Antibiotic Resistance Genes Occurrence in Conventional and Antibiotic-Free Poultry Farming, Italy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Activities
2.2. Extraction of Nucleic Acids and ARG Screening
2.3. Statistical Analysis
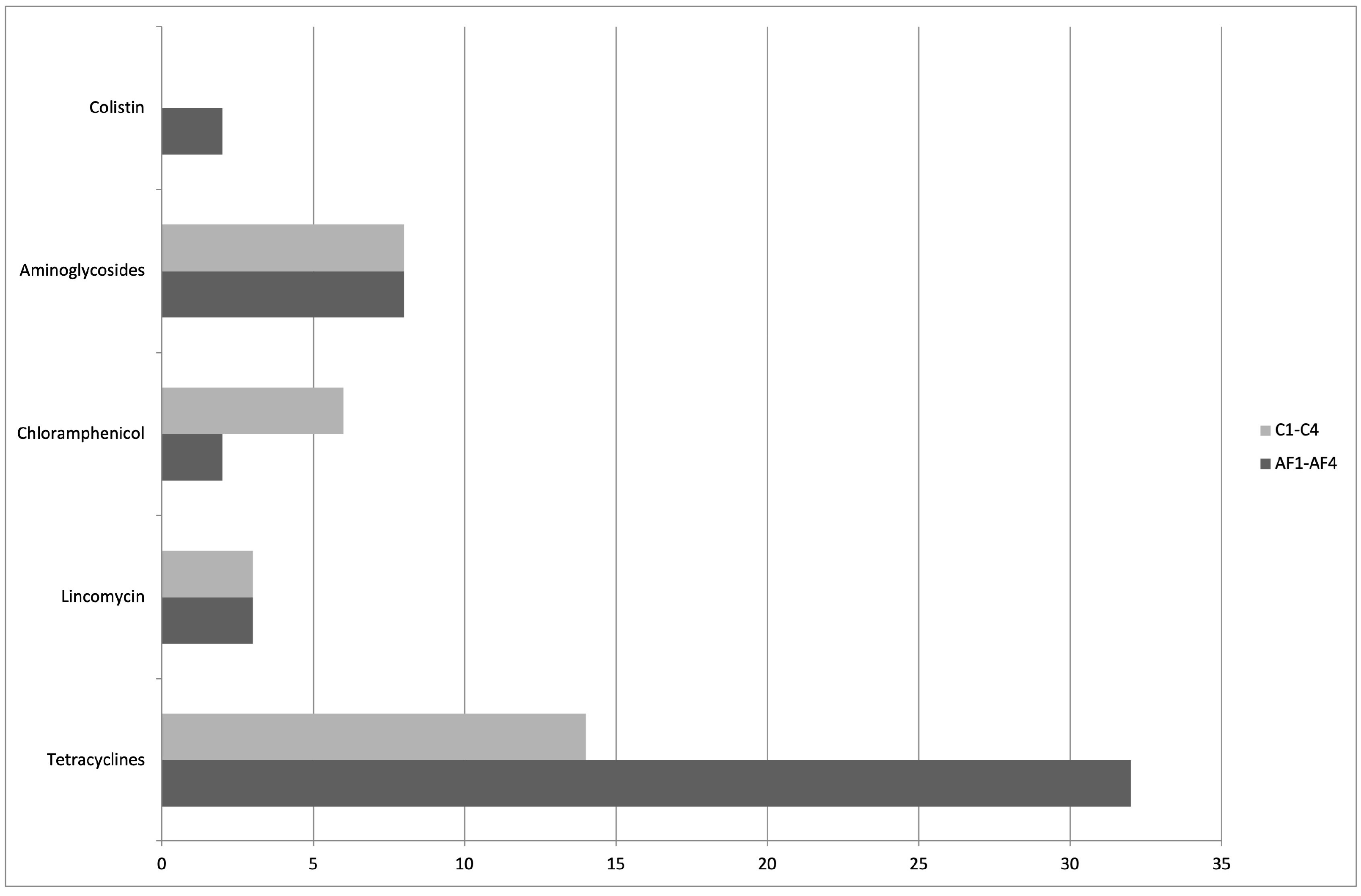
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [PubMed]
- de Mesquita Souza Saraiva, M.; Lim, K.; do Monte, D.F.M.; Givisiez, P.E.N.; Alves, L.B.R.; de Freitas Neto, O.C.; Kariuki, S.; Júnior, A.B.; de Oliveira, C.J.B.; Gebreyes, W.A. Antimicrobial resistance in the globalized food chain: A One Health perspective applied to the poultry industry. Braz. J. Microbiol. 2022, 53, 465–486. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Multi-Partner Trust Fund Annual Report 2021; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization; Food and Agriculture Organization of the United Nations; United Nations Environment Programme and World Organisation for Animal Health: Geneva, Switzerland, 2022.
- EFSA Panel on Animal Health and Welfare (AHAW); Nielsen, S.S.; Bicout, D.J.; Calistri, P.; Canali, E.; Drewe, J.A.; Garin-Bastuji, B.; Rojas, J.L.G.; Schmidt, C.G.; Herskin, M.; et al. Assessment of animal diseases caused by bacteria resistant to antimicrobials: Poultry. EFSA J. 2021, 19, e07114. [Google Scholar] [CrossRef]
- ISMEA, Istituto di Servizi per il Mercato Agricolo Alimentare. Il Biologico nel 2021 e il Futuro del Settore. Available online: https://www.ismea.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/11843 (accessed on 1 July 2022).
- Van Loo, E.; Caputo, V.; Nayga, R.M., Jr.; Meullenet, J.-F.; Crandall, P.G.; Ricke, S.C. Effect of organic poultry purchase frequency on consumer attitudes toward organic poultry meat. J. Food Sci. 2010, 75, S384–S397. [Google Scholar] [CrossRef] [PubMed]
- Kassem, I.I.; Kehinde, O.; Kumar, A.; Rajashekara, G. Antimicrobial-Resistant Campylobacter in Organically and Conventionally Raised Layer Chickens. Foodborne Pathog. Dis. 2017, 14, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, H.S.; Yim, J.H.; Kim, Y.J.; Kim, D.H.; Chon, J.W.; Kim, H.; Om, A.S.; Seo, K.H. Comparison of the isolation rates and characteristics of Salmonella isolated from antibiotic-free and conventional chicken meat samples. Poult. Sci. 2017, 96, 2831–2838. [Google Scholar] [CrossRef]
- Bailey, M.A.; Taylor, R.M.; Brar, J.S.; Corkran, S.C.; Velásquez, C.; Novoa Rama, E.; Oliver, H.F.; Singh, M. Prevalence and antimicrobial resistance of Campylobacter from antibiotic-free broilers during organic and conventional processing. Poult. Sci. 2019, 98, 1447–1454. [Google Scholar] [CrossRef]
- Musa, L.; Casagrande Proietti, P.; Branciari, R.; Menchetti, L.; Bellucci, S.; Ranucci, D.; Marenzoni, M.L.; Franciosini, M.P. Antimicrobial Susceptibility of Escherichia coli and ESBL-Producing Escherichia coli Diffusion in Conventional, Organic and Antibiotic-Free Meat Chickens at Slaughter. Animals 2020, 10, 1215. [Google Scholar] [CrossRef]
- Di Francesco, C.E.; Smoglica, C.; Profeta, F.; Farooq, M.; Di Giannatale, E.; Toscani, T.; Marsilio, F. Research Note: Detection of antibiotic-resistance genes in commercial poultry and turkey flocks from Italy. Poult. Sci. 2021, 100, 101084. [Google Scholar] [CrossRef]
- Laconi, A.; Mughini-Gras, L.; Tolosi, R.; Grilli, G.; Trocino, A.; Carraro, L.; Di Cesare, F.; Cagnardi, P.; Piccirillo, A. Microbial community composition and antimicrobial resistance in agricultural soils fertilized with livestock manure from conventional farming in Northern Italy. Sci. Total Environ. 2021, 760, 143404. [Google Scholar] [CrossRef]
- Zhang, H.N.; Dong, M.J.; Zhou, Y.F.; Sun, J.X.; Chang, M.J.; Zhai, Z.Z. Animal Manure Fertilization Promotes Antibiotic Resistance Gene Dissemination Among Manure, Soil, and Vegetables. Huan Jing Ke Xue 2021, 42, 2080–2088. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- De Cesare, A.; Oliveri, C.; Lucchi, A.; Savini, F.; Manfreda, G.; Sala, C. Pilot Study on Poultry Meat from Antibiotic Free and Conventional Farms: Can Metagenomics Detect Any Difference? Foods 2022, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Salerno, B.; Furlan, M.; Sabatino, R.; Di Cesare, A.; Leati, M.; Volanti, M.; Barco, L.; Orsini, M.; Losasso, C.; Cibin, V. Antibiotic resistance genes load in an antibiotic free organic broiler farm. Poult. Sci. 2022, 101, 101675. [Google Scholar] [CrossRef]
- European Medicines Agency. Sales of veterinary antimicrobial agents in 31 European countries in 2019 and 2020. In European Surveillance of Veterinary Antimicrobial Consumption; EMA/58183/2021; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Critically Important Antimicrobials for Human Medicine, 6th Revision; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2019.
- Stata Statistical Software: Release 16; StataCorp LLC: College Station, TX, USA, 2019.
- White, A.; Hughes, J.M. Critical Importance of a One Health Approach to Antimicrobial Resistance. Ecohealth 2019, 16, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Ruffini, F.; (Gesco Cons. Coop a r.l., Teramo, Italy). Personal communication, 2022.
- Khine, N.O.; Lugsomya, K.; Niyomtham, W.; Pongpan, T.; Hampson, D.J.; Prapasarakul, N. Longitudinal Monitoring Reveals Persistence of Colistin-Resistant Escherichia coli on a Pig Farm Following Cessation of Colistin Use. Front. Vet. Sci. 2022, 9, 845746. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar] [CrossRef] [PubMed]
- Storey, N.; Cawthraw, S.; Turner, O.; Rambaldi, M.; Lemma, F.; Horton, R.; Randall, L.; Duggett, N.A.; AbuOun, M.; Martelli, F.; et al. Use of genomics to explore AMR persistence in an outdoor pig farm with low antimicrobial usage. Microb. Genom. 2022, 8, 000782. [Google Scholar] [CrossRef] [PubMed]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Duarte, I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics 2019, 8, 23. [Google Scholar] [CrossRef]
- Di Francesco, A.; Salvatore, D.; Sakhria, S.; Catelli, E.; Lupini, C.; Abbassi, M.S.; Bessoussa, G.; Ben Yahia, S.; Ben Chehida, N. High Frequency and Diversity of Tetracycline Resistance Genes in the Microbiota of Broiler Chickens in Tunisia. Animals 2021, 11, 377. [Google Scholar] [CrossRef]
- Ghosh, S.; LaPara, T.M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007, 1, 191–203. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Guo, X.; Yan, Z.; Wang, W.; Chen, B.; Ge, F.; Ye, B. A Comprehensive Analysis on Spread and Distribution Characteristic of Antibiotic Resistance Genes in Livestock Farms of Southeastern China. PLoS ONE 2016, 11, e0156889. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dyall-Smith, M.; Marenda, M.; Hu, H.W.; Browning, G.; Billman-Jacobe, H. Antibiotic Resistance Genes in Antibiotic-Free Chicken Farms. Antibiotics 2020, 9, 120. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, M.J.; Min, B.R.; Castleberry, L.; Waldrip, H.; Parker, D.; Brauer, D.; Pitta, D.; Nagaraju, I. Antibiotic resistance, antimicrobial residues, and bacterial community diversity in pasture-raised poultry, swine, and beef cattle manures. J. Anim. Sci. 2021, 99, skab144. [Google Scholar] [CrossRef]
- Juricova, H.; Matiasovicova, J.; Kubasova, T.; Cejkova, D.; Rychlik, I. The distribution of antibiotic resistance genes in chicken gut microbiota commensals. Sci. Rep. 2021, 11, 3290. [Google Scholar] [CrossRef] [PubMed]
- Agersø, Y.; Jensen, L.B.; Givskov, M.; Roberts, M.C. The identification of a tetracycline resistance gene tet(M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 2002, 214, 251–256. [Google Scholar] [CrossRef]
- de Vries, L.E.; Christensen, H.; Skov, R.L.; Aarestrup, F.M.; Agersø, Y. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 2009, 64, 490–500. [Google Scholar] [CrossRef]
- Semedo-Lemsaddek, T.; Cota, J.B.; Ribeiro, T.; Pimentel, A.; Tavares, L.; Bernando, F.; Oliveira, M. Resistance and virulence distribution in enterococci isolated from broilers reared in two farming systems. Ir. Vet. J. 2021, 74, 22. [Google Scholar] [CrossRef]
- Pesciaroli, M.; Magistrali, C.F.; Filippini, G.; Epifanio, E.M.; Lovito, C.; Marchi, L.; Maresca, C.; Massacci, F.R.; Orsini, S.; Scoccia, E.; et al. Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 2020, 314, 108391. [Google Scholar] [CrossRef]
- Cavicchio, L.; Dotto, G.; Giacomelli, M.; Giovanardi, D.; Grilli, G.; Franciosini, M.P.; Trocino, A.; Piccirillo, A. Class 1 and class 2 integrons in avian pathogenic Escherichia coli from poultry in Italy. Poult. Sci. 2015, 94, 1202–1208. [Google Scholar] [CrossRef]
- Roy, P.; Dhillon, A.S.; Lauerman, L.H.; Schaberg, D.M.; Bandli, D.; Johnson, S. Results of Salmonella isolation from poultry products, poultry, poultry environment, and other characteristics. Avian Dis. 2002, 46, 17–24. [Google Scholar] [CrossRef]
- Obeng, A.S.; Rickard, H.; Sexton, M.; Pang, Y.; Peng, H.; Barton, M. Antimicrobial susceptibilities and resistance genes in Campylobacter strains isolated from poultry and pigs in Australia. J. Appl. Microbiol. 2012, 113, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Bendary, M.M.; Abd El-Hamid, M.I.; El-Tarabili, R.M.; Hefny, A.A.; Algendy, R.M.; Elzohairy, N.A.; Ghoneim, M.M.; Al-Sanea, M.M.; Nahari, M.H.; Moustafa, W.H. Clostridium perfringens Associated with Foodborne Infections of Animal Origins: Insights into Prevalence, Antimicrobial Resistance, Toxin Genes Profiles, and Toxinotypes. Biology 2022, 11, 551. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Chaslus-Dancla, E. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 2001, 32, 201–225. [Google Scholar] [CrossRef] [PubMed]
- Falgenhauer, L.; Ghosh, H.; Guerra, B.; Yao, Y.; Fritzenwanker, M.; Fischer, J.; Helmuth, R.; Imirzalioglu, C.; Chakraborty, T. Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Vet. Microbiol. 2017, 200, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Arsène, M.M.J.; Davares, A.K.L.; Viktorovna, P.I.; Andreevna, S.L.; Sarra, S.; Khelifi, I.; Sergueïevna, D.M. The public health issue of antibiotic residues in food and feed: Causes, consequences, and potential solutions. Vet. World. 2022, 15, 662–671. [Google Scholar] [CrossRef]
- Samper-Cativiela, C.; Diéguez-Roda, B.; Trigo da Roza, F.; Ugarte-Ruiz, M.; Elnekave, E.; Lim, S.; Hernández, M.; Abad, D.; Collado, S.; Sáez, J.L.; et al. Genomic characterization of multidrug-resistant Salmonella serovar Kentucky ST198 isolated in poultry flocks in Spain (2011–2017). Microb. Genom. 2022, 8, 000773. [Google Scholar] [CrossRef]
- Esperón, F.; Sacristán, C.; Carballo, M.; Torre, A. Antimicrobial Resistance Genes in Animal Manure, Manure-Amended and Nonanthropogenically Impacted Soils in Spain. Adv. Biosci. Biotechnol. 2018, 9, 469–480. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 2020, 318, 108478. [Google Scholar] [CrossRef]
- Vounba, P.; Rhouma, M.; Arsenault, J.; Bada Alambédji, R.; Fravalo, P.; Fairbrother, J.M. Prevalence of colistin resistance and mcr-1/mcr-2 genes in extended-spectrum β-lactamase/AmpC-producing Escherichia coli isolated from chickens in Canada, Senegal and Vietnam. J. Glob. Antimicrob. Resist. 2019, 19, 222–227. [Google Scholar] [CrossRef]
- Gagliotti, C.; Bolzoni, L.; Carretto, E.; Sarti, M.; Ricchizzi, E.; Ambretti, S.; Barozzi, A.; Bracchi, C.; Confalonieri, M.; Menozzi, I.; et al. Reduction trend of mcr-1 circulation in Emilia-Romagna Region, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2585–2592. [Google Scholar] [CrossRef]
- Clemente, L.; Manageiro, V.; Correia, I.; Amaro, A.; Albuquerque, T.; Themudo, P.; Ferreira, E.; Caniça, M. Revealing mcr-1-positive ESBL-producing Escherichia coli strains among Enterobacteriaceae from food-producing animals (bovine, swine and poultry) and meat (bovine and swine), Portugal, 2010–2015. Int. J. Food Microbiol. 2019, 296, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Bastidas-Caldes, C.; de Waard, J.H.; Salgado, M.S.; Villacís, M.J.; Coral-Almeida, M.; Yamamoto, Y.; Calvopiña, M. Worldwide Prevalence of mcr-mediated Colistin-Resistance Escherichia coli in Isolates of Clinical Samples, Healthy Humans, and Livestock—A Systematic Review and Meta-Analysis. Pathogens 2022, 11, 659. [Google Scholar] [CrossRef] [PubMed]
- Maron, D.F.; Smith, T.J.; Nachman, K.E. Restrictions on antimicrobial use in food animal production: An international regulatory and economic survey. Glob. Health 2013, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Alexander, J.; Bierbaum, G.; Hammerl, J.A.; Hembach, N.; Schwartz, T.; Schmithausen, R.M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria, antibiotic resistance genes, and antibiotic residues in wastewater from a poultry slaughterhouse after conventional and advanced treatments. Sci. Rep. 2021, 11, 16622. [Google Scholar] [CrossRef]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect. 2018, 24, 1241–1250. [Google Scholar] [CrossRef] [Green Version]

| Farms | ARG Litter Samples |
|---|---|
| C1 | tetA, tetB, tetL, catA1, aadA2 |
| C2 | tetA, tetB, tetL, catA1, aadA2, lnuA |
| C3 | tetK, tetM, tetA(P), catA1, aadA2, lnuB |
| C4 | tetK, tetM, tetA(P), aadA2 |
| AF1 | tetA, tetB, tetC, tetK, tetL, tetM, lnuA, catA1, aadA2, aac(3)IV, mcr-1 |
| AF2 | tetA, tetB, tetK, tetL, tetM, tetA(P), tetB(P), catA1, aadA2 |
| AF3 | tetA, tetB, tetM, tetA(P), tetB(P), aadA2, aac(3)IV |
| AF4 | tetA, tetB, tetC, tetM, InuA, aadA2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooq, M.; Smoglica, C.; Ruffini, F.; Soldati, L.; Marsilio, F.; Di Francesco, C.E. Antibiotic Resistance Genes Occurrence in Conventional and Antibiotic-Free Poultry Farming, Italy. Animals 2022, 12, 2310. https://doi.org/10.3390/ani12182310
Farooq M, Smoglica C, Ruffini F, Soldati L, Marsilio F, Di Francesco CE. Antibiotic Resistance Genes Occurrence in Conventional and Antibiotic-Free Poultry Farming, Italy. Animals. 2022; 12(18):2310. https://doi.org/10.3390/ani12182310
Chicago/Turabian StyleFarooq, Muhammad, Camilla Smoglica, Fausto Ruffini, Lidia Soldati, Fulvio Marsilio, and Cristina E. Di Francesco. 2022. "Antibiotic Resistance Genes Occurrence in Conventional and Antibiotic-Free Poultry Farming, Italy" Animals 12, no. 18: 2310. https://doi.org/10.3390/ani12182310






