Methylation Status of the Telomerase Reverse Transcriptase Promoter in Parotid Tumours and Adjacent Parotid Gland Tissue: A Pilot Study on the Implications for Recurrence and Development of Malignancy
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Tissue Extraction
2.2.2. DNA Isolation
2.2.3. THOR Methylation Analysis
2.2.4. TERT Promoter Mutations (TPMs) Analysis
2.2.5. Statistical Approach, Programme, and Sample Size Calculations
3. Results
3.1. Characteristics of the Population
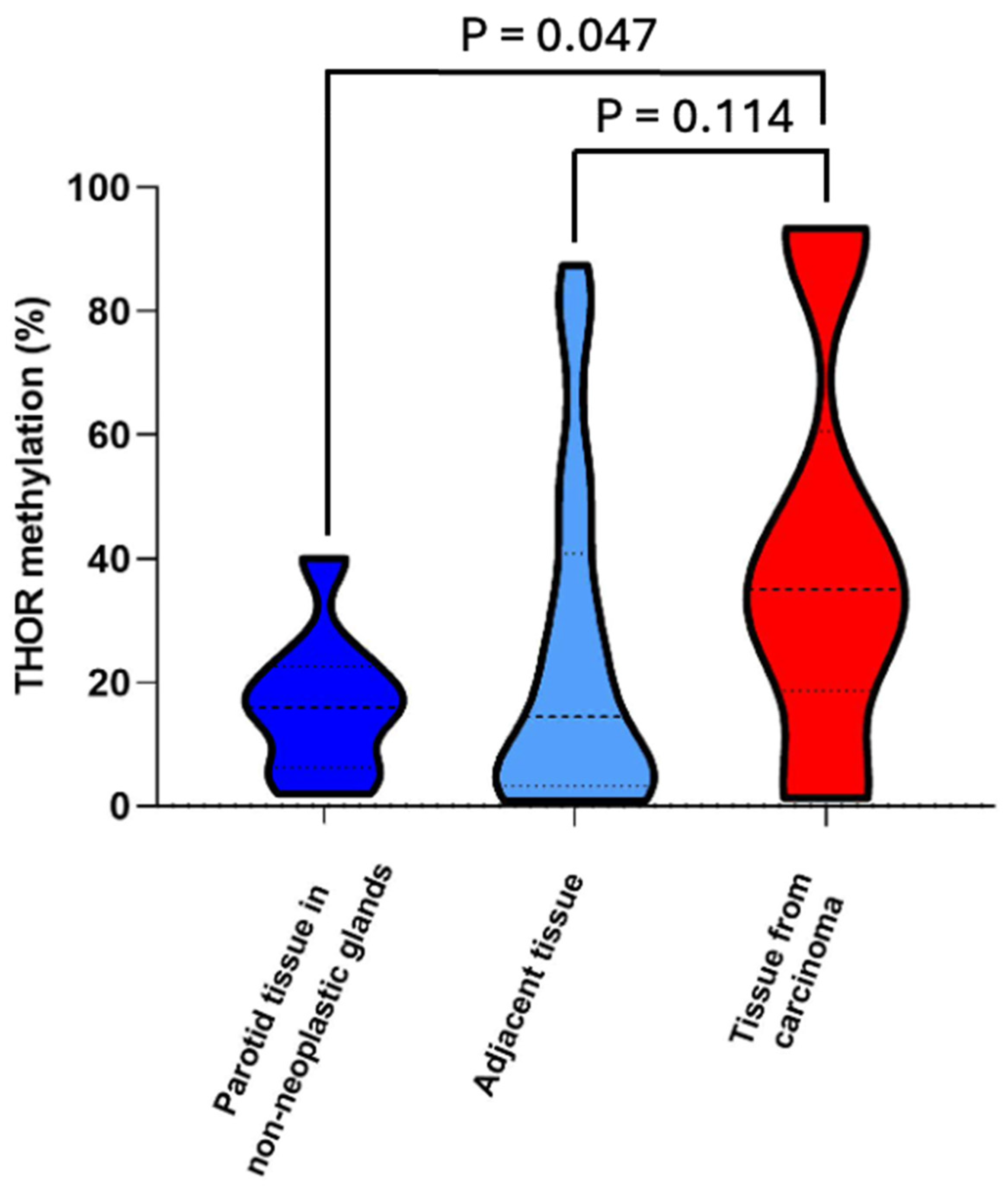
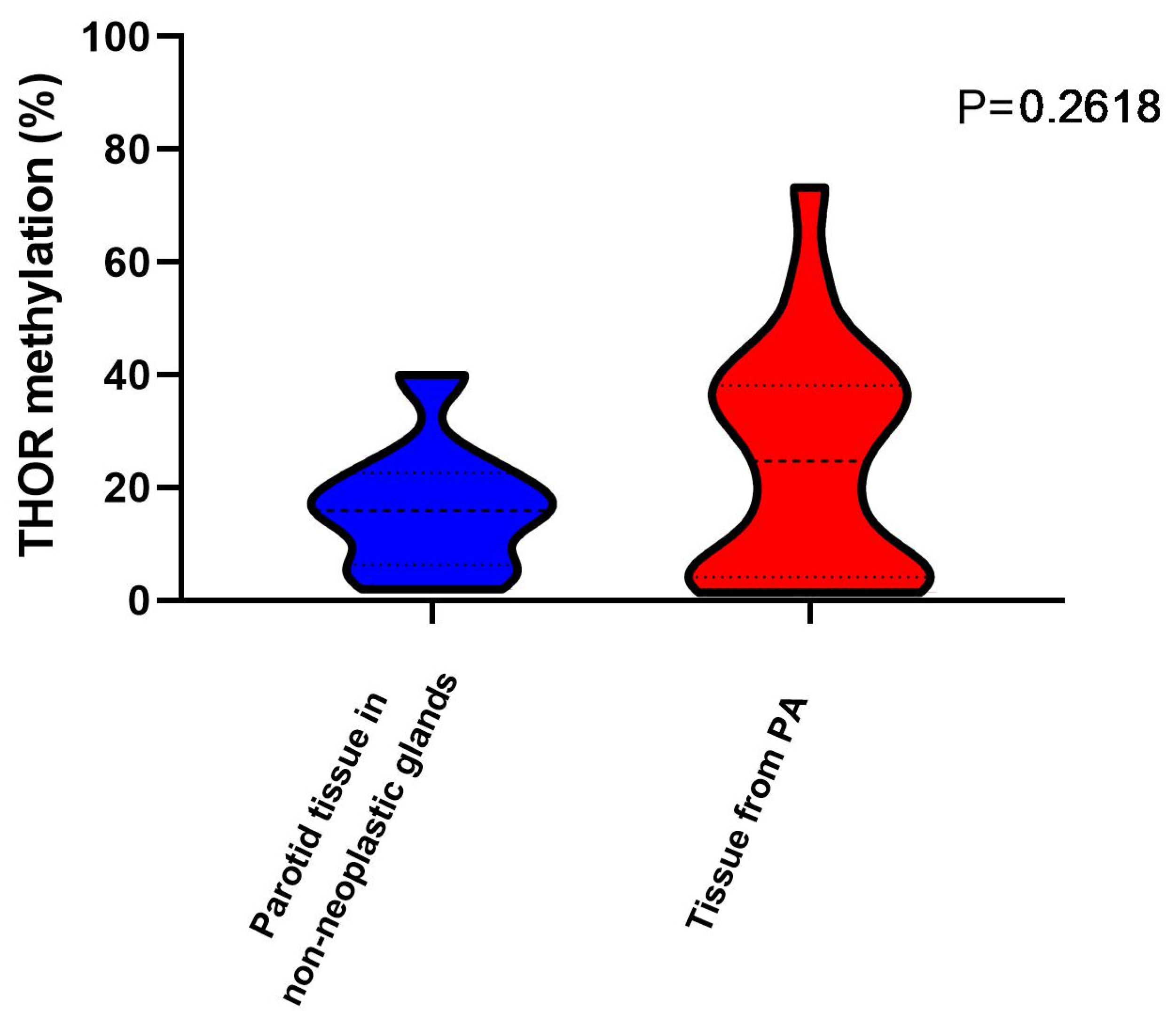
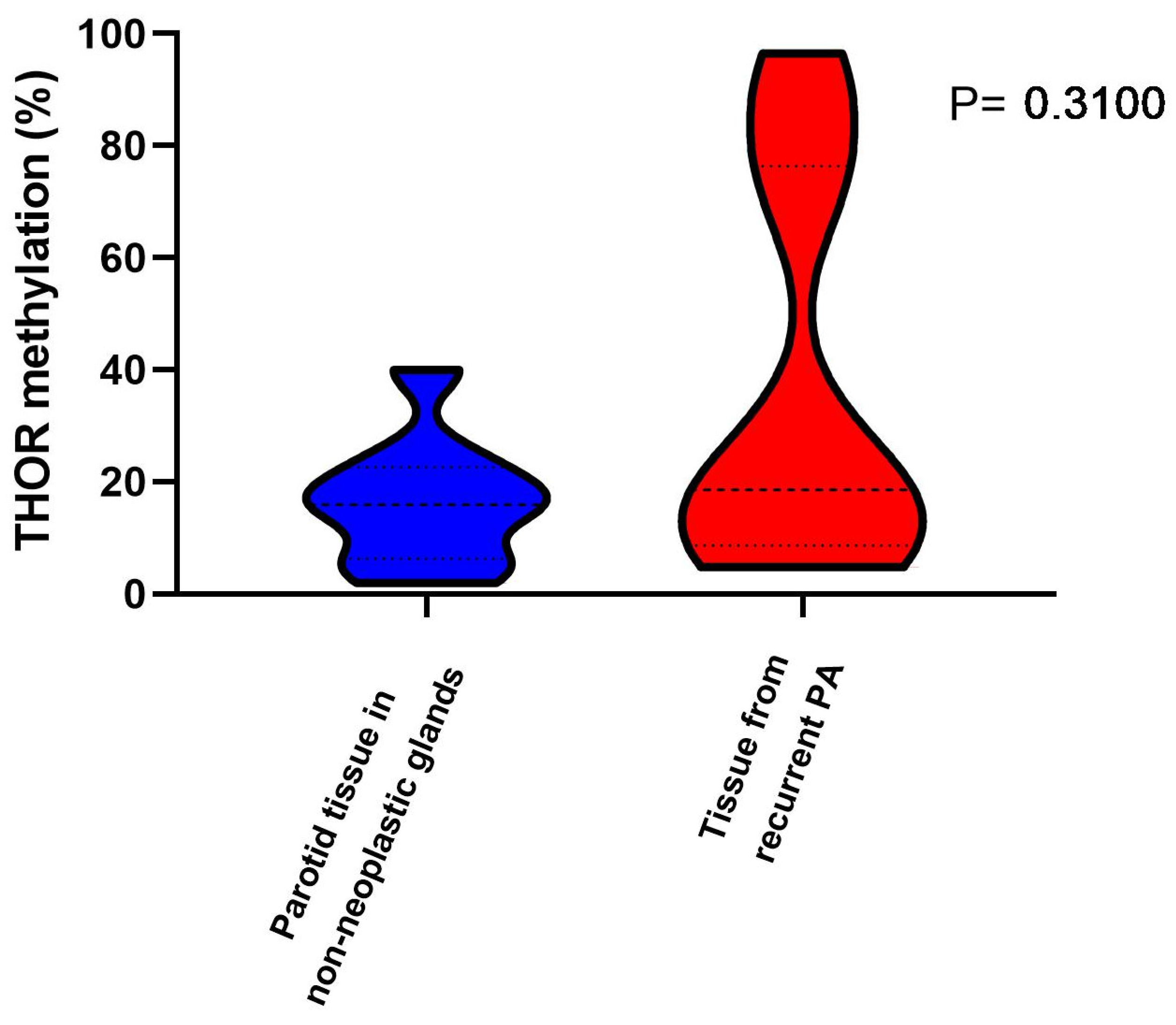
3.2. THOR Methylation
3.3. TERT Promoter Mutations (TPMs) Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| CxPA | carcinoma ex-pleomorphic adenoma |
| hTERT | human telomerase reverse transcriptase |
| PA | pleomorphic adenoma |
| rPA | recurrent pleomorphic adenoma |
| SGT | salivary gland tumour |
| THOR | TERT hypermethylated oncological region |
| TPM | TERT promoter mutations |
References
- Blackburn, E.H.; Gall, J.G. A tandemly repeated sequence at the termini of the extrachromossomal ribosomal RNA in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T.; Serrano, M.; Blasco, M.A. The common biology of cancer and ageing. Nature 2007, 448, 767–774. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Choufani, S.; Mack, S.; Gallagher, D.; Zhang, C.; Lipman, T.; Lipman, T.; Zhukova, N.; Walker, E.J.; Martin, D.; et al. Differentially methylated region of the TERT promoter and risk for stratification of childhood brain tumours: An integrative genomic and molecular study. Lancet Oncol. 2013, 14, 534–542. [Google Scholar] [CrossRef]
- Faleiro, I.; Apolonio, J.D.; Price, A.J.; De Mello, R.A.; Roberto, V.P.; Tabori, U.; Castelo-Branco, P. The TERT hypermethylated oncologic region predicts recurrence and survival in pancreatic cancer. Future Oncol. 2017, 13, 2045–2051. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Leão, R.; Lipman, T.; Campbell, B.; Lee, D.; Price, A.; Zhang, C.; Heidari, A.; Stephens, D.; Boerno, S.; et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: A retrospective cohort study. Oncotarget 2016, 7, 57726–57736. [Google Scholar] [CrossRef]
- Apolónio, J.D.; Dias, J.S.; Fernandes, M.; Komosa, M.; Lipman, T.; Zhang, C.; Leão, R.; Lee, D.; Nunes, N.M.; Maia, A.-T.; et al. THOR is a targetable epigenetic biomarker with clinical implications in breast cancer. Clin. Epigenetics 2022, 14, 178. [Google Scholar] [CrossRef]
- Lee, D.D.; Leão, R.; Komosa, M.; Gallo, M.; Zhang, C.H.; Lipman, T.; Remke, M.; Heidari, A.; Nunes, N.M.; Apolónio, J.D.; et al. DNA hypermethylation with TERT promoter upregulates TERT expression in cancer. J. Clin. Investig. 2019, 129, 223–229. [Google Scholar] [CrossRef]
- Dulguerov, P.; Todic, J.; Pusztaszeri, M.; Alotaibi, N.H. Why Do Parotid Pleomorphic Adenomas Recur? A Systematic Review of Pathological and Surgical Variables. Front. Surg. 2017, 4, 26. [Google Scholar] [CrossRef]
- Mantsopoulos, K.; Iro, H. Tumour spillage of the pleomorphic adenoma of the parotid gland: A proposal for intraoperative measures. Oral Oncol. 2021, 112, 104986. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218. [Google Scholar] [CrossRef] [PubMed]
- Vasuri, F.; Chillotti, S.; Maloberti, T.; Albertini, E.; Germinario, G.; Cescon, M.; Ravaioli, M.; de Biase, D.; D’Errico, A. Beyond histology: A tissue algorithm predictive of post-surgical recurrence in hepatocellular carcinomas, including TERT promoter mutation. Virchows Arch. 2025, 486, 365–372. [Google Scholar] [CrossRef]
- Hellquist, H.; Paiva-Correia, A.; Vander Poorten, V.; Quer, M.; Hernandez-Prera, J.C.; Andreasen, S.; Zbären, P.; Skalova, A.; Rinaldo, A.; Ferlito, A. Analysis of the Clinical Relevance of Histological Classification of Benign Epithelial Salivary Gland Tumours. Adv. Ther. 2019, 36, 1950–1974. [Google Scholar] [CrossRef]
- Johnson, A.A.; Akman, K.; Calimport, S.R.G.; Wuttke, D.; Stolzing, A.; Magalhães, J.P. The Role of DNA Methylation in Aging, Rejuvenation, and Age-Related Disease. Rejuvenation Res. 2012, 15, 483–494. [Google Scholar] [CrossRef]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Wang, Z.; Ling, S.; Rettig, E.; Sobel, R.; Tan, M.; Fertig, E.J.; Considine, M.; El-Naggar, A.K.; Brait, M.; Fakhry, C.; et al. Epigenetic screening of salivary gland mucoepidermoid carcinoma identifies hypomethylation of CLIC3 as a common alteration. Oral Oncol. 2015, 51, 1120–1125. [Google Scholar] [CrossRef]
- Nikolic, N.; Carkic, J.; Dimitrijevic, I.I.; Eljabo, N.; Radunovic, M.; Anicic, B.; Tanic, N.; Falk, M.; Milasin, J. P14 methylation: An epigenetic signature of salivary gland mucoepidermoid carcinoma in the Serbian population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 52–58. [Google Scholar] [CrossRef]
- Mitani, Y.; Roberts, D.B.; Fatani, H.; Weber, R.S.; Kies, M.S.; Lippman, S.M.; El-Naggar, A.K. MicroRNA profiling of salivary adenoid cystic carcinoma: Association of miR-17-92 upregulation with poor outcome. PLoS ONE 2013, 8, e66778. [Google Scholar] [CrossRef]
- Jurmeister, P.; Leitheiser, M.; Arnold, A.; Capilla, E.P.; Mochmann, L.H.; Zhdanovic, Y.; Schleich, K.; Jung, N.; Chimal, E.C.; Jung, A.; et al. DNA Methylation Profiling of Salivary Gland Tumors Supports and Expands Conventional Classification. Mod. Pathol. 2024, 37, 100625. [Google Scholar] [CrossRef]
- Jerónimo, C.; Costa, I.; Martins, M.C.; Monteiro, P.; Lisboa, S.; Palmeira, C.; Henrique, R.; Teixeira, M.R.; Lopes, C. Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clin. Cancer Res. 2003, 9, 3413. [Google Scholar] [PubMed]
- Zare-Mirzaie, A.; Mollazadehghomi, S.; Heshmati, S.; Mehrtash, A.; Mollazadehghomi, S. TERT Promoter Mutation in Benign and Malignant Salivary Gland Tumors; A Cross-Sectional Study. Iran. J. Patho 2023, 18, 64–74. [Google Scholar] [CrossRef]
- Kim, H.; Ahn, D.; Sohn, J.; Kim, Y.H.; Lee, J.H.; Lee, H. TERT Promoter Mutation and Telomere Length in Salivary Gland Tumors. Pathol. Oncol. Res. 2017, 24, 697–698. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F. Histone Deacetylases and Histone Deacetylase Inhibitors: Molecular Mechanisms of Action in Various Cancers. Adv. Biomed. Res. 2019, 8, 63–74. [Google Scholar]
| Characteristics | Study Population (n = 57) |
|---|---|
| Gender | |
| Female | 35 (61%) |
| Male | 22 (39%) |
| Group | |
| 9 (9%) |
| 22 (23%) |
| 17 (18%) |
| 12 (13%) |
| 9 (9%) |
| 14 (15%) |
| 13 (14%) |
| Age, years | 47.0 (35.5, 61.5) |
| Follow-up, months | 131 (51.2, 159) |
| Treatment | |
| SPE | 37 (65%) |
| PR | 12 (21%) |
| Parotidectomy | 7 (12%) |
| RT | 10 (18%) |
| ENU | 1 (2%) |
| Sample | Diagnosis | Age * (Years) | Sex * (F/M) | Treatment * | Follow-Up (Months) * | THOR Methylation (%) |
|---|---|---|---|---|---|---|
| 49 | Cyst | 51 | F | SPE | 60, NED | 20.66 |
| 80 | Infl | 44 | F | SPE | 60, NED | 1.91 |
| 83 | Cyst | 71 | M | SPE | 36, NED | 7.96 |
| 84 | Cyst | 65 | M | SPE | 84, NED | 4.58 |
| 85 | Cyst | 16 | F | SPE | 82, NED | 1.94 |
| 86 | Cyst | 64 | F | SPE | 36, NED | 24.53 |
| 88 | Cyst | 47 | M | SPE | 60, NED | 40.04 |
| 110 | Cyst | 64 | M | SPE | 66, NED | 17.44 |
| 111 | Cyst | 53 | M | PR | 96, NED | 14.44 |
| Adjacent Tissue Sample Code | THOR Methylation (%) | PA Tissue Sample Code | THOR Methylation (%) | Age * (Years) | Sex * (F/M) | Treatment * | Follow-Up (Months) * |
|---|---|---|---|---|---|---|---|
| No AT | - | 21 | 43.56 | 15 | F | SPE | 240, NED |
| 22 | 51.45 | 23 | 33.10 | 61 | F | SPE | 240, NED |
| 24 | 83.73 | 25 | 36.32 | 33 | F | SPE, RT | 96, NED |
| 26 | 24.60 | 27 | 56.98 | 17 | M | SPE | 250, NED |
| 28 | 8.04 | 29 | 45.86 | 46 | F | SPE | 200, NED |
| 30 | 87.43 | 31 | 30.22 | 34 | F | SPE | 240, NED |
| 32 | 3.77 | 33 | 2.95 | 58 | M | SPE | 70, DOC |
| 34 | 0.83 | 35 | 1.34 | 58 | M | SPE | 90, NED |
| 36 | 10.67 | 37 | 18.41 | 56 | F | SPE | 220, NED |
| No AT | - | 38 | 73.27 | 20 | M | SPE | 250, NED |
| No AT | - | 39 | 3.65 | 25 | F | SPE | 250, NED |
| 40 | 1.54 | 41 | 4.66 | 28 | M | SPE | L |
| 44 | 2.89 | 45 | 8.04 | 38 | M | SPE | 250, NED |
| No AT | - | 46 | 39.93 | 47 | M | SPE | 130, NED |
| 53 | 2.02 | 52 | 2.93 | 38 | F | SPE | 132, NED |
| 55 | 55.81 | 54 | 27.98 | 36 | M | PR | 60, NED |
| 69 | 16.48 | 70 | 21.37 | 38 | F | SPE | 37, NED |
| No AT | - | 72 | 37.48 | 50 | F | SPE | 37, NED |
| 75 | 23.79 | 76 | 4.29 | 48 | F | SPE | 21, NED |
| No AT | - | 82 | 1.61 | 41 | F | ENU | 132, NED |
| 89 | 12.07 | 90 | 36.47 | 39 | M | SPE | 192, NED |
| 107 | 1.85 | 108 | ** | 29 | F | ENU | 228, NED |
| 120 | 6.59 | 121 | 13.27 | 24 | F | SPE | 252, NED |
| Adjacent Tissue Sample Code | THOR Methylation (%) | rPA Tissue Sample Code | THOR Methylation (%) | Age * (Years) | Sex * (F/M) | Treatment * | Follow-Up * (Months) |
|---|---|---|---|---|---|---|---|
| 57 | 80.84 | 56 | 88.99 | 78 | F | SPE | 84, NED |
| 91 | 5.81 | 92 | 4.77 | 32 | F | PR | 192, NED |
| 93 | 19.36 | 94 | 16.88 | 41 | F | PE | 240, NED |
| 97 | 77.85 | 98 | 19.74 | 59 | F | SPE | 216, NED |
| 101 | 42.19 | 102 | ** | 36 | F | PR | 144, NED |
| 103 | 15.79 | 104 | 5.08 | 33 | F | SPE | 36, R, L |
| 105 | 22.10 | 106 | 17.26 | 64 | F | PR | 36, AWD |
| 113 | 6.34 | 114 | 78.33 | 55 | F | PR | 12, R, L |
| 118 | 4.14 | 119 | 7.96 | 26 | F | PR | 228, NED |
| No AT | - | 81 | 96.58 | 51 | F | P | 30, R |
| No AT | - | 112 | 29.33 | 41 | M | PR | 24, R, L |
| No AT | - | 117 | 70.37 | 35 | F | PR | 156, NED |
| No AT | - | 122 | 10.75 | 56 | F | PR | 12, R, L |
| Adjacent Tissue Sample Code | THOR Methylation (%) | Carcinoma Tissue Sample Code | THOR Methylation (%) | Age * (Years) | Sex * (F/M) | Treatment * | Follow-Up (Months) |
|---|---|---|---|---|---|---|---|
| No AT | - | 2 (ACC) | 38.54 | 15 | F | PR | 140, NED |
| 5 | 67.64 | 6 (MEC) | 2.77 | 53 | F | SPE, RT | 127, NED |
| 7 | 1.74 | 8 (AdCC) | 1.29 | 44 | M | PR | 84, NED |
| 9 | 1.57 | 10 (EMC) | 1.29 | 64 | M | PE | 126, NED |
| 11 | 51.84 | 12 (ACC) | 25.57 | 75 | M | PE, RT | 130, DOC, NED |
| 13 | 33.36 | 14 (EMC) | ** | 76 | M | SPE | 90, DOC, NED |
| 16 | 30.91 | 17 (MEC) | 50.35 | 63 | F | SPE | 116, NED |
| No AT | - | 18 (ACC) | 42.07 | 41 | M | SPE | 134, NED |
| 19 | 14.44 | 20 (CxPA) | 91.29 | 76 | M | PR | L |
| No AT | - | 47 (AdCC) | 91.70 | 41 | F | SPE, RT | 144, NED |
| 51 | 3.23 | 50 (CxPA) | 23.91 | 70 | F | SPE | 24, NED |
| 59 | 4.55 | 60 (ACC) | ** | 75 | F | PE, RT | 109, NED |
| 61 | 40.83 | 62 (EMC) | 36.51 | 43 | F | PR, RT | 137, NED |
| 63 | 1.48 | 64 (EMC) | 93.44 | 61 | F | PE, RT | 136, NED |
| 65 | 29.01 | 66 (BCA) | ** | 68 | F | PE | 24, DOC |
| No AT | - | 68 (MEC) | 33.56 | 72 | M | SPE, RT | 56, NED |
| 95 | 1.63 | 96 (CxPA) | 27.68 | 62 | M | PR | 168, NED, DOC |
| Non-Neoplastic | PAs and rPAs | Carcinomas * | |
|---|---|---|---|
| >20% | 18 | 17 | 11 |
| <20% | 30 | 17 | 3 |
| p = 0.025 | |||
| Non-neoplastic + PAs and rPAs | Carcinomas * | ||
| >20% | 35 | 11 | |
| <20% | 47 | 3 | |
| p = 0.013 | |||
| Non-neoplastic | Neoplastic * | ||
| >20% | 18 | 28 | |
| <20% | 30 | 20 | |
| p = 0.041 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paiva-Correia, A.; Apolónio, J.; Nadal, A.; Brandão, J.R.; Silva, N.; Machado, B.; Archilla, I.; Castelo-Branco, P.; Hellquist, H. Methylation Status of the Telomerase Reverse Transcriptase Promoter in Parotid Tumours and Adjacent Parotid Gland Tissue: A Pilot Study on the Implications for Recurrence and Development of Malignancy. Curr. Oncol. 2025, 32, 312. https://doi.org/10.3390/curroncol32060312
Paiva-Correia A, Apolónio J, Nadal A, Brandão JR, Silva N, Machado B, Archilla I, Castelo-Branco P, Hellquist H. Methylation Status of the Telomerase Reverse Transcriptase Promoter in Parotid Tumours and Adjacent Parotid Gland Tissue: A Pilot Study on the Implications for Recurrence and Development of Malignancy. Current Oncology. 2025; 32(6):312. https://doi.org/10.3390/curroncol32060312
Chicago/Turabian StylePaiva-Correia, António, Joana Apolónio, Alfons Nadal, José Ricardo Brandão, Nádia Silva, Bianca Machado, Ivan Archilla, Pedro Castelo-Branco, and Henrik Hellquist. 2025. "Methylation Status of the Telomerase Reverse Transcriptase Promoter in Parotid Tumours and Adjacent Parotid Gland Tissue: A Pilot Study on the Implications for Recurrence and Development of Malignancy" Current Oncology 32, no. 6: 312. https://doi.org/10.3390/curroncol32060312
APA StylePaiva-Correia, A., Apolónio, J., Nadal, A., Brandão, J. R., Silva, N., Machado, B., Archilla, I., Castelo-Branco, P., & Hellquist, H. (2025). Methylation Status of the Telomerase Reverse Transcriptase Promoter in Parotid Tumours and Adjacent Parotid Gland Tissue: A Pilot Study on the Implications for Recurrence and Development of Malignancy. Current Oncology, 32(6), 312. https://doi.org/10.3390/curroncol32060312






