1. Introduction
Microalgae cultivation has recently piqued the interest of researchers due to their ability to synthesize a variety of biologically active substances, rapid biomass growth, and the ability to adjust their biochemical composition depending on cultivation conditions. Unlike heterotrophic microorganisms, which need various organic compounds for growth, unicellular photosynthetic organisms produce biomass from fully oxidized inorganic substances and mineral elements due to light energy converted during cultivation photosynthesis. Furthermore, microalgae biomass production technologies do not pollute the environment, use carbon dioxide while releasing oxygen, consume a relatively small amount of water, and can be produced using land resources unsuitable for crop cultivation [
1,
2,
3].
There are currently two main applications for microalgae: biomass production as a biologically active additive (BAA) and cultivation for the subsequent isolation of biologically active substances (BAS) from the biomass [
4].
Microalgae are rich in nutrients and biologically active substances, such as proteins, carbohydrates, lipids, polyunsaturated fatty acids, vitamins, pigments, phycobiliproteins, enzymes, etc. Biologically active substances from microalgae can exhibit antioxidant, immunostimulating, antibacterial, antiviral, antitumor, antihypertensive, regenerative, and neuroprotective effects. These compounds are in high demand in medicine, cosmetics, the food industry, fish farming, energy, agriculture, feed, and functional food production [
5,
6,
7,
8].
Only a few microalgae species (
Arthrospira (Spirulina) platensis,
Chlorella or
Chlorella vulgaris,
Dunaliella,
Aphanizomenon, and
Nostoc) are currently approved for human consumption. These microalgae are a promising object for large-scale cultivation due to the high content of biologically active substances and the relatively cheap production process. Other microalgae species, such as
Chlamydomonas sp.,
Chlorococcum sp.,
Scenedescmus sp.,
Tetraselmis chuii, and
Nanochloropsis sp., have demonstrated their value as ingredients in feed, fertilizers, cosmetics, and aquaculture but do not yet have GRAS status [
9,
10,
11,
12,
13].
Finding new, unstudied strains of microalgae can broaden the scope of their industrial application and create new opportunities for use. Because of the wide variety of microalgae, their high metabolic flexibility, and the variety of cultivation conditions, their true potential has yet to be fully evaluated. Innovative developments in microalgae production optimization will make their use economically viable and in demand in the future.
Marine microalgae are microalgae that are used in many parts of the world as food, feed, and fertilizer, as well as a potential renewable resource in medicine and commercial activities. The biostimulatory properties of marine microalgae are being investigated for potential applications in the development of new antibiotics. Many metabolites isolated from marine microalgae have bioactive properties [
14]. Bioactive natural products are widely distributed in the plant world, and extracts of various plants, as well as red, green, and brown macro- and microalgae, can be used as natural products [
15]. Marine algae are a never-ending source of raw materials for pharmaceuticals, medicine, food processing, and cosmetics [
16]. The need for compounds with biological activity for potential pharmaceutical applications or other potentially significant economic properties has resulted in a sharp increase in research into the chemistry of marine microalgae in recent years [
17]. Marine microalgae are an important source of bioactive natural substances [
18]. Particular attention was paid to the antibacterial activity of marine microalgae against several pathogens [
19]. It was demonstrated that various marine microalgae extracts and compounds have antibacterial activity against both Gram-positive and Gram-negative bacteria [
19,
20]. Antimicrobial compounds derived from marine microalgae are composed of a diverse group of chemical compounds [
21,
22].
The microalgae
Chlorella vulgaris and
Arthrospira platensis are of particular interest because they have high potential for use and grow rapidly (doubling time up to 6 h), are more productive at small scales than plants, do not require agriculturally significant land (do not compete with the food industry), use very basic mineral components for growth, use salty sea water, and can grow on wastewater while treating it, use only solar energy, absorb carbon dioxide when growing, contain a large amount of proteins, fats, and carbohydrates [
22].
This study sought to identify the antimicrobial metabolites present in microalgae by analyzing their FTIR spectra. The novelty of this study lies in the fact that the antimicrobial activity of biologically active chemical groups identified by IR spectroscopy and found in the proteins, lipids, and carbohydrates of microalgae Chlorella vulgaris and Arthrospira platensis isolated from the Baltic Sea was investigated for the first time.
4. Discussion
Moderate antimicrobial activity was detected when three concentrations of organic complexes that were isolated and purified from microalgae were analyzed. Taking into account the revealed activity only in the carbohydrate and lipid complexes, tests were carried out with various concentrations of these substances.
Sea microalgae were selected based on morphological features characteristic of the edible microalgae A. platensis and C. vulgaris, which were additionally identified using DNA sequences. Only edible microalgae were studied because, in accordance with the proposed hypothesis, the authors intended to obtain a feed additive based on these microalgae for livestock and poultry if the antibacterial activity of the lipid and carbohydrate complexes is confirmed. It is not possible to feed animals with inedible microalgae species.
Lipids play an important role in the metabolism and growth of microalgae, acting as a reservoir of energy and carbon [
28,
29]. Microalgae contain both polar and neutral lipids. Polar lipids such as phospholipids and glycolipids generated by chloroplasts dominate the cell wall and organellar membranes, while neutral lipids (mono-, di-, and triacylglycerol) are stored in cell organelles [
29,
30]. The lipid profile of microalgae is wide and ranges from 2 to 77% depending on the species and environment. In
C. vulgaris, it ranges from 5 to 58% of dry matter.
C. vulgaris produces more lipids (60–68%) when cultivated under mixotrophic conditions [
29,
31].
Arthrospira platensis was grown on Zarrouk medium, and Chlorella vulgaris was grown on Tamiya medium. The biomass yield of each microalga after 7–14 days of cultivation was 30–40 million cells/mL. Protein extraction from Arthrospira platensis biomass was 60.03 ± 1.80%; from Chlorella vulgaris biomass, 56.20 ± 1.68%; the extraction of lipids from the biomass of Arthrospira platensis was 7.23 ± 0.21%; from the biomass of Chlorella vulgaris, 16.24 ± 0.48%; the extraction amounts of carbohydrates from microalgae biomass were 11.44 ± 0.34% and 11.22 ± 0.83% for Arthrospira platensis and Chlorella vulgaris, respectively.
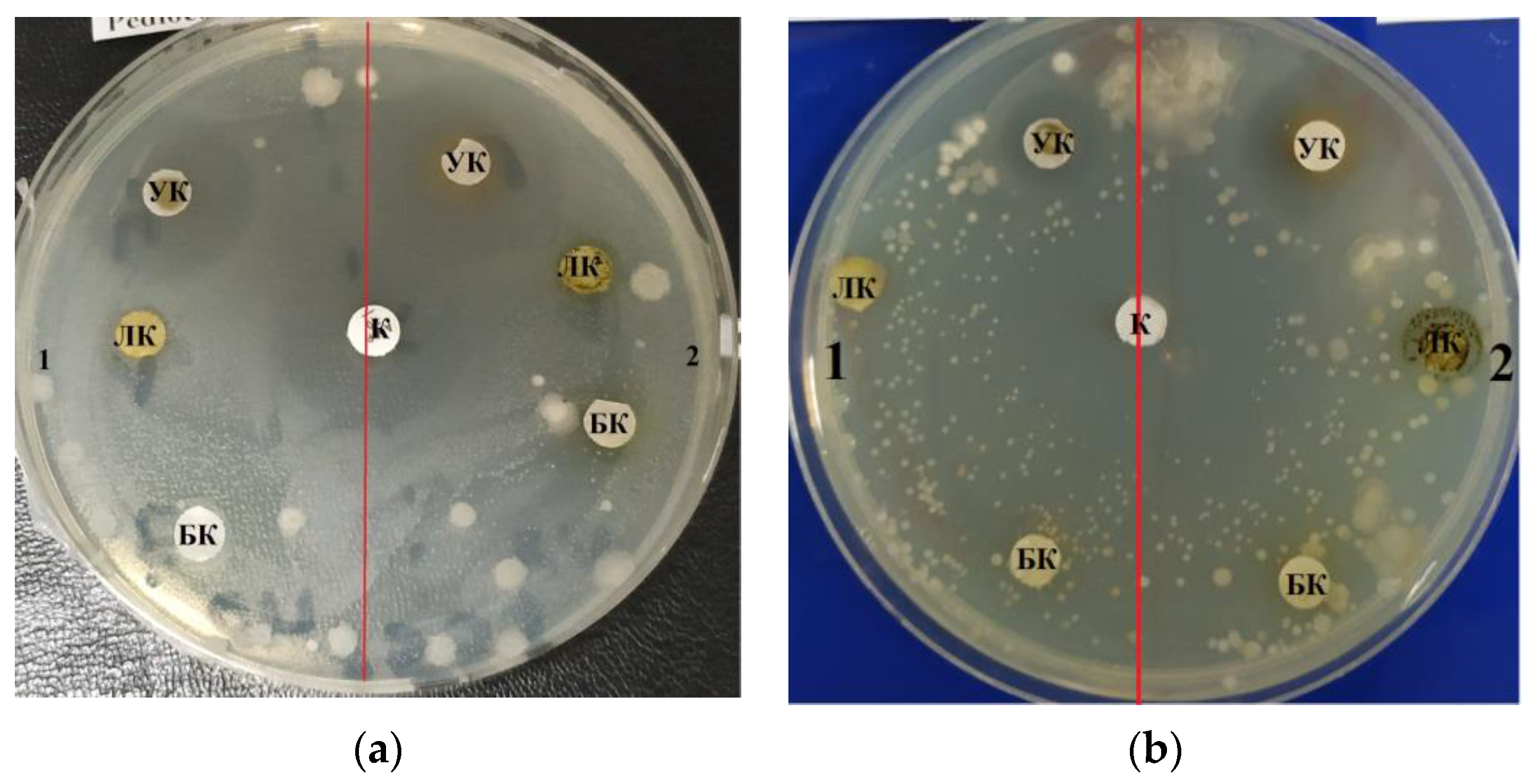
The purified lipid complex isolated from
C. vulgaris demonstrated antimicrobial activity against a Gram-negative strain of the bacterium
E. coli, as evidenced by the zones of inhibition results shown in
Table 1. The maximum effective inhibitory concentration of the lipid complex samples against
E. coli was observed at a concentration of 10.0–1.0 mg/mL; the area of the zone was 1.3 ± 0.06 cm
2. At a higher concentration (100.0 mg/mL), inhibition was less pronounced and amounted to 1.0 ± 0.05 cm (
Figure 1a).
C. vulgaris lipid complex samples also inhibited the growth of Gram-positive
B. subtilis and
B. pumilus strains; the maximum zone of inhibition was 2.2 ± 0.11 cm at a concentration of 100.0 mg/mL (
Table 1).
Table 2 shows the results of measuring zones of inhibition at different concentrations of the purified lipid complex obtained from
A. platensis samples. The lipid complex was found to be effective against E. coli across the entire concentration range examined, 100.0–10.0–1.0 mg/mL. The lipid complex of microalgae samples also suppressed the growth of the Gram-positive strain B. subtilis; the maximum zone of inhibition was 0.7 ± 0.03 cm at all concentrations used. No increase in the inhibition zone area was found with increasing concentration of the extract (
Figure 1b).
Carbohydrates such as starch, glucose, sugar, and polysaccharides are commonly used as energy and carbon storage in microalgae. The most common polysaccharides of
C. vulgaris are starch, composed of amylose and amylopectin, followed by polysaccharide cellulose in the cell wall. The total carbohydrate content of
C. vulgaris can reach 12–55% of dry weight when grown under unfavorable conditions, especially those with limited nitrogen [
29,
32].
Arthrospira platensis contains proteins (60%), carbohydrates (15%), lipids, phycobiliproteins, carotenoids, vitamins, and minerals [
33].
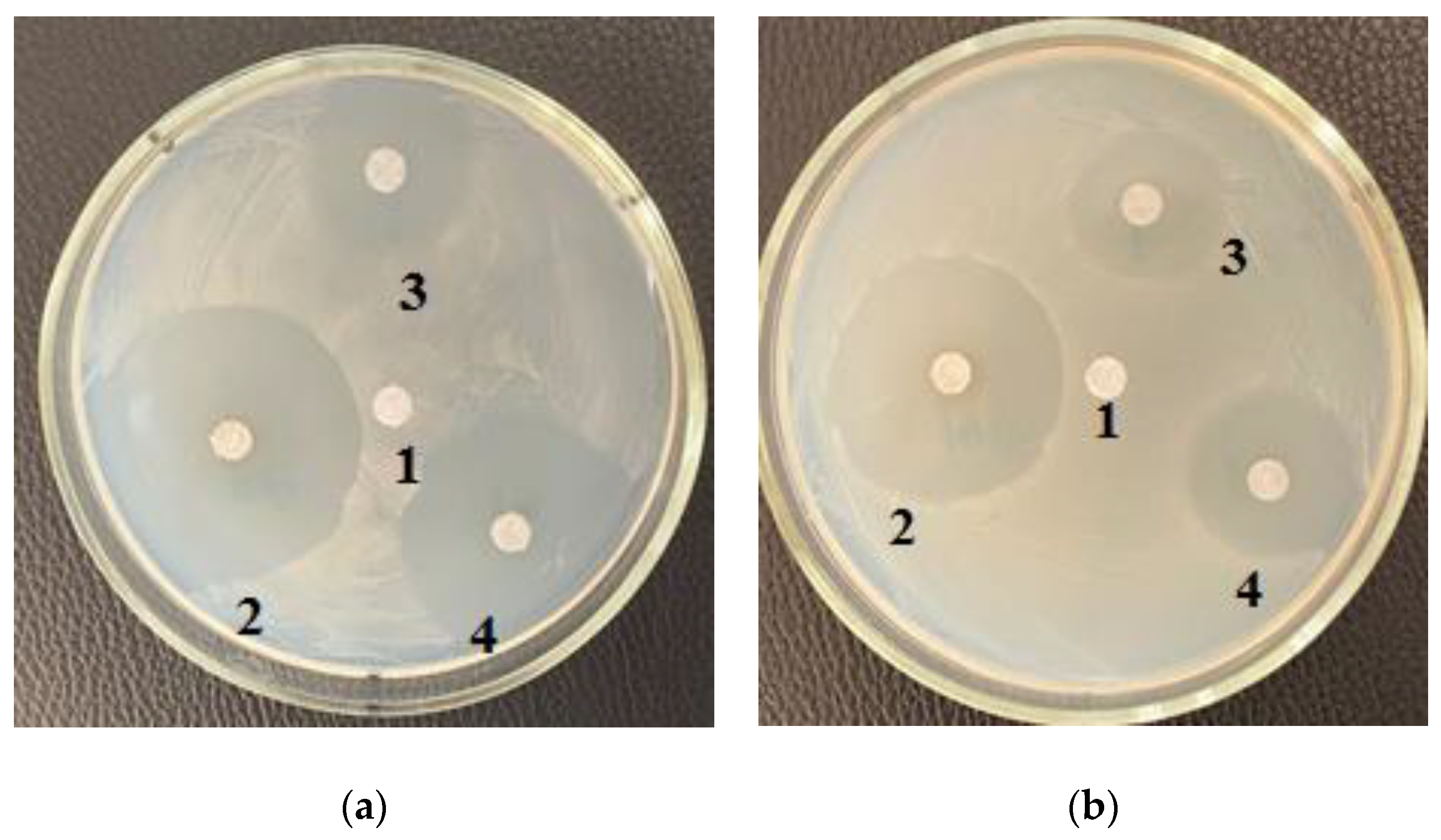
Table 3 shows the zones of inhibition results of various concentrations of purified carbohydrate complex derived from
C. vulgaris. Samples of the purified carbohydrate complex were effective against
E. coli at concentrations of 100.0–10.0–1.0 mg/mL. Samples of the purified carbohydrate complex of
C. vulgaris exhibited antibacterial activity against the Gram-positive
B. subtilis strain; the maximum zone of inhibition was 3.5 ± 0.17 cm at a concentration of 1.0 mg/mL (
Figure 3a). A similar effect was also observed against
B. pumilus; the maximum zone of inhibition was 3.5 ± 0.17 cm at a concentration of 100.0 mg/mL.
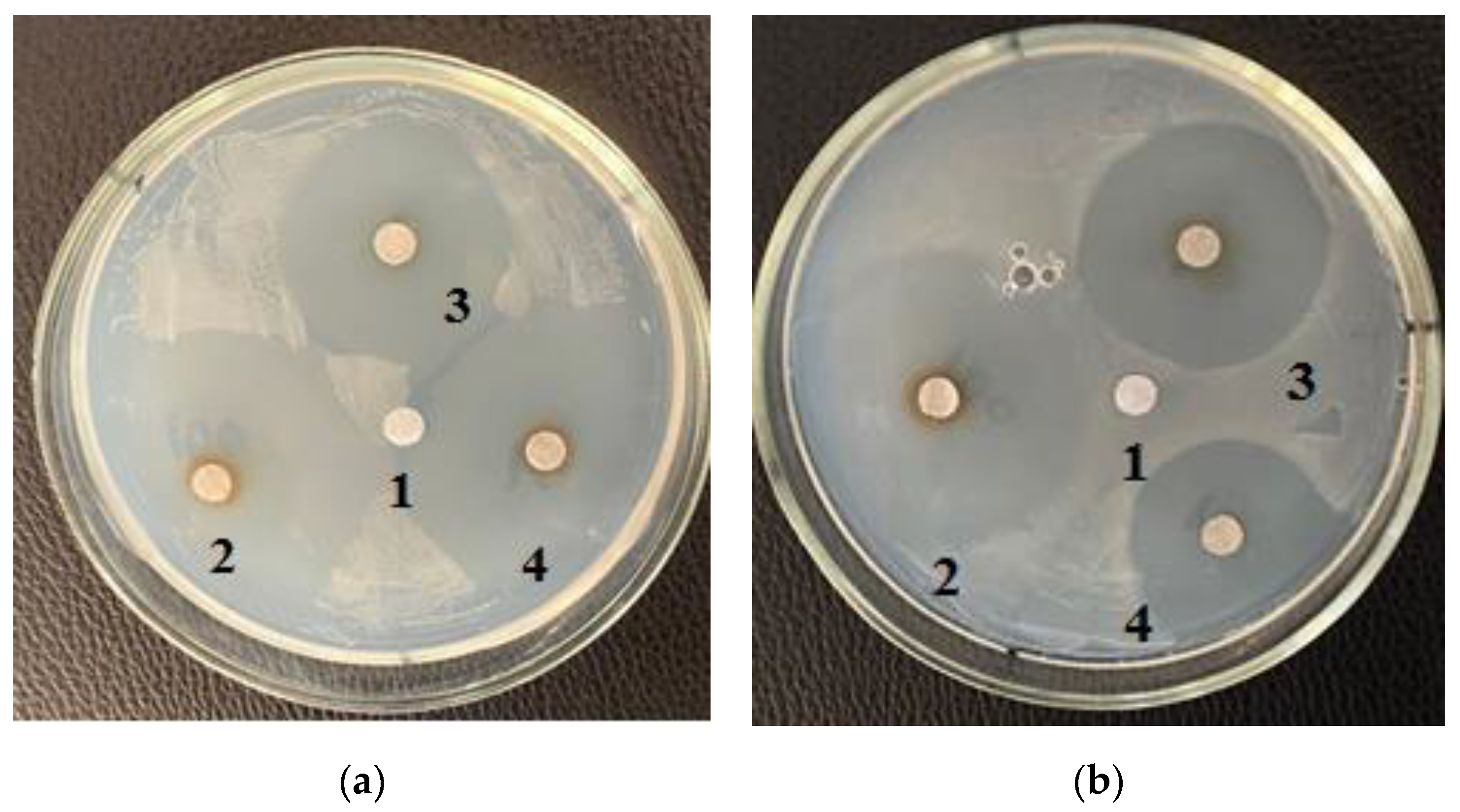
Table 4 shows the zones of inhibition results of various concentrations of purified carbohydrate complex obtained from
A. platensis. Antimicrobial activity against
E. coli,
B. pumilus, and
B. subtilis was revealed. The greatest inhibitory effect of samples of the carbohydrate complex of microalgae was observed at a concentration of 100.0 mg/mL, and the inhibition zone was 3.0–4.2 cm.
Other studies have also shown the antimicrobial activity of various extracts from
C. vulgaris biomass [
34,
35]. In this study [
34], an aqueous extract of C. vulgaris showed antibacterial activity against both Gram-negative (
E. coli) and Gram-positive (
S. aureus) bacteria. It was found that an aqueous extract at a concentration of 150 mg/mL exhibited antimicrobial activity against E. coli, and the diameter of the inhibition zone was 2.4 cm. The highest antimicrobial activity against E. coli had a protein fraction obtained by TCA from an aqueous extract, with MICs in the range of 32.5–65 mg/mL [
34]. In other studies, extracts of green unicellular algae showed pronounced antagonistic activity against numerous opportunistic and pathogenic bacteria [
33,
34,
35,
36,
37].
It has been established that
Chlorella vulgaris synthesizes silver nanoparticles in the dark, for which antimicrobial activity was studied on three pathogenic microorganisms: Gram-negative bacterium
Erwinia, Gram-positive bacterium
Bacillus sp. and pathogenic fungus
Candida. The specific antibiotics penicillin (10 mg), tetracycline (30 mg), and streptomycin (10 mg) were used as controls. The synthesized AgNP solutions had an inhibitory effect on all tested microorganisms, with the following order of
Bacillus,
Erwinia,
Candida in terms of increasing the radius of the zone of inhibition. The effect of nanoparticles on all test organisms was more pronounced than the effect of silver nitrate and penicillin. This effect can be explained by the fact that silver nanoparticles penetrated the bacterial cell wall and damaged them due to their interactions with compounds containing phosphorus and sulfur, including DNA [
38].
Previous studies have reported that chlorellin (a mixture of fatty acids) from
C. vulgaris exhibited inhibitory activity against both Gram-positive and Gram-negative bacteria [
29,
39]. Linolenic acid in ethanolic extracts of
C. vulgaris also showed antibacterial activity against
Staphylococcus aureus (a common cause of skin infections) and
Salmonella typhi (causative agent of typhoid fever). This property suggests there is a possibility to use
C. vulgaris as a natural antibiotic and that that it could be a promising alternative to traditional synthetic drugs with a wider spectrum of action against pathogenic infections [
29,
40]. One of the most important polysaccharides found in
C. vulgaris samples, β-1,3-glucan, has recently gained popularity among researchers due to its dietary qualities and therapeutic properties on human health, such as scavenging free radicals and lowering blood lipids [
29,
41].
C. vulgaris polysaccharides have also been found to exhibit other health benefits, such as antitumor, antiviral, and strong immunomodulatory effects, indicating potential medical applications [
42].
In this regard, our results confirm and reveal the potential of
C. vulgaris extracts for the production of natural fungicides and bactericides. The use of
C. vulgaris extracts with antimicrobial properties as an alternative to antibiotics in animal husbandry reduces the risk of emergence of antibiotic-resistant bacteria and transmission of antibiotic-resistant pathogens to humans [
43,
44,
45]. Moreover,
C. vulgaris is a source of valuable biomass suitable for use in the production of various bioproducts [
46].
It is also known that
A. platensis has the potential to act as an antimicrobial agent. The antimicrobial activity of
Arthrospira platensis against four types of Gram-positive (
Micrococcus luteus,
Staphylococcus aureus,
Bacillus cereus,
Listeria monocytogene sp.) and two Gram-negative (
Salmonella typhi,
Pseudomonas aeruginosa) bacteria was shown by determining zones of inhibition.
A. platensis showed antibacterial potential against all the studied microorganisms. The inhibition zones of the tested strains varied from 0.9 to 1.3 cm [
33].
Extracts of
A. platensis can be effectively used in aquaculture to combat bacterial diseases [
47]. Cyanobacteria (blue-green algae) have unique biochemical properties and are a potential source of biologically active secondary metabolites. Cyanobacteria produce intracellular and extracellular metabolites with antialgae, antibacterial, antifungal, and antiviral activity [
33,
47]. Spirulina is an ideal bioresource due to its richness in protein, phycocyanin, essential amino acids, polysaccharides, carotenoids, minerals, vitamins, and essential fatty acids [
33,
47]. It is also rich in vitamins, minerals, carbohydrates, and gamma-linolenic acid. Spirulina is of interest not only because of its nutritional value, but also because it has the potential to be used in the development of pharmaceutical preparations. Spirulina has therapeutic effects as a growth promoter, probiotic, and immune system enhancer in animals, including fish [
47]. Phycocyanin is the main biologically active substance in spirulina, and its content ranges from 10 to 15% of dry weight. Spirulina samples exhibit antiviral and antioxidant properties against human pathogens [
47].
Thus, algae and cyanobacteria, in addition to their nutritional value, have a wide range of other properties and characteristics, including antimicrobial ones. The study [
35] showed that
Ascophyllum nodosum had the highest inhibitory effect on the growth of E. coli compared to other algal species. The inhibitory effect of
A. nodosum on the growth of
E. coli is most likely due to functional algae compounds such as phlorotannins, which are polyphenols with bacteriostatic and bactericidal activity [
35,
48].
Lithothamnium calcareum also showed antimicrobial activity due to the ability of algae to produce metabolites of antimicrobial drugs, such as diterpenes [
49], monoterpenes [
50], phenolic compounds [
51], sterols [
52], polysaccharides [
53], and fatty acids [
52,
54]. Studies have confirmed the antimicrobial activity of natural extracts derived from algae and cyanobacteria [
55].
A. nodosum and
C. vulgaris, at the highest concentration tested (1:4), have been found to have significant antibacterial activity.
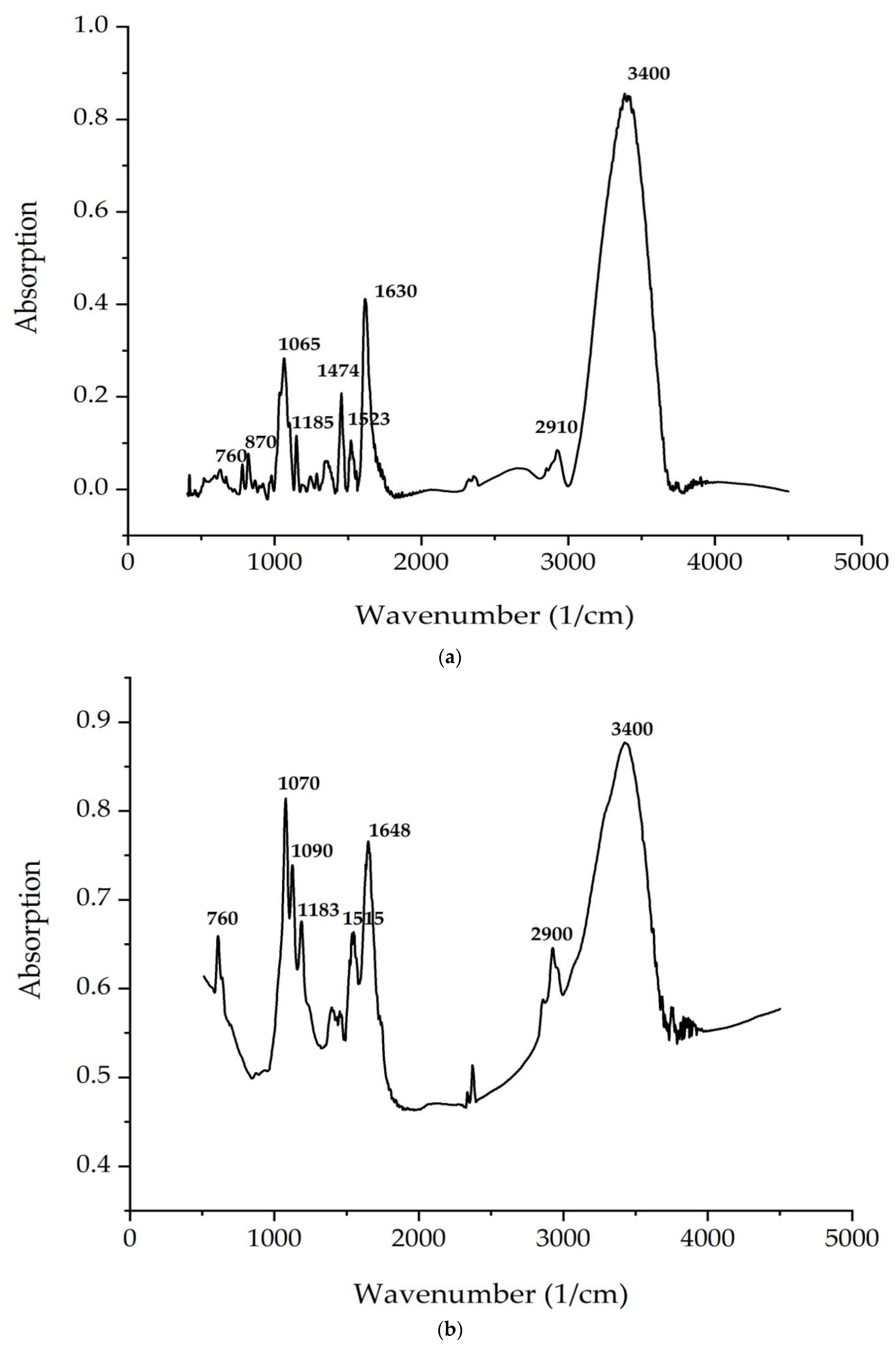
According to
Figure 4 and
Figure 5, the bands correspond to asymmetric and symmetric stretching vibrations of the methylene groups in the range from 2938 to 2835 cm
–1. Asymmetric and symmetric bands were designated as Ʋ (CH2) and Ʋ s(CH2), respectively.
At 1680 cm
−1 (
C. vulgaris) and 1700 cm
−1 (
A. platensis), there is a very important band associated with the stretching vibrations of the carbonyl group (C = O), which is part of the ester bond between fatty acids and glycerol in lipid molecules. However, this type of bond can also be formed by the peroxidation of fatty acid chains. Therefore, it can be assumed that an increase in the intensity of this band indicates an increase in lipid oxidation in the sample. The following bands can be noted: a series of bands from 1474 to 1395 cm
−1 (
A. platensis,
C. vulgaris) (deformation vibrations of the methylene groups of fatty acids), 1183 cm
−1 (
A. platensis) and 1185 cm
−1 (
C. vulgaris) (stretching vibration of group C–O ester bond of lipids), and 1044 cm
–1 (
A. platensis) and 1045 cm
–1 (
C. vulgaris) (stretching vibration of the phosphate group PO
2) [
56].
The analysis of the IR spectra of the
C. vulgaris and
A. platensis lipid complexes confirms the presence of neutral lipids, glyco- and phospholipids, as well as unsaturated fatty acids, such as γ-linolenic acid, in the composition. As shown in
Figure 5a,b, spectrum peaks are typical of saccharides. Absorption in the region of 1200–650 cm
−1 is attributed to the stretching vibrations of the C–O-group of carbohydrates, a number of authors consider the bands at 1090 and 1044 cm
−1 to be absorption bands of cellulose, and bands at 2940 cm
−1 are attributed to starch. The band at 850 cm
−1 indicates type I glycosidic bonds [
57]. Absorption in the region 1040–1200 is associated with the presence of pyranose rings (configurations) [
58].
The peaks recorded at 3400 cm
−1 and 2940 cm
−1 indicate the stretching vibrations of the hydroxyl groups that make up carbohydrates, and the asymmetric stretching vibrations –CH2– of their molecules, respectively. The identified organic compounds showed good antimicrobial activity. In the present study, both algae contained phenolic and alcohol compounds that were responsible for antibacterial activity [
59]. Our results correlate well with data reported in other studies [
60,
61,
62].
The study [
60] used Fourier transform infrared spectroscopy (FTIR) to analyze a lipid extract from a natural isolate of
C. vulgaris. To assess the productivity of the strain, a reference strain was obtained. Lipids were extracted using the Bligh and Dyer method, and the samples were subjected to FTIR analysis to examine the absorption spectra in the range from 2000 to 3000 cm
−1 to confirm the presence of lipids. For a natural
C. vulgaris isolate, lipid samples were analyzed using FTIR. As a result, they had eight clear bands in the range of wavenumbers from 4000 to 5000 cm
−1. These bands were previously identified based on reference standards [
61] and published FTIR spectra for specific molecular groups. The results were interpreted. The absorption peak at 3949.70 cm
−1 and 3840.10 cm
−1 indicates the presence of primary amines and very weak secondary amines. Peaks at 3383.39 cm
−1, which indicate OH group absorption, reveal the presence of a strong alcohol group.
The absorption of C–H appearing at 2144.99 cm−1 indicated the presence of lipids. Absorption at 2920 cm−1 indicated an alkyne group, and that at 1638.05 cm−1 and 1523.62 cm−1 indicated C=C absorption, which is an alkene group.
In particular, the absorption at 1273.16 cm−1 indicated the presence of a strong acid. An absorption at 1149.44 cm−1 indicates an ester group; the remaining absorption range at 589.99 cm−1, 699.12 cm−1, and 1032.75 cm−1 indicates the presence of a strong alkyl halide. The presence of peaks at 3383.39 cm−1, indicating the absorption of OH groups and indicating the presence of a strong alcohol group, determines the presence of antimicrobial properties of the C. vulgaris lipid complex under study.
All
Chlorella microalgae species are very important for lipid and biomass production, are adaptable to a variety of environmental conditions, are tolerant of high CO
2 concentrations, and are useful in industrial wastewater treatment and wastewater treatment systems due to the phenolic compounds found in the microalgae lipid fraction exhibiting significant antimicrobial activity [
20].
The study [
62] showed that the FTIR profile of the
C. vulgaris lipid fraction revealed various chemical functional groups. The FTIR spectrogram of the lipid complex is dominated by a broad strong absorption band at 3465 cm
−1 attributed to the hydroxyl group (-OH absorption), whereas the spectral peaks located in the range of 4000–3400 cm
–1 can be attributed to alcohol and acids [
62], which exhibit the highest antimicrobial properties. Strong peaks in the range from 1628 to 1428 cm
−1 indicate asymmetric and symmetric stretching vibrations, which are related to carboxylate anions (COO-) [
62]. These significant spectral peaks may help in elucidating the structure of microalgae lipid complexes for recognizing metal–carboxylate interactions, according to Flórez-Fernández et al. [
63]. The spectral band at about 2923–2854 cm
–1 is consistent with the band obtained by Aprilliza [
64] and is associated with aliphatic –CH absorption, as well as symmetric and asymmetric absorption (C–H) CH
2, in addition to aromatic and/or vinyl C–H absorption and (CH)-anomer absorption [
65].
The FTIR spectrogram of the
C. vulgaris lipid complex also illustrates the peak of C–H stretching vibrations recognizing alkanes, C=O indicates a carbonyl group, and COO- stretching vibrations are attributed to carboxylate, as well as C–O-C stretching vibrations. Peaks reaching the range of 1090–1030 cm
−1 are related to the C–O absorption of the pyranosyl ring, asymmetric C–O-C absorption (glycosidic bond), and C–C absorption, which are attributed to the structure of the alginate saccharide [
66,
67]. In addition, absorbances of the C=O group were recorded at 1734 cm
−1, as reported by Carpenter and Saharan [
67]. The present results are consistent with those of Cardenas-Jiron et al. [
68] and Bouissil et al. [
69]. According to Gomaa et al. [
70], the spectral band at 848 cm
−1 confirms the presence of sulfate groups in the fucoidan of phenolic compounds. Peaks at around 600 cm
−1 may be associated with symmetric and asymmetric O=S=O deformation, as reported by Flórez-Fernández et al. [
63].
Noura El-Ahmady El-Naggar et al. investigated the production and properties of
Chlorella vulgaris carbohydrates (polysaccharides) [
71]. FT-IR analysis of the extracted polysaccharides showed the presence of N–H, O–H, C–H, –CH3,>CH2, COO–1, S=O, and C=O functional groups. Spectral analysis showed the presence of proteins, nucleic acids, and chemical groups (ether, carbonyl, carboxyl, and amine) [
72].
The study [
72] used the IR spectrum to identify the functional group of active components based on the peak value in the infrared region. The crude powder of
U. lactuca was passed through an IR spectrometer, and the functional groups of the components were separated based on the ratio of their peaks. The results of the FTIR analysis of microalgae showed different peaks at 620.15 and 848.99, with the functional group being alkyl halides; 928.18—the functional group is carboxylic acid; 1086.14 and 1144.12—the functional group is aliphatic amine; 1384.42—the functional group is nitromethane; 1428.88—the functional group is a group representing aromatic compounds; 1634.33—the functional group is amide; 2295.04—the functional group is nitrile; 3406.60—the functional group is alcohol.
U. lactuca sample contained phenols. Similarly, crude
G. corticata powder was passed through the IR spectrum and the functional groups of the components were separated based on the ratio of their peaks. The results of FTIR analysis showed, as in
G. corticata, different peak values: 3321.46—the functional groups are alcohols and phenols; 2925.49—the functional groups are alkanes; 2084.83—the functional groups are allenes, ketenes, cyanates, and isothiocyanates; 1647.84—the functional groups are isamides; 1471.30—the functional groups are isaromatic compounds. The
G. corticata sample contained isaliphatic amines with the functional group (1116.62), primary and secondary amines with the functional group (874.41), and alkyl halides with the functional group (750.98, 712.56, 657.10, and 617.26) [
72].