 in Gingival Crevicular Fluid after Scaling and Root Planing Associated with Periodontal Pocket Healing. Open J. Stomatol. 2017, 7, 361–376. [Google Scholar] [CrossRef]
- Kasai, S.; Onizuka, S.; Katagiri, S.; Nakamura, T.; Hanatani, T.; Kudo, T.; Sugata, Y.; Ishimatsu, M.; Usui, M.; Nakashima, K. Associations of Cytokine Levels in Gingival Crevicular Fluid of Mobile Teeth with Clinical Improvement after Initial Periodontal Treatment. J. Oral Sci. 2020, 62, 189–196. [Google Scholar] [CrossRef] [Green Version]
| Sex | Age (Years) Mean (SD) | Probing Depth (mm) Mean (SD) | BOP (%) Mean (SD) | Number of Red Complex Pathogens in Tongue Coat Mean (SD) | |
|---|---|---|---|---|---|
| Healthy (60) | Male: 12 | 26.9 (5.2) | 1.4 (0.3) | 2.8 (2.0) | 27,439 (82,438) |
| Female: 16 | |||||
| Periodontitis (28) | Male: 19 | 52.1 (11.3) * | 2.3 (1.3) * | 8.2 (5.3) * | 405,243 * (506,060) |
| Female: 41 |
| Bacterial Name | Strain No. | cfu/mL | Bacterial Name | Strain No. | cfu/mL | ||
|---|---|---|---|---|---|---|---|
| 1 | Bordetella pertussis | NBRC107857 | 1.69 × 108 | 21 | Streptococcus mutans | NBRC13955 | 1.13 × 108 |
| 2 | Candida albicans | NBRC1385 | 5.17 × 108 | 22 | Streptococcus oralis | JCM12997 | 8.60 × 107 |
| 3 | Corynebacterium diphtheriae | JCM1310 | 5.25 × 106 | 23 | Streptococcus pneumoniae | NBRC102642 | 6.45 × 109 |
| 4 | Enterococcus durans | NBRC100479 | 3.75 × 108 | 24 | Streptococcus pyrogenes | JCM5674 | 1.07 × 108 |
| 5 | Enterococcus faecalis | NBRC100480 | 6.00 × 108 | 25 | Streptococcus intermedius | ATCC9895 | 6.50 × 108 |
| 6 | Haemophilus infuluenzae | ATCC9006 | 1.56 × 108 | 26 | Streptococcus sanguis | JCM5708 | 2.85 × 107 |
| 7 | Listeria monocytogenes | JCM7671 | 1.07 × 109 | 27 | Neisseria gonorrhoeae | ATCC19424 | 1.02 × 109 |
| 8 | Moraxella catarrhalis | ATCC8176 | 2.14 × 108 | 28 | Neisseria meningitidis | ATCC13077 | 4.20 × 109 |
| 9 | Mycoplasma orale | NBRC14477 | 2.48 × 107 | 29 | Neisseria sicca | ATCC9913 | 6.20 × 108 |
| 10 | Mycoplasma pneumoniae | NBRC14401 | 1.81 × 107 | 30 | Neisseria subflava | ATCC19243 | 2.11 × 108 |
| 11 | Mycoplasma salivarium | NBRC14478 | 1.02 × 106 | 31 | Streptococcus pyogenes | ATCC12353(T12) | 2.35 × 107 |
| 12 | Mycoplasma hominis | NBRC14850 | 1.34 × 105 | 32 | Streptococcus pyogenes | ATCC12962(T28) | 4.00 × 106 |
| 13 | Proteus vulgaris | NBRC3045 | 4.25 × 109 | 33 | Streptococcus pyogenes | BAA-1066(M4) | 2.30 × 107 |
| 14 | Pseudomonas aeruginosa | NBRC12689 | 3.85 × 109 | 34 | Escherichia coli | ATCC11775 (JCM1649) | 2.40 × 109 |
| 15 | Serratia marcescens | NBRC3046 | 1.04 × 109 | 35 | Klebsiella pneumoniae | ATCC13883 (JCM1662) | 1.76 × 108 |
| 16 | Staphylococcus aureus | NBRC102135 | 3.17 × 109 | 36 | Citrobacter freundii | JCM1657 | 2.24 × 109 |
| 17 | Staphylococcus epidermidis | NBRC100911 | 1.38 × 108 | 37 | Salmonella enteritidis | IFO3313 | 1.15 × 109 |
| 18 | Streptococcus agalactiae | JCM5671 | 1.70 × 108 | 38 | Salmonella typhimurium | IFO13245 | 8.2 × 109 |
| 19 | Streptococcus anginosus | JCM12993 | 2.08 × 108 | 39 | Streptococcus dysgalactiae subsp. equisimilis | ATCC12388 | 3.7 × 108 |
| 20 | Streptococcus dysgalactiae subsp. dysgalactiae | JCM5673 | 4.04 × 108 | 40 | Streptococcus constellatus subsp. constellatus | JCM12994 | 5.05 × 108 |
| Real-Time PCR | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. gingivalis | T. forsythia | T. denticola | Red Complex | Sensitivity | Specificity | PPV | NPV | Accuracy | ||||||||||
| POS | NEG | SUM | POS | NEG | SUM | POS | NEG | SUM | POS | NEG | SUM | |||||||
| ADCHECK | POS | 15 | 16 | 31 | 29 | 2 | 31 | 13 | 18 | 31 | 30 | 1 | 31 | 83% | 98% | 97% | 90% | 92% |
| NEG | 9 | 48 | 57 | 9 | 48 | 57 | 3 | 54 | 57 | 6 | 51 | 57 | ||||||
| SUM | 24 | 64 | 88 | 38 | 50 | 88 | 16 | 72 | 88 | 36 | 52 | 88 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usui, M.; Iwasaki, M.; Ariyoshi, W.; Kobayashi, K.; Kasai, S.; Yamanaka, R.; Nakashima, K.; Nishihara, T. The Ability of a Novel Trypsin-like Peptidase Activity Assay Kit to Detect Red-Complex Species. Diagnostics 2022, 12, 2172. https://doi.org/10.3390/diagnostics12092172
Usui M, Iwasaki M, Ariyoshi W, Kobayashi K, Kasai S, Yamanaka R, Nakashima K, Nishihara T. The Ability of a Novel Trypsin-like Peptidase Activity Assay Kit to Detect Red-Complex Species. Diagnostics. 2022; 12(9):2172. https://doi.org/10.3390/diagnostics12092172
Chicago/Turabian StyleUsui, Michihiko, Masanori Iwasaki, Wataru Ariyoshi, Kaoru Kobayashi, Shingo Kasai, Rieko Yamanaka, Keisuke Nakashima, and Tatsuji Nishihara. 2022. "The Ability of a Novel Trypsin-like Peptidase Activity Assay Kit to Detect Red-Complex Species" Diagnostics 12, no. 9: 2172. https://doi.org/10.3390/diagnostics12092172
APA StyleUsui, M., Iwasaki, M., Ariyoshi, W., Kobayashi, K., Kasai, S., Yamanaka, R., Nakashima, K., & Nishihara, T. (2022). The Ability of a Novel Trypsin-like Peptidase Activity Assay Kit to Detect Red-Complex Species. Diagnostics, 12(9), 2172. https://doi.org/10.3390/diagnostics12092172









