Abstract
Crop breeding for high nitrogen use efficiency (NUE) or tolerance to low nitrogen fertilization is thought to be an ideal solution to reduce the cost, carbon footprint, and other environmental problems caused by the excess use of nitrogen fertilizers. As a model plant for cereal crops, barley has many advantages, including good adaptability, a short growth period, and high natural stress resistance or tolerance. Therefore, research on improving NUE in barley is not only beneficial for nitrogen-efficient barley breeding but will also inform NUE improvement in other cereal crops. In this review, recent progress in understanding barley’s response to nitrogen nutrition, evaluation of NUE or low-nitrogen tolerance, quantitative trait loci (QTL) mapping and gene cloning associated with improving NUE, and breeding of nitrogen-efficient barley is summarized. Furthermore, several biotechnological tools that could be used for revealing the molecular mechanisms of NUE or breeding for improving NUE in barley are introduced, including GWAS, omics, and gene editing. The latest research ideas in unraveling the molecular mechanisms of improving NUE in other crops are also discussed. Thus, this review provides a better understanding of improving the NUE of barley and some directions for future research in this area.
1. Introduction
Nitrogen (N) is one of the most important components of many biological large molecules, such as nucleic acids, proteins, chlorophylls, and phospholipids. It is also an essential macro element for plant growth and development. Therefore, chemical N fertilizers are produced and applied to ensure maximum productivity because the usable soil N is usually limited [1,2]. The application of chemical synthetic N fertilizers has increased rapidly over the past 50 years, while the growth rate of chemical nitrogen fertilizer application is much higher than that of crop yield [3,4]. In addition, the overall efficient use rate of N fertilizers is still no more than 50%, and up to 75% of applied N is lost by leaching into the soil, causing environmental problems and escalating global warming [5,6,7,8]. The application of synthetic N fertilizers also causes excessive energy consumption and a higher cost of crop production. It is estimated that the production of 1 tonne of ammonia consumes 873 m3 of natural gas [9]. Moreover, the N fertilizer-dependent breeding strategy to increase crop production has reached a plateau [3]. Therefore, it is necessary to use new solutions to improve crop yield while decreasing or maintaining the application of N fertilizers, and one such solution is improving the N use efficiency (NUE) of crops.
There are various definitions of NUE in different studies, while the most common and widely-accepted definition is grain production per unit of N available in the soil (including the residual N in the soil and the N fertilizer) [10]. NUE is determined by the efficiencies of two distinct processes: N uptake and N utilization (NUpE and NUtE) [1,10,11]. These processes are mainly related to N absorption, translocation, assimilation, and redistribution [1,12,13,14]. However, considering the difficulties of field trials, hydroponic experiments and seedling biomass-related traits tend to be chosen for the evaluation of NUE [15].
The NUE of crops could be improved by both N management practices and breeding [16,17]. In crop breeding, traditional breeding (or conventional breeding) and genetic engineering are the primary methods used to produce crop varieties with high NUE. Both methods rely on identifying and exploiting NUE-related genes [2,18]. Traditional breeding can only utilize genes from closely related species or the same species, whereas genetic engineering can obtain useful genes from a variety of sources. NUE is very complicated and is a quantitative trait determined by multiple genes. Therefore, it is imperative to conduct research related to the molecular mechanisms of high NUE and obtain as many useful genes as possible. Thus far, there has been great progress in rice [19,20,21,22,23,24], but few significant developments have been made in barley.
Barley is one of the most important cereal crops in the world and an important genetic model for cereal crops. Its ability to adapt to various environments suggests that it may have many genes for stress resistance and efficient utilization of nutrients. Thus, the study of the molecular mechanisms of NUE in barley will be beneficial for high NUE breeding in barley and other crops. Here, the progress of low-N tolerance and high NUE-related research in barley over the last 20 years is summarized. We mainly focus on barley responses to N fertilizer, the evaluation of barley varieties with high NUE, barley QTL mapping and gene cloning for high NUE, barley breeding with high NUE, and new technologies and perspectives for revealing the molecular mechanisms of NUE in barley.
2. Effects of N Fertilizers on Barley Growth, Yield, and Quality
N has a great impact on barley growth, development, yield, and quality. McKenzie et al. [25] showed that the yield of barley grains increased with additional application of N fertilizers and then plateaued or decreased if N fertilization was in excess. Thus, there was always an optimal fertilization amount for barley production, but it varied for different barley genotypes. Grain protein concentration also rose with increased N fertilization, showing an obvious linear relationship, although it too was affected by the barley genotype. The grain protein concentration is the most important quality characteristic for malting barley and is normally 10–13%. Therefore, effects on both grain yield and quality must be considered in the application of N fertilizers to achieve the best balance in malting barley production.
De et al. [26] found that many barley traits were affected by different levels of N fertilizer application at the flowering stage, including chlorophyll content, net photosynthetic rate (Pn), stomatal conductance (Gs), transpiration rate (Tr), initial fluorescence (Fo), maximum fluorescence (Fm), maximal photochemical efficiency (Fv/Fm), photochemical quenching coefficient (qP), N accumulation rate after anthesis, contribution rate of N accumulation to grain after anthesis, leaf N translocation rate, stem N translocation rate, contribution rate of leaf N translocation to grain, contribution rate of stem N translocation to grain, N fertilizer production efficiency (NGPE) (NGPE = Grain yield/N accumulation in above ground of barley plants), N agriculture efficiency (NAE) (NAE = (Grain yield with N fertilization − Grain yield without N fertilization)/N fertilization application), N fertilizer partial productivity (PEP) (PEP = Grain yield/N fertilizer application), and N physiological efficiency (NPE) (NPE = (Grain yield with N fertilization-grain yield without N fertilization)/(N accumulation in above ground of barley plants with N fertilization − N accumulation in above ground of barley plants without N fertilization)). Among these traits, some were improved but then reduced upon increasing N fertilizer application, e.g., chlorophyll content, Pn, Gs, Tr, Fo, Fm, Fv/Fm, qP, and grain yield, indicating that there was an optimal N fertilizer application rate for barley production. Other traits were initially increased but then reached a plateau, e.g., N accumulation rate after anthesis and contribution rate of N accumulation in the grain after anthesis, indicating that there was a limitation for N absorption in barley. Conversely, other traits, such as leaf N translocation rate, stem N translocation rate, the contribution rate of leaf N translocation to grain, and the contribution rate of stem N translocation to grain, NGPE and NPE were initially decreased and then plateaued or re-increased, indicating their variable responses to different low N treatments. Furthermore, the traits of NAE and PEP were decreased with the increase of N fertilizer application, indicating that the increase of N fertilizer application would reduce the NUE of barley. There were also differences between the barley varieties with high NUE and low NUE, with the N-efficient barley varieties showing advantages in photosynthesis-related traits, leaf and stem N translocation rate, the contribution rate of leaf and stem N translocation to grain, NAE, PEP, and grain yield.
The hydroponic experiments showed that the restriction of barley seedlings was mainly in shoots when considering the dry biomass under low-N treatment, while the opposite trend was observed in roots [15,27,28]. Furthermore, the uninhibited shoots were usually accompanied by significant growth promotion of the roots, while the opposite was not necessarily true [27]. This indicated that the inhibition of shoots might be more accurate for distinguishing barley genotypes with different low-N tolerances. However, low-N stress also caused chlorosis in barley, indicating that low-N stress or N deficiency could affect or impair chlorophyll synthesis and photosynthesis. The effects of low-N treatment in barley are briefly summarized in Figure 1.
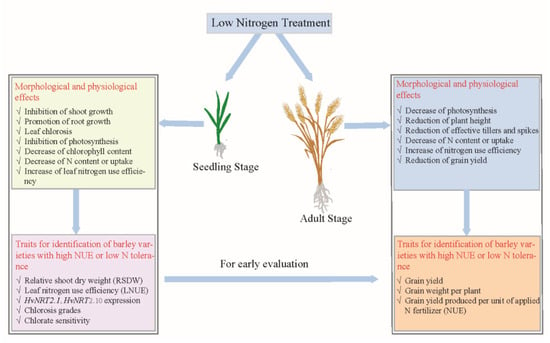
Figure 1.
The effects of low nitrogen treatment on barley at seedling and adult stages and the related traits for identification of barley varieties with high NUE or low nitrogen tolerance.
3. Methods for Identification and Evaluation of Barley with High NUE
NUE comprises two processes, namely NUpE and NutE, but its evaluation is still complex and difficult to measure. Therefore, a variety of traits have been used to evaluate NUE. Grain yield in the field is the best trait for screening barley cultivars with high NUE, although it requires more time and space to conduct the experiments. These experiments are also greatly limited by both their cost and efficiency. Therefore, simple and rapid methods for identifying low-N tolerance are needed, especially those related to the early stages of plant growth [29].
Chen et al. [30,31] analyzed the relationship of related traits between the seedling stage by using a hydroponic experiment and the adult stage by using a field trial under different N treatments to establish a method for early screening and identification of barley genotypes with low-N tolerance. In those studies, traits of relative nitrogen uptake per shoot (RNU), relative shoot dry weight (RSDW), relative tiller number (RTN), and relative grain yield (RGY) were more suitable for screening barley varieties with different responses to low N treatment, and the RUN and RTN were thought to be used for the evaluation and identification of barley varieties with low-N tolerance. Subsequently, Yang et al. [15] also used the RSDW for identifying barley genotypes with low-N tolerance and further classified them into three types: RSDW > 0.9 indicated low-N tolerance; 0.7 < RSDW < 0.9 indicated moderately low-N tolerance; and RSDW < 0.7 indicated low N sensitivity. However, some modifications have been made. Karunarathne et al. [32] used a similar criterion based on the RSDW for the evaluation of low-N tolerance in barley: RSDW > 0.75 indicated low-N tolerance; 55% < RSDW < 75% indicated moderately low-N tolerance; and RSDW < 55% indicated low N sensitivity. The use of RSDW initially established an easy and rapid method for screening barley genotypes with low-N tolerance at the seedling stage and had advantages for the large-scale identification of barley genotypes with low-N tolerance. However, the values of RSDW are not suitable for statistical analysis, and they can also vary significantly if the concentration or the duration of low N treatment changes. Therefore, Jiang et al. [28] and Chen et al. [33] further developed methods for screening barley genotypes with low-N tolerance based on the direct comparison of shoot dry weight (SDW) between a certain low-N treatment and control conditions with a certain treatment period.
Recently, Karunarathne et al. [34] just established a method for the identification of barley varieties with different NUE according to leaf chlorosis by the combined treatment of low-N and chlorate. They were divided into five grades: 0 for green, 1 for 1–20% leaf chlorosis, 2 for 20–40% leaf chlorosis, 3 for 40–60% leaf chlorosis, 4 for 60–80% leaf chlorosis, and 5 for 80–100% leaf chlorosis. Furthermore, our lab also established a method for discrimination of barley varieties with high NUE by chlorate treatment, but the response to chlorate was estimated according to chlorate sensitivity based on traits of seedling height or shoot dry weight (SDW) [35,36]. Decouard et al. [37] established a method for the classification of barley varieties with different low-nitrogen tolerance based on leaf nitrogen use efficiency (LNUE).
Transporters play pivotal roles in the uptake of N from soils, and Quan et al. [38] conducted transcriptome analysis according to the expression of the HvNRT2.1 gene. Thus, this gene might be used as a marker gene for the evaluation of NUE, and its function was also verified in Arabidopsis [39]. In addition, the HvNRT2.10 gene was commonly up-regulated in the three types of barley varieties with different low-nitrogen tolerance, suggesting that this gene might be used as a marker gene [37]. The traits for the identification of barley varieties with high NUE or low N tolerance were also briefly summarized in Figure 1.
4. Quantitative Trait Loci (QTL) Mapping and Gene Cloning Related to NUE in Barley
There are two main genetic approaches for improving NUE in modern crop breeding: marker-assisted selection (MAS) and genetic transformation [1,40,41]. However, genetic transformation in barley is restricted in many regions because of the requirement for risk assessment and related policies covering genetically modified plants, and transformation with genes cloned based on homologous has not consistently improved NUE in many studies, especially in those performed in the field [41]. Therefore, researchers have high expectations for genetic mapping based on gene cloning. Markers linked with QTL/genes could also be used for MAS breeding to speed up the breeding process and improve breeding efficiency. There are also many studies related to QTL mapping for high NUE; these studies were recently reviewed by Karunarathne et al. [2].
QTL mapping for NUE has primarily been conducted under different N supplies, typically including a low-N treatment. A variety of traits have been used for the evaluation of NUE, including agronomic, morphological, and physiological traits. Using a doubled haploid (DH) population of barley, Saal et al. [42] detected 82 QTL for agronomic characteristics under two N fertilization conditions in six different environments. Schnaithmann and Pillen [43] found 65 QTL for 13 traits under low and high N fertilization by using wild barley introgression lines, and QTL detected only under low N fertilization might be beneficial for improving low-N tolerance in barley. Hoffmann et al. [44] detected 58 QTL for 10 traits under two different N supplies using wild barley introgression lines at the seedling stage and found that some were consistent with those detected at the adult stage. This also provides a strategy to identify QTL related to NUE at an early stage. Thus, QTL mapping at the seedling stage was thought to be a fast and cost-effective method for MAS or breeding with high NUE in barley. Kindu et al. [45] detected 41 QTL for 18 traits using a recombinant inbred line (RIL) population, and most QTL for NUE or its components were detected under low-N treatments. Han et al. [41] conducted a combined analysis by using predicted genes in barley identified according to similarity with genes with potential roles for improving NUE in other plants and QTL mapping based on NUE-related traits in barley; then, they attempted to obtain the QTL or genes of high NUE with the greatest possibilities. Specific QTL under low-nitrogen supply are summarized in Table 1.

Table 1.
Summary of potential QTL related to NUE.
With the development of next-generation sequencing technology and its greatly reduced cost, QTL mapping based on SNP markers has become very convenient, and it can be accomplished by re-sequencing DNA from two bulked DNA pools [46]. The constraints of bi-parental mapping population construction have also been greatly reduced because of the high density of SNP markers. There have been some reports of this kind of work on barley, such as studies involving plant height [47], net blotch disease [48], yield and grain plumpness [49], grain amylopectin content [50], and grain size and weight [51]. However, there are no reports on NUE. Furthermore, different mapping populations often give rise to different QTL because quantitative traits are controlled by multiple genes; thus, QTL mapping based on bi-parental populations still has many limitations in gene discovery [52]. Genome-wide association study (GWAS) analysis based on DNA sequencing or SNP markers provides a possible solution for resolving this problem, and there are several reports on the identification of candidate genes for NUE using GWAS in barley [32,34,53]. This will be further described in detail in the following section.
Substantial progress has been made in gene identification for improving NUE in other crops, especially in rice [23,24], and this has been reviewed by Li et al. [54]. In barley, however, although many potential candidate genes have been predicted, their real functions have not yet been identified [32,41]. There are a few reports about the identified N efficiency genes in barley, such as one describing a barley gene encoding alanine aminotransferase (HvAlaAT) that significantly improved NUE in rice [55,56], and another two recent reports showing that HvNRT2.1 and HvNLP2 might also be beneficial for improving the NUE [39,57]. It was shown that the biomass and grain yield of HvAlaAT overexpression transgenic rice lines were significantly higher than sibling nulls or wild-type controls under different N application rates. The seeds of transgenic Arabidopsis thaliana overexpressing the HvNRT2.1 gene were significantly enlarged, including seed length and width, resulting in increased thousand-kernel weight. In the HvNLP2 mutants, not only the expressions of the nitrate-responsive genes were suppressed under nitrate treatment, but other traits were also inhibited, such as biomass, seed yield and NUE. It suggested that the HvNLP2 gene might play an important role in improving NUE.
5. Breeding of Barley Varieties with High NUE
Since the first ‘Green Revolution’, semi-dwarf crop breeding has become an important method of increasing crop yields using the same amount of chemical N fertilizers; however, crop breeding that is dependent on chemical N fertilizers has a gene selection bias, causing the loss of N-efficiency genes [19]. The excessive use of chemical N fertilizers has also caused numerous problems, including environmental pollution [58]. With the need to further increase crop yield, the demand for decreased chemical N fertilizer application to reduce the carbon footprint of agriculture becomes challenging. Crop breeding with high NUE seems to provide the best choice for increasing or maintaining crop yield while reducing chemical N fertilizer application [3].
Based on the difficulties or limitations of traits for determining barley varieties with high NUE, high NUE breeding in barley has mainly focused on screening or selecting varieties with high yields under a low application of N fertilizers in the present barley varieties [40,59]. ‘Vivar’, ‘Xena’, H97097001001, and H96014002 were thought to have a superior NUE according to their grain yield under low N fertilization [40]. In addition, several reports focused on rapidly creating new barley germplasm with high NUE by using hybridization or mutagenesis combined with microspore culture [60,61]. Germplasms could be used directly as N-efficient barley cultivars or strains for breeding new cultivars with high NUE.
With the identification of N-efficient genes in barley, HvAlaAT and HvNRT2.1 can be used to improve NUE in barley breeding by transgenic technology [39,55,56,62]. NRT1.1B, a special allelic gene for indica rice varieties that only carries one SNP, greatly enhanced the NUE [20]. Thus, NUE improvements in barley breeding could be made by clustered regularly interspaced short palindromic repeat (CRISPR) technology, but the public is still divided about the safety of genome editing; this topic is further discussed in the review by Karunarathne et al. [2].
6. New Techniques and Methods for Improving the NUE of Barley
The improvement of NUE in barley requires many genes involved in N uptake, transport, assimilation, and so on [54,63]. Therefore, it is vital to seek and identify as many genes related to NUE as possible.
With the publication of the high-quality barley reference genome and the drastic reduction of sequencing costs, it has become easier to explore genome-wide genetic variations and conduct high-throughput genotyping [64,65]. Compared to QTL mapping, GWAS is very powerful. Karunarathne et al. [32] conducted a GWAS analysis of ten traits related to NUE among 282 barley accessions under optimal and low-N treatments by using SNP and DArTseq markers. A total of 299 markers associated with those traits were obtained; then, it was narrowed down to 136 marker-trait associations (MTAs) based on the association with at least two traits under low-N, and 66 MTA regions were determined based on those MTAs. Further, 140 candidate genes were identified from the 66 MTA regions, of which 47 were considered more likely associated with high NUE of barley. Karunarathne et al. [34] also conducted a GWAS analysis according to leaf chlorosis among 180 barley RILs and identified nine MTAs. Therefore, GWAS analysis for the identification of NUE genes is not limited by the number of QTL identified from bi-parental populations and can still quickly identify NUE-related genes.
Omics analysis, such as transcriptomics, metabolomics, and ionomics, provides a comprehensive view of molecular changes related to NUE. These molecules could be used as candidates for improving NUE in barley. Quan et al. [38] conducted a comparative transcriptome analysis between a low-N tolerant barley genotype and a low-N sensitive genotype in the roots under low-N treatment and found that 533 genes were up-regulated and 446 genes were down-regulated in the low-N tolerant genotype ‘XZ149’. Chen et al. [58] conducted whole transcriptome analysis on the shoots of a barley landrace with low-N tolerance and identified 498 lncRNAs (487 of them were new) at the whole genome level; of these, 31 were up-regulated and 25 were down-regulated in response to low-N stress. Subsequently, Quan et al. [66,67] further analyzed the molecular mechanisms in adaptation to low-N stress by comparing the differences in both shoots and roots of two different barley genotypes using metabolomics and ionomics.
As a kind of gene editing technology, CRISPR technology shows advantages compared to the other two gene editing technologies based on zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). It can be used for the quick, precise, and efficient editing of targeted genes [68]. Thus, it will be of great value in the functional verification of N-efficient related barley genes and barley breeding for improving NUE. The gene editing technology based on CRISPR/Cas9 was first applied in barley by Lawrenson et al. [69], and it was subsequently tested by other researchers [70,71,72]. Although it could be used for gene function validation, the donor materials used in these technology systems are mainly based on immature embryos of the barley genotype ‘Golden Promise’. This is because genome editing currently requires an initial genetic transformation to incorporate Cas9 and gRNA genes into the genome (although alternative methods are being developed) and genetic modification of barley is highly genotype-dependent, greatly restricting the utilization of this kind of technology in barley breeding. Recently, however, Han et al. [73] developed a highly efficient gene editing technology using callus from another culture, and it succeeded in several commercial barley varieties. This provides further opportunities for the use of this technology to improve NUE in barley breeding.
7. Challenges and Perspectives
NUE is controlled by multiple genes, and the evaluation of barley NUE is very complex. Phenotypic, physiological, and agronomic traits are typically used for the evaluation of NUE in barley varieties. However, there is still no uniform standard system to determine barley NUE, except for crop yield. Although RSDW and SDW are considered to be determination indices for evaluating NUE in barley at an early stage by several researchers, this has not been demonstrated in the final grain yield [15,28,31,32]. Moreover, field experiments are also affected by many factors, such as ecological habits, extreme climates, and disease resistance. Thus, grain yield cannot always accurately reflect differences in NUE. Hu et al. [20] used a chlorate-sensitivity test to evaluate nitrate absorption and used this trait for NUE evaluation; based on this process, the NUE gene NRT1.1B in rice was obtained through map-based cloning. The map-based cloning of the OsNR2 gene, which increased the effective tiller number, grain yield, and NUE, was also based on the chlorate-sensitivity test [74]. Tang et al. [21] used the effective panicle number ratio (EPNR) for association analysis and obtained the OsNPF6.1 gene for improving NUE in rice by GWAS analysis. Therefore, traits based on the response to chlorate and EPNR could also be good choices for NUE evaluation in barley. In addition, high-throughput phenomics is being developed in the estimation of responses to nitrogen treatment in crops by using non-invasive imaging systems, spectroscopy, image analysis, robotics, enzyme-based sensors, etc. [75,76,77,78]. These methods can provide a large amount of real-time data, which will have a revolutionary impact on the estimation of phenotypes in the study of NUE in barley and will also be an important research direction.
Although many studies on NUE have been carried out in plants thus far, its molecular mechanisms are still not clear [1]. Furthermore, many NUE genes are only effective in the laboratory. Research on the molecular mechanisms of NUE in plants is still not enough; however, elucidation of these mechanisms might require many genes together to improve NUE in the field. These problems are even more serious in barley, as only a few barley genes have been demonstrated to improve NUE [39,55,56,62]. Therefore, it is important to continue to research the molecular mechanisms of barley NUE to identify more NUE genes.
Recently, the NRT1.1B gene of rice was shown to recruit special bacteria in the rhizosphere, and these bacteria were related to N metabolism functions; one of these bacterial strains even improved rice growth in an inoculation experiment. Improving N efficiency or low-N tolerance in barley through the interaction of plant and rhizosphere microorganisms is also a direction worthy of further research [22,79]. In addition, significant progress has been made in the study of mycorrhizal symbiosis in rice, providing possibilities for biological N fixation in cereal crops [80,81]. Furthermore, a homozygous rhizopine producing (RhiP) barley line and a hybrid rhizopine uptake system were recently developed for a synthetic plant-controlled symbiosis and N fixation in barley [82]. Further research in these areas will also be beneficial for improving the nitrogen use efficiency of barley.
Author Contributions
Z.C. conceived and rewrote the manuscript. L.L. performed the literature search and wrote the draft. N.G.H. revised the manuscript. H.X., L.H., R.G. and R.L. gave some useful information about nitrogen efficiency research in barley. C.L. provided some guidance on preparing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by China Agriculture Research System of MOF and MARA (Grant No. CARS-05-01A-02) and the Climbing Plan of Shanghai Academy of Agricultural Sciences (Grant No. PG22211). N.G.H. is supported at Rothamsted Research by the Biotechnology and Biological Sciences Research Council (BBSRC) via the Designing Future Wheat Programme (BB/P016855/1).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank LetPub (accessed on 28 September 2021) for its linguistic assistance during the primary preparation of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunarathne, S.D.; Han, Y.; Zhang, X.Q.; Li, C. Advances in understanding the molecular mechanisms and potential genetic improvement for nitrogen use efficiency in barley. Agronomy 2020, 10, 662. [Google Scholar] [CrossRef]
- Han, M.; Okamoto, M.; Beatty, P.H.; Rothstein, S.J.; Good, A.G. The genetics of nitrogen use efficiency in crop plants. Annu. Rev. Genet. 2015, 49, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.K.; Bali, S.K. A review of methods to improve nitrogen use efficiency in agriculture. Sustainability 2018, 10, 51. [Google Scholar] [CrossRef] [Green Version]
- Beman, J.M.; Arrigo, K.R.; Matson, P.A. Agricultural runoff fuels large phytoplankton blooms in vulnerable areas of the ocean. Nature 2005, 434, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Giles, J. Nitrogen study fertilizes fears of pollution. Nature 2005, 433, 791. [Google Scholar] [CrossRef] [Green Version]
- Camargo, J.A.; Alonso, A. Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: A global assessment. Environ. Int. 2006, 32, 831–849. [Google Scholar] [CrossRef]
- Hirel, B.; Tétu, T.; Lea, P.J.; Dubois, F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 2011, 3, 1452–1485. [Google Scholar] [CrossRef]
- UNEP. Part 1: The fertilizer industry’s manufacturing processes and environmental issues. In Mineral Fertilizer Production and the Environment, 2nd ed.; United Nations: Paris, France, 1998; p. 39. ISBN 92-807-1640-9. [Google Scholar]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron. J. 1982, 74, 562–564. [Google Scholar] [CrossRef]
- Good, A.G.; Shrawat, A.K.; Muench, D.G. Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 2004, 9, 597–605. [Google Scholar] [CrossRef]
- Lam, H.M.; Coschigano, K.T.; Oliveira, I.C.; Melo-Oliveira, R.; Coruzzi, G.M. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 569–593. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Fan, X.R.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.N.; Hu, H.L.; Zhu, B.; Jin, X.L.; Wu, F.B.; Zhang, G.P. Genotypic variations of N use efficiency in Tibetan wild and cultivated barleys. J. Zhejiang Univ. 2014, 2, 155–164, (In English with Chinese Abstract). [Google Scholar]
- Raun, W.R.; Johnson, G.V. Improving nitrogen use efficiency for cereal production. Agron. J. 1999, 91, 357–363. [Google Scholar] [CrossRef] [Green Version]
- Anbessa, Y.; Juskiw, P. Review: Strategies to increase nitrogen use efficiency of spring barley. Can. J. Plant Sci. 2012, 92, 617–625. [Google Scholar] [CrossRef]
- Teng, W.; He, X.; Tong, Y.P. Transgenic approaches for improving use efficiency of nitrogen, phosphorus and potassium in crops. J. Integr. Agric. 2017, 16, 2657–2673. [Google Scholar] [CrossRef]
- Sun, H.; Qian, Q.; Wu, K.; Luo, J.; Wang, S.; Zhang, C.; Ma, Y.; Liu, Q.; Huang, X.; Yuan, Q.; et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 2014, 46, 652–656. [Google Scholar] [CrossRef]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef]
- Tang, W.; Ye, J.; Yao, X.; Zhao, P.; Xuan, W.; Tian, Y.; Zhang, Y.; Xu, S.; An, H.; Chen, G.; et al. Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 2019, 10, 5279. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liu, Y.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Wang, S.; Song, W.; Zhang, J.; Wang, Y.; Liu, Q.; Yu, J.; Ye, Y.; Li, S.; Chen, J.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, eaaz2046. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef]
- McKenzie, R.; Middleton, A.B.; Hall, L.; DeMulder, J.; Bremer, E. Fertilizer response of barley grain in south and central Alberta. Can. J. Soil Sci. 2004, 84, 513–523. [Google Scholar] [CrossRef] [Green Version]
- De, M.; Liu, Z.; Wang, L.; Wang, J.; Qi, H.; Guo, C.; Lu, E.; Ba, T.; Xu, S. Effects of different nitrogen levels on photosynthetic performance, nitrogen accumulation and translocation of barley. Acta Agric. Boreali-Sin. 2020, 3, 126–135, (In Chinese with English Abstract). [Google Scholar]
- Chen, Z.; Liu, C.; Wang, Y.; He, T.; Gao, R.; Xu, H.; Guo, G.; Li, Y.; Zhou, L.; Lu, R.; et al. Expression analysis of nitrogen metabolism-related genes reveals differences in adaptation to low-nitrogen stress between two different barley cultivars at seedling stage. Int. J. Genom. 2018, 2018, 8152860. [Google Scholar] [CrossRef]
- Jiang, Q.; Chen, Z.; Liu, C.; He, T.; Guo, G.; Gao, R.; Xu, H.; Li, Y.; Lu, R.; Huang, J. Screening and identification indices of low-nitrogen tolerance for barley landraces at seedling stage. Acta Agric. Boreali-Sin. 2019, 1, 148–155, (In Chinese with English Abstract). [Google Scholar]
- Beatty, P.H.; Anbessa, Y.; Juskiw, P.; Carroll, R.T.; Wang, J.; Good, A.G. Nitrogen use efficiencies of spring barley grown under varying nitrogen conditions in the field and growth chamber. Ann. Bot. 2010, 105, 1171–1182. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; He, T.; Lu, R.; Wang, Y.; Zou, L.; Du, Z.; Shan, L.; He, Y.; Huang, J. Biological responses of different-genotype barleys to low nitrogen stress at seedling stage. Acta Agric. Shanghai 2010, 26, 28–32, (In Chinese with English Abstract). [Google Scholar]
- Chen, Z.; Zou, L.; Lu, R.; Wang, Y.; He, T.; Du, Z.; Zhang, Y.; Huang, J. Study on the relationship between the traits for low-nitrogen tolerance of different barley genotypes at seedling stage and grain yield. J. Triticeae Crops 2010, 1, 158–162, (In Chinese with English Abstract). [Google Scholar]
- Karunarathne, S.D.; Han, Y.; Zhang, X.Q.; Zhou, G.; Hill, C.B.; Chen, K.; Angessa, T.; Li, C. Genome-wide association study and identification of candidate genes for nitrogen use efficiency in barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 571912. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Jiang, Q.; Xu, J.; Zhang, W.; He, T.; Guo, G.; Wang, Y.; Ma, Y.; Huang, J.; Liu, C.; et al. Effects of different low nitrogen stresses on low nitrogen tolerance of barley landraces at seedling stage. Plant Physiol. J. 2019, 5, 642–648, (In Chinese with English Abstract). [Google Scholar]
- Karunarathne, S.D.; Han, Y.; Zhang, X.Q.; Dang, V.H.; Angessa, T.T.; Li, C. Using chlorate as an analogue to nitrate to identify candidate genes for nitrogen use efficiency in barley. Mol. Breed. 2021, 41, 47. [Google Scholar] [CrossRef]
- Teng, S.; Tian, C.; Chen, M.; Zeng, D.; Guo, L.; Zhu, L.; Han, B.; Qian, Q. QTLs and candidate genes for chlorate resistance in rice (Oryzasativa L.). Euphytica 2006, 152, 141–148. [Google Scholar] [CrossRef]
- Li, L.; Zong, Y.; Yang, H.; Xu, H.; Gao, R.; Lu, R.; Liu, C.; Chen, Z. Effects of chlorate treatment on traits related to different barley landraces at the seedling stage. Acta Agric. Boreali-Sin. 2022; accepted. (In Chinese with English Abstract). [Google Scholar]
- Decouard, B.; Bailly, M.; Rigault, M.; Marmagne, A.; Arkoun, M.; Soulay, F.; Caïus, J.; Paysant-Le Roux, C.; Louahlia, S.; Jacquard, C.; et al. Genotypic variation of nitrogen use efficiency and amino acid metabolism in barley. Front. Plant Sci. 2022, 12, 807798. [Google Scholar] [CrossRef]
- Quan, X.; Zeng, J.; Ye, L.; Chen, G.; Han, Z.; Shah, J.M.; Zhang, G. Transcriptome profiling analysis for two Tibetan wild barley genotypes in responses to low nitrogen. BMC Plant Biol. 2016, 16, 30. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Li, Y.; Wang, S.; Li, D.; Lv, C.; Xu, R. Characterization of the nitrate transporter gene family and functional identification of HvNRT2.1 in barley (Hordeum vulgare L.). PLoS ONE 2020, 15, e0232056. [Google Scholar] [CrossRef]
- Anbessa, Y.; Juskiw, P.; Good, A.; Nyachiro, J.; Helm, J. Genetic variability in nitrogen use efficiency of spring barley. Crop Sci. 2009, 49, 1259–1269. [Google Scholar] [CrossRef]
- Han, M.; Wong, J.; Su, T.; Beatty, P.H.; Good, A.G. Identification of nitrogen use efficiency genes in barley: Searching for QTLs controlling complex physiological traits. Front. Plant Sci. 2016, 7, 1587. [Google Scholar] [CrossRef] [Green Version]
- Saal, B.; von Korff, M.; Léon, J.; Pillen, K. Advanced-backcross QTL analysis in spring barley: IV. Localization of QTL × nitrogen interaction effects for yield-related traits. Euphytica 2011, 177, 223–239. [Google Scholar] [CrossRef]
- Schnaithmann, F.; Pillen, K. Detection of exotic QTLs controlling nitrogen stress tolerance among wild barley introgression lines. Euphytica 2013, 189, 67–88. [Google Scholar] [CrossRef]
- Hoffmann, A.; Maurer, A.; Pillen, K. Detection of nitrogen deficiency QTL in juvenile wild barley introgression lines growing in a hydroponic system. BMC Genet. 2012, 13, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindu, G.A.; Tang, J.; Yin, X.; Struik, P.C. Quantitative trait locus analysis of nitrogen use efficiency in barley (Hordeum vulgare L.). Euphytica 2014, 199, 207–221. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Tan, C.; Wang, J.; Zhang, X.; Zhu, J.; Luo, H.; Yang, J.; Westcott, S.; Broughton, S.; Moody, D.; et al. Marker development using SLAF-seq and whole-genome shotgun strategy to fine-map the semi-dwarf gene ari-e in barley. BMC Genom. 2016, 17, 911. [Google Scholar] [CrossRef] [Green Version]
- Hisano, H.; Sakamoto, K.; Takagi, H.; Terauchi, R.; Sato, K. Exome QTL-seq maps monogenic locus and QTLs in barley. BMC Genom. 2017, 18, 125. [Google Scholar] [CrossRef] [Green Version]
- Obsa, B.T.; Eglinton, J.; Coventry, S.; March, T.; Guillaume, M.; Le, T.P.; Hayden, M.; Langridge, P.; Fleury, D. Quantitative trait loci for yield and grain plumpness relative to maturity in three populations of barley (Hordeum vulgare L.) grown in a low rain-fall environment. PLoS ONE 2017, 12, e0178111. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Zhu, J.; Dong, W.; Sun, Y.; Lv, C.; Guo, B.; Xu, R. Comparative mapping and candidate gene analysis of SSIIa associated with grain amylopectin content in barley (Hordeum vulgare L.). Front. Plant Sci. 2017, 8, 1531. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Zhang, X.; Zhang, X.; Tong, T.; Zhang, Z.; Wu, G.; Hou, L.; Zheng, J.; Niu, C.; Li, J.; et al. A high-density genetic linkage map of SLAFs and QTL analysis of grain size and weight in barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 620922. [Google Scholar] [CrossRef]
- Pauli, D.; Muehlbauer, G.J.; Smith, K.P.; Cooper, B.; Hole, D.; Obert, D.E.; Ullrich, S.E.; Blake, T.K. Association mapping of agronomic QTLs in U.S. spring barley breeding germplasm. Plant Genome 2014, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Falcon, C.M.; Horsley, R.; Hu, G.; Blake, T.; Smith, K.P. Mapping QTLs for grain protein concentration and agronomic traits under different nitrogen levels in barley. Crop Sci. 2019, 59, 68–83. [Google Scholar] [CrossRef]
- Li, H.; Hu, B.; Chu, C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Shrawat, A.K.; Carroll, R.T.; DePauw, M.; Taylor, G.J.; Good, A.G. Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 2008, 6, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, M.G.; Valencia, M.O.; Ogawa, S.; Lu, Y.; Wu, L.; Downs, C.; Skinner, W.; Lu, Z.; Kridl, J.C.; Ishitani, M.; et al. Development and field performance of nitrogen use efficient rice lines for Africa. Plant Biotechnol. J. 2017, 15, 775–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Quan, S.; Lyu, B.; Tian, T.; Liu, Z.; Nie, Z.; Qi, S.; Jia, J.; Shu, J.; Groot, E.; et al. Barley transcription factor HvNLP2 mediates nitrate signaling and affects nitrogen use efficiency. J. Exp. Bot. 2022, 73, 770–783. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Jiang, Q.; Jiang, P.; Zhang, W.; Huang, J.; Liu, C.; Halford, N.G.; Lu, R. Novel low-nitrogen stress-responsive long non-coding RNAs (lncRNA) in barley landrace B968 (Liuzhutouzidamai) at seedling stage. BMC Plant Biol. 2020, 20, 142. [Google Scholar] [CrossRef]
- Tadesse, D.; Assefa, A. Genetic variation in N-use efficiency of malting barley genotypes. Intern. J. Agric. Forest. 2013, 7, 303–308. [Google Scholar]
- Gao, R.; Guo, G.; Fang, C.; Huang, S.; Chen, J.; Lu, R.; Huang, J.; Liu, C. Rapid generation of barley mutant lines with high nitrogen uptake efficiency by microspore mutagenesis and field screening. Front. Plant Sci. 2018, 9, 450. [Google Scholar] [CrossRef]
- Xu, H.; Li, Y.; Gao, R.; Xu, R.; Guo, G.; Lu, R.; Halford, N.G.; Chen, Z.; Liu, C. Rapid generation and analysis of a barley doubled haploid line with higher nitrogen use efficiency than parental lines by F1 microspore embryogenesis. Plants 2021, 10, 1588. [Google Scholar] [CrossRef]
- Tiong, J.; Sharma, N.; Sampath, R.; MacKenzie, N.; Watanabe, S.; Metot, C.; Lu, Z.; Skinner, W.; Lu, Y.; Kridl, J.; et al. Improving nitrogen use efficiency through overexpression of alanine aminotransferase in rice, wheat, and barley. Front. Plant Sci. 2021, 12, 628521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gao, S.; Chu, C. Improvement of nutrient use efficiency in rice: Current toolbox and future perspectives. Theor. Appl. Genet. 2020, 133, 1365–1384. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Jayakodi, M.; Padmarasu, S.; Haberer, G.; Bonthala, V.S.; Gundlach, H.; Monat, C.; Lux, T.; Kamal, N.; Lang, D.; Himmelbach, A.; et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 2020, 588, 284–289. [Google Scholar] [CrossRef]
- Quan, X.; Qian, Q.; Ye, Z.; Zeng, J.; Han, Z.; Zhang, G. Metabolic analysis of two contrasting wild barley genotypes grown hydroponically reveals adaptive strategies in response to low nitrogen stress. J. Plant Physiol. 2016, 206, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Zeng, J.; Han, Z.; Zhang, G. Ionomic and physiological responses to low nitrogen stress in Tibetan wild and cultivated barley. Plant Physiol. Biochem. 2017, 111, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Ostergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [Green Version]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [Green Version]
- Holme, I.B.; Wendt, T.; Gil-Humanes, J.; Deleuran, L.C.; Starker, C.G.; Voytas, D.F.; Brinch-Pedersen, H. Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol. Biol. 2017, 95, 111–121. [Google Scholar] [CrossRef]
- Kapusi, E.; Corcuera-Gómez, M.; Melnik, S.; Stoger, E. Heritable genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Front. Plant Sci. 2017, 8, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Broughton, S.; Liu, L.; Zhang, X.Q.; Zeng, J.; He, X.; Li, C. Highly efficient and genotype-independent barley gene editing based on anther culture. Plant Commun. 2020, 2, 100082. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Wang, Y.; Chen, G.; Zhang, A.; Yang, S.; Shang, L.; Wang, D.; Ruan, B.; Liu, C.; Jiang, H.; et al. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 2019, 10, 5207. [Google Scholar] [CrossRef] [PubMed]
- Kefauver, S.C.; Vicente, R.; Vergara-Díaz, O.; Fernandez-Gallego, J.A.; Kerfal, S.; Lopez, A.; Melichar, J.; Serret Molins, M.D.; Araus, J.L. Comparative UAV and Field Phenotyping to Assess Yield and Nitrogen Use Efficiency in Hybrid and Conventional Barley. Front. Plant Sci. 2017, 8, 1733. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.N.; Maharjan, P.; Maphosa, L.; Vakani, J.; Thoday-Kennedy, E.; Kant, S. A Robust Automated Image-Based Phenotyping Method for Rapid Vegetative Screening of Wheat Germplasm for Nitrogen Use Efficiency. Front. Plant Sci. 2019, 10, 1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prey, L.; Hu, Y.; Schmidhalter, U. High-Throughput Field Phenotyping Traits of Grain Yield Formation and Nitrogen Use Efficiency: Optimizing the Selection of Vegetation Indices and Growth Stages. Front. Plant Sci. 2020, 10, 1672. [Google Scholar] [CrossRef] [Green Version]
- Liang, T.; Duan, B.; Luo, X.; Ma, Y.; Yuan, Z.; Zhu, R.; Peng, Y.; Gong, Y.; Fang, S.; Wu, X. Identification of High Nitrogen Use Efficiency Phenotype in Rice (Oryza sativa L.) Through Entire Growth Duration by Unmanned Aerial Vehicle Multispectral Imagery. Front. Plant Sci. 2021, 12, 740414. [Google Scholar] [CrossRef]
- Zhang, Z.; Chu, C. Nitrogen-Use divergence between Indica and Japonica rice: Variation at nitrate assimilation. Mol. Plant 2020, 13, 6–7. [Google Scholar] [CrossRef]
- He, J.; Zhang, C.; Dai, H.; Liu, H.; Zhang, X.; Yang, J.; Chen, X.; Zhu, Y.; Wang, D.; Qi, X.; et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 2019, 12, 1561–1576. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; He, J.; Dai, H.; Wang, G.; Zhang, X.; Wang, C.; Shi, J.; Chen, X.; Wang, D.; Wang, E. Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2023738118. [Google Scholar] [CrossRef]
- Haskett, T.L.; Paramasivan, P.; Mendes, M.D.; Green, P.; Geddes, B.A.; Knights, H.E.; Jorrin, B.; Ryu, M.; Brett, P.; Voigt, C.; et al. Engineered plant control of associative nitrogen fixation. Proc. Natl. Acad. Sci. USA 2022, 119, e2117465119. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).