Lymph Node Metastasis in Gastrointestinal Carcinomas: A View from a Proteomics Perspective
Abstract
1. Introduction
2. Lymphangiogenesis and the Role of Growth Factors and Chemokines
3. Proteome Profiling in GICs
3.1. Colorectal Carcinoma
3.2. Proteomics in LN Metastatic Colorectal Carcinoma
| (A) Tissue/serum-based studies | ||||||
| Reference | Proteomics Approach/Samples | DEPs | Validation Method/Samples | Validated Proteins | Result | Functional Characterization |
| [46] | 2DE LC MS/MS FFr tissue (primary tumor/ 5 LN positive, 5 LN negative) | 25 (Mascot score > 63, p < 0.05) | Western blot and IHC/ FFPE primary tumor 40 LN negative and 40 LN positive | HSP-27, GST, Annexin II, L-FABP | HSP-27, GST and Annexin II upregulated in LN-positive patients L-FABP downregulated in LN-negative patients | NA |
| [47] | 2D DIGE, MS/FFr tissue (primary tumor) 12 LN negative and 12 LN positive | 6 (FC > 2) | IHCs using TMAs/FFPE primary tumor 48 LN negative and 46 LN positive | Transgelin | Upregulated in LNM | miRNA-mediated knockdown in two cell lines. Decreased invasion and metastatic characteristics of cells and reduced the clonogenic survival and the percentage of viable cells. |
| [59] | 2 DE, MALDI-TOF MS/MS/FFr tissue (primary tumor) 6 LN negative and 6 LN positive | 12 (FC > 1.5) | Western blot and IHCs/FFPE primary tumor 46 LN negative 37 LN positive | TCPZ and PPIB | TCPZ was downregulated PPIB was upregulated | siRNA mediated knockdown of PP1B in SW480 cells. Inhibited cell migration, invasion and the inhibition of closure rate |
| [52] | 2DE, MALDI-TOF-MS/FFr tissue (lymph node) 62 normal LN and 126 sentinel lymph nodes from 43 patients | 40 (FC > 2) | Western blot, IHC/FFPE lymph nodes 62 normal LN and 126 sentinel lymph nodes from 43 patients | Annexin A1, hnRNP A1, ezrin, Tubulin b-2C | All 4 proteins upregulated in sentinel lymph nodes | NA |
| [48] | 2DE, MALDI-TOF-MS/FFr tissue (primary tumor) FFr, 5 LN positive and 5 LN negative | 33 (FC ≥ 2) | Western blots and IHCs/FFPE 27 normal colorectal mucosa, 65 primary CRC and 26 positive LNs | Cathepsin D, UCH-L1 and ferritin heavy chain (FHC) | Cathepsin D, UCH-L1, upregulated in LNM FHC downregulated in LNM | In vitro Overexpression of UCH-L1, resulting in increased invasiveness of HCT8 cells. |
| [60] | LFQ, MS/FFr tissue (primary tumor) 3 LN negative and 3 LN positive | 28 (FC > 2, p < 0.05) | IHCs/FFPE 168 primary colon cancer (87 LN negative and 81 LN positive) | FXYD3, S100A11, GSTM3 | FXYD3, S100A11, GSTM3 Upregulated in LNM | NA |
| [61] | LFQ LC-MS/MS/FFr tissue (primary tumor) 9 LN negative and 10 LN positive | 29 Using R package Local FDR < 0.15 | IHCs/FFPE primary 20 LN negative and 20 LN positive | MX1, IGF1-R and IRF2BP1 | MX1 and IGF1-R upregulated in LNM; IRF2BP1 downregulated in LNM | In vitro siRNA mediated knockdown of MX1. MX1 knockdown strongly inhibits wound healing of DLD1 cells. |
| [62] | iTRAQ LC-MS/MS/FFr tissue (primary tumor) 5 LN positive, 5 LN negative | 60 (FC < 0.5) | Western blots and IHCs/FFPE primary 54 LN positive 103 LN negative | HSP47 | HSP47 upregulated in LNM | NA |
| [49] | iTRAQ LC-MS/MS/FFr tissue (primary tumor) 5 LN positive, 5 LN negative | 55 (FC < 0.75) | IHCs and RTPCR Cohort 1 IHC FFPE 82 LN positive and 113 LN negative Cohort 2 RTPCR FFr primary (63 LN positive and 107 LN negative) | Ezrin | Ezrin upregulated in LNM | In vitro siRNA mediated ezrin knockdown in DLD1 and LoVo cells. Ezrin contributes to the migration and invasion capacity of CRC cells. |
| [50] | 2D-DIGE MS/ FFr tissue (primary tumor) 8 LN negative 8 LN positive | 18 (FC > 1.5) | IHCs/FFPE primary (18 LN negative and 22 LN positive) | Gelsolin, peroxiredoxin 4 | Both proteins are overexpressed in LNM | In vitro and in vivo Silencing of GSN and PRDX4 by lentiviral shRNA induces cell cycle arrest and decreases migration and invasion of DLD-1 cells. |
| [51] | iTRAQ 2D LC-MS/MS FFr tissue (primary tumor) 12 LN negative 12 LN positive | 48 (FC > 1.5) | IHCs/FFPE primary 60 LN negative 56 LN positive | Ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCH-L1) chromogranin A e (CHGA) | Upregulated in LNM | In vitro and in vivo Silencing of UCH-L1 and CHGA induced cell cycle arrest and decreased migration and invasion. Suppressed tumorigenesis in vivo. |
| [53] | 2D, MS/serum 32 LN negative; 40 LN positive | 8 (FC > 2, p < 0.05) | ELISA/86 serum samples | Transthyretin (TTR) | Downregulated in LNM | NA |
| (B). Cell line-based studies | ||||||
| Reference | Proteomics Approach/Samples | DEPs | Validation Method/Samples | Validated Proteins | Result | Functional Characterization |
| [55] | 2D MALDI TOF/Cell lines SW 480 AND SW620 | 11 (FC > 2, p < 0.05) | Western blot, RT PCR and IHCs/FFPE primary: 30 LN positive and 38 LN negative | HSP27 | HSP27 overexpression in LNM patients | NA |
| [56] | LC-MS/MS/Secretome of SW 480 and SW620 | 145 (FC > 1.5) | ELISA and IHCs/ serum of 76 LN positive, 68 LN negative and 156 healthy For IHCs, 31 FFPE LN negative 38 LN positive | ELISA: TFF3 AND GDF15 IHC: TFF3 AND GDF15 For ELISA | Elevated expression of GDF15 and TFF3 in LN-positive CRC patients | NA |
| [57] | iTRAQ-based LC-MS/MS/ cell lysate SW480 and SW620 | 147 (FC 1.5, p < 0.05) | Western blot RT-PCR/ cell lines | CacyBP and β-Catenin | CacyBP upregulated in SW620 β-Catenin downregulated in SW480 | Overexpression of CacyBP in primary colon cancer cell lines showed downregulated levels of cellular β-Catenin and significant reduction in cellular adhesion. |
| [58] | LFQ LC-MS/MS/ EVof SW480 and SW620 | Only profiling has been carried out SW480 EV-enriched proteins: 368 SW620 EV-enriched proteins: 359 (FC > 1.5). No validations | Gene ontology studies undertaken | NA | ||
3.3. Gastric Carcinoma
3.4. Proteomics in LN Metastatic Gastric Carcinoma
3.5. Hepatocellular Carcinoma
3.6. Esophageal Carcinoma
3.7. Pancreatic Carcinoma
3.8. Protoeomics in LN Metastatic Pancreatic Carcinoma
3.9. Other Rare GICs
3.10. Proteomics in LN Metastatic Rare GICs
4. Discussion and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gastrointestinal Tract. Available online: https://en.wikipedia.org/wiki/Gastrointestinal_tract (accessed on 15 October 2023).
- Zemła, P.; Stelmach, A.; Jabłońska, B.; Gołka, D.; Mrowiec, S. A Retrospective Study of Postoperative Outcomes in 98 Patients Diagnosed with Gastrointestinal Stromal Tumor (GIST) of the Upper, Middle, and Lower Gastrointestinal Tract Between 2009 and 2019 at a Single Center in Poland. Med. Sci. Monit. 2021, 27, e932809-1. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020, 159, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, A.E.; Moravan, V.; Christian, F. A competing risk analysis of colorectal cancer recurrence after curative surgery. BMC Gastroenterol. 2022, 22, 95. [Google Scholar] [CrossRef]
- Liu, D.; Lu, M.; Li, J.; Yang, Z.; Feng, Q.; Zhou, M.; Zhang, Z.; Shen, L. The patterns and timing of recurrence after curative resection for gastric cancer in China. World J. Surg. Oncol. 2016, 14, 305. [Google Scholar] [CrossRef] [PubMed]
- Papaconstantinou, D.; Tsilimigras, D.I.; Pawlik, T.M. Recurrent Hepatocellular Carcinoma: Patterns, Detection, Staging and Treatment. J. Hepatocell. Carcinoma 2022, 9, 947–957. [Google Scholar] [CrossRef]
- Lou, F.; Sima, C.S.; Adusumilli, P.S.; Bains, M.S.; Sarkaria, I.S.; Rusch, V.W.; Rusch, V.W.; Rizk, N.P. Esophageal cancer recurrence patterns and implications for surveillance. J. Thorac. Oncol. 2013, 8, 1558–1562. [Google Scholar] [CrossRef] [PubMed]
- Groot, V.P.; Gemenetzis, G.; Blair, A.B.; Rivero-Soto, R.J.; Yu, J.; Javed, A.A.; Burkhart, R.A.; Rinkes, I.H.M.B.; Molenaar, I.Q.; Cameron, J.L.; et al. Defining and Predicting Early Recurrence in 957 Patients with Resected Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2019, 269, 1154–1162. [Google Scholar] [CrossRef]
- Yuan, Z.; Shui, Y.; Liu, L.; Guo, Y.; Wei, Q. Postoperative recurrent patterns of gallbladder cancer: Possible implications for adjuvant therapy. Radiat. Oncol. 2022, 17, 118. [Google Scholar] [CrossRef]
- Duong, T.; Koopman, P.; Francois, M. Tumor lymphangiogenesis as a potential therapeutic target. J. Oncol. 2012, 2012, 204946. [Google Scholar] [CrossRef]
- Zong, J.; Guo, C.; Liu, S.; Sun, M.Z.; Tang, J. Proteomic research progress in lymphatic metastases of cancers. Clin. Transl. Oncol. 2012, 14, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Torres, V.C.; Tichauer, K.M. Noninvasive detection of cancer spread to lymph nodes: A review of molecular imaging principles and protocols. J. Surg. Oncol. 2018, 118, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Li, M.; Bao, Y.; Zhang, L.; Lu, P.; Tong, T.; Peng, J. Size and number of lymph nodes were risk factors of recurrence in stage II colorectal cancer. BMC Cancer 2023, 23, 518. [Google Scholar] [CrossRef]
- Bembenek, A.; Gretschel, S.; Schlag, P.M. Sentinel lymph node biopsy for gastrointestinal cancers. J. Surg. Oncol. 2007, 96, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, X.; Du, F.; Zhu, X.; Ren, H.; Ye, C.; Liu, Z.; Zhao, Y.; Yu, X.; Zhang, C.; et al. A narrative review of intraoperative use of indocyanine green fluorescence imaging in gastrointestinal cancer: Situation and future directions. J. Gastrointest. Oncol. 2023, 14, 1095–1113. [Google Scholar] [CrossRef]
- Wei, J.; Bu, Z. Sentinel lymph node detection for gastric cancer: Promise or pitfall? Surg. Oncol. 2020, 33, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Pan, M.; Chen, B. A Systematic Review and Meta-Analysis of Sentinel Lymph Node Biopsy in Gastric Cancer, an Optimization of Imaging Protocol for Tracer Mapping. World J. Surg. 2021, 45, 1126–1134. [Google Scholar] [CrossRef]
- Kamiya, S.; Takeuchi, H.; Fukuda, K.; Kawakubo, H.; Takahashi, N.; Mitsumori, N.; Terashima, M.; Tsujimoto, H.; Kinami, S.; Natsugoe, S.; et al. A multicenter non-randomized phase III study of sentinel node navigation surgery for early gastric cancer. Jpn. J. Clin. Oncol. 2021, 51, 305–309. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Takeuchi, H.; Takagi, Y.; Natsugoe, S.; Terashima, M.; Murakami, N.; Fujimura, T.; Tsujimoto, H.; Hayashi, H.; Yoshimizu, N.; et al. Sentinel node mapping for gastric cancer: A prospective multicenter trial in Japan. J. Clin. Oncol. 2013, 10, 3704–3710. [Google Scholar] [CrossRef]
- Mao, X.; Mei, R.; Yu, S.; Shou, L.; Zhang, W.; Li, K.; Qiu, Z.; Xie, T.; Sui, X. Emerging Technologies for the Detection of Cancer Micrometastasis. Technol. Cancer Res. Treat. 2022, 21, 15330338221100355. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Natsugoe, S.; Arigami, T.; Uenosono, Y.; Yanagita, S.; Nakajo, A.; Matsumoto, M.; Okumura, H.; Kijima, Y.; Sakoda, M.; Mataki, Y.; et al. Lymph node micrometastasis in gastrointestinal tract cancer--a clinical aspect. Int. J. Clin. Oncol. 2013, 18, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Luo, G.; Liu, C.; Wu, C.; Liu, L.; Liu, Z.; Ni, Q.; Long, J.; Yu, X. Molecular mechanism underlying lymphatic metastasis in pancreatic cancer. Biomed Res. Int. 2014, 2014, 925845. [Google Scholar] [CrossRef] [PubMed]
- Langheinrich, M.C.; Schellerer, V.; Perrakis, A.; Lohmüller, C.; Schildberg, C.; Naschberger, E.; Stürzl, M.; Hohenberger, W.; Croner, R.S. Molecular mechanisms of lymphatic metastasis in solid tumors of the gastrointestinal tract. Int. J. Clin. Exp. Pathol. 2012, 5, 614–623. [Google Scholar] [PubMed]
- Stacker, S.A.; Williams, S.P.; Karnezis, T.; Shayan, R.; Fox, S.B.; Achen, M.G. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 2014, 14, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, N.; Sun, Y.; Guo, T.; Zhu, X.; Guo, J. Expression of VEGF with tumor incidence, metastasis and prognosis in human gastric carcinoma. Cancer Biomark. 2018, 22, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, Y. Lymphangiogenesis and colorectal cancer. Saudi Med. J. 2017, 38, 237–244. [Google Scholar] [CrossRef]
- Schwartzberg, L.S.; Rivera, F.; Karthaus, M.; Fasola, G.; Canon, J.L.; Hecht, J.R.; Yu, H.; Oliner, K.S.; Go, W.Y. PEAK: A randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J. Clin. Oncol. 2014, 32, 2240–2247. [Google Scholar]
- Kabbinavar, F.F.; Hambleton, J.; Mass, R.D.; Hurwitz, H.I.; Bergsland, E.; Sarkar, S. Combined analysis of efficacy: The addition of bevacizumab to fluorouracil/leucovorin improves survival for patients with metastatic colorectal cancer. J. Clin. Oncol. 2005, 23, 3706–3712. [Google Scholar] [CrossRef]
- Murakami, T.; Kawada, K.; Iwamoto, M.; Akagami, M.; Hida, K.; Nakanishi, Y.; Kanda, K.; Kawada, M.; Seno, H.; Taketo, M.M.; et al. The role of CXCR3 and CXCR4 in colorectal cancer metastasis. Int. J. Cancer 2013, 132, 276–287. [Google Scholar] [CrossRef]
- Chen, G.; Zhou, Z.; Jin, J.; Zhou, Y.; Liu, Y.; Wang, W. CXCR4 is a prognostic marker that inhibits the invasion and migration of gastric cancer by regulating VEGF expression. Oncol. Lett. 2013, 22, 587. [Google Scholar] [CrossRef]
- Schulz, P.; Fischer, C.; Detjen, K.M.; Rieke, S.; Hilfenhaus, G.; von Marschall, Z.; Böhmig, M.; Koch, I.; Kehrberger, J.; Hauff, P.; et al. Angiopoietin-2 drives lymphatic metastasis of pancreatic cancer. FASEB J. 2011, 25, 3325–3335. [Google Scholar] [CrossRef]
- Rudno-Rudzińska, J.; Kielan, W.; Frejlich, E.; Kotulski, K.; Hap, W.; Kurnol, K.; Dzierżek, P.; Zawadzki, M.; Hałoń, A. A review on Eph/ephrin, angiogenesis and lymphangiogenesis in gastric, colorectal and pancreatic cancers. Chin. J. Cancer Res. 2017, 29, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.; Yamada, S.; Koizumi, K.; Sakurai, H.; Saiki, I. Tumour-derived fibroblast growth factor-2 exerts lymphangiogenic effects through Akt/mTOR/p70S6kinase pathway in rat lymphatic endothelial cells. Eur. J. Cancer 2007, 43, 1748–1754. [Google Scholar] [CrossRef] [PubMed]
- Da, M.X.; Wu, Z.; Tian, H.W. Tumor lymphangiogenesis and lymphangiogenic growth factors. Arch. Med. Res. 2008, 39, 365–372. [Google Scholar] [CrossRef]
- Qiu, M.Q.; Wang, H.J.; Ju, Y.F.; Sun, L.; Liu, Z.; Wang, T.; Kan, S.F.; Yang, Z.; Cui, Y.Y.; Ke, Y.Q.; et al. Fatty Acid Binding Protein 5 (FABP5) Promotes Aggressiveness of Gastric Cancer Through Modulation of Tumor Immunity. J. Gastric Cancer. 2023, 23, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Jiang, L.; Wang, X.; Wang, M.; She, F.; Chen, Y. Tumor necrosis factor-α promotes the lymphangiogenesis of gallbladder carcinoma through nuclear factor-κB-mediated upregulation of vascular endothelial growth factor-C. Cancer Sci. 2014, 105, 1261–1271. [Google Scholar] [CrossRef]
- Itatani, Y.; Kawada, K.; Yamamoto, T.; Sakai, Y. Resistance to Anti-Angiogenic Therapy in Cancer-Alterations to Anti-VEGF Pathway. Int. J. Mol. Sci. 2018, 19, 1232. [Google Scholar] [CrossRef]
- Van der Jeught, K.; Xu, H.C.; Li, Y.J.; Lu, X.B.; Ji, G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. 2018, 24, 3834–3848. [Google Scholar] [CrossRef]
- Li, S.; Li, Q. Cancer stem cells, lymphangiogenesis, and lymphatic metastasis. Cancer Lett. 2015, 357, 438–447. [Google Scholar] [CrossRef]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef] [PubMed]
- Krogue, J.D.; Azizi, S.; Tan, F.; Flament-Auvigne, I.; Brown, T.; Plass, M.; Reihs, R.; Müller, H.; Zatloukal, K.; Richeson, P.; et al. Predicting lymph node metastasis from primary tumor histology and clinicopathologic factors in colorectal cancer using deep learning. Commun. Med. 2023, 3, 59. [Google Scholar] [CrossRef]
- Ong, M.L.; Schofield, J.B. Assessment of lymph node involvement in colorectal cancer. World J. Gastrointest. Surg. 2016, 8, 179–192. [Google Scholar] [CrossRef]
- Mochizuki, K.; Kudo, S.E.; Ichimasa, K.; Kouyama, Y.; Matsudaira, S.; Takashina, Y.; Maeda, Y.; Ishigaki, T.; Nakamura, H.; Toyoshima, N.; et al. Left-sided location is a risk factor for lymph node metastasis of T1 colorectal cancer: A single-center retrospective study. Int. J. Colorectal Dis. 2020, 35, 1911–1919. [Google Scholar] [CrossRef]
- Pei, H.; Zhu, H.; Zeng, S.; Li, Y.; Yang, H.; Shen, L.; Chen, J.; Zeng, L.; Fan, J.; Li, X.; et al. Proteome analysis and tissue microarray for profiling protein markers associated with lymph node metastasis in colorectal cancer. J. Proteome Res. 2007, 6, 2495–2501. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Buckhaults, P.J.; Lee, J.R.; Xiong, H.; Farrell, C.; Podolsky, R.H.; Schade, R.R.; Dynan, W.S. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia 2009, 11, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhao, M.; Zhong, J.; Shi, L.; Luo, Q.; Liu, J.; Wang, J.; Yuan, X.; Huang, C. Proteomic profiling of proteins associated with lymph node metastasis in colorectal cancer. J. Cell Biochem. 2010, 110, 1512–1519. [Google Scholar] [CrossRef]
- Mori, K.; Toiyama, Y.; Otake, K.; Ide, S.; Imaoka, H.; Okigami, M.; Okugawa, Y.; Fujikawa, H.; Saigusa, S.; Hiro, J.; et al. Successful identification of a predictive biomarker for lymph node metastasis in colorectal cancer using a proteomic approach. Oncotarget 2017, 8, 106935–106947. [Google Scholar] [CrossRef]
- Huang, C.Y.; Lee, K.C.; Tung, S.Y.; Huang, W.S.; Teng, C.C.; Lee, K.F.; Hsieh, M.C.; Kuo, H.C. 2D-DIGE-MS Proteomics Approaches for Identification of Gelsolin and Peroxiredoxin 4 with Lymph Node Metastasis in Colorectal Cancer. Cancers 2022, 14, 3189. [Google Scholar] [CrossRef]
- Lee, K.C.; Chen, H.H.; Cheng, K.C.; Liu, T.T.; Lee, K.F.; Teng, C.C.; Huang, C.Y.; Hsieh, M.C.; Kuo, H.C. Use of iTRAQ-based quantitative proteomic identification of CHGA and UCHL1 correlated with lymph node metastasis in colorectal carcinoma. J. Cell Mol. Med. 2023, 27, 2004–2020. [Google Scholar] [CrossRef]
- He, Z.Y.; Wen, H.; Shi, C.B.; Wang, J. Up-regulation of hnRNP A1, Ezrin, tubulin β-2C and Annexin A1 in sentinel lymph nodes of colorectal cancer. World J. Gastroenterol. 2010, 16, 4670–4676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, Y.; Sun, X.; Peng, K.; Ding, Y. Serum proteome analysis for profiling protein markers associated with lymph node metastasis in colorectal carcinoma. J. Comp. Pathol. 2011, 144, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Leibovitz, A.; Stinson, J.C.; McCombs WB 3rd McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar]
- Ghosh, D.; Yu, H.; Tan, X.F.; Lim, T.K.; Zubaidah, R.M.; Tan, H.T.; Chung, M.C.; Lin, Q. Identification of key players for colorectal cancer metastasis by iTRAQ quantitative proteomics profiling of isogenic SW480 and SW620 cell lines. J. Proteome Res. 2011, 10, 4373–4387. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liu, L.; Wang, S.; Zhang, Y.F.; Yu, L.; Ding, Y.Q. Differential proteomic analysis of human colorectal carcinoma cell lines metastasis-associated proteins. J. Cancer Res. Clin. Oncol. 2007, 133, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Lü, B.; Zhang, J.; Wu, M.; Huang, Q.; Wu, Q.; Sheng, H.; Wu, D.; Hu, J.; Lai, M. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J. Proteome Res. 2010, 9, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.S.; Choi, D.Y.; Hong, B.S.; Jang, S.C.; Kim, D.K.; Lee, J.; Kim, Y.K.; Kim, K.P.; Gho, Y.S. Quantitative proteomics of extracellular vesicles derived from human primary and metastatic colorectal cancer cells. J. Extracell. Vesicles 2012, 1, 18704. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Wang, L.S.; Xia, L.; Wang, X.L.; Feng, B.; Lu, A.G.; Chen, G.Q.; Zheng, M.H. Modulated T-complex protein 1 ζ and peptidyl-prolyl cis-trans isomerase B are two novel indicators for evaluating lymph node metastasis in colorectal cancer: Evidence from proteomics and bioinformatics. Proteom. Clin. Appl. 2009, 3, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Meding, S.; Balluff, B.; Elsner, M.; Schöne, C.; Rauser, S.; Nitsche, U.; Maak, M.; Schäfer, A.; Hauck, S.M.; Ueffing, M.; et al. Tissue-based proteomics reveals FXYD3, S100A11 and GSTM3 as novel markers for regional lymph node metastasis in colon cancer. J. Pathol. 2012, 228, 459–470. [Google Scholar] [CrossRef]
- Croner, R.S.; Stürzl, M.; Rau, T.T.; Metodieva, G.; Geppert, C.I.; Naschberger, E.; Lausen, B.; Metodiev, M.V. Quantitative proteome profiling of lymph node-positive vs. -negative colorectal carcinomas pinpoints MX1 as a marker for lymph node metastasis. Int. J. Cancer 2014, 135, 2878–2886. [Google Scholar] [CrossRef]
- Mori, K.; Toiyama, Y.; Otake, K.; Fujikawa, H.; Saigusa, S.; Hiro, J.; Kobayashi, M.; Ohi, M.; Tanaka, K.; Inoue, Y.; et al. Proteomics analysis of differential protein expression identifies heat shock protein 47 as a predictive marker for lymph node metastasis in patients with colorectal cancer. Int. J. Cancer 2017, 140, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Polom, K.; Williams, C.; de Oliveira, F.M.S.; Guergova-Kuras, M.; Lisacek, F.; Karlsson, N.G.; Roviello, F.; Kamali-Moghaddam, M. A targeted proteomics approach reveals a serum protein signature as diagnostic biomarker for resectable gastric cancer. eBioMedicine 2019, 44, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.Y.; Liang, H. Clinical significance of lymph node metastasis in gastric cancer. World J. Gastroenterol. 2014, 20, 3967–3975. [Google Scholar] [CrossRef]
- Jung, J.H.; Kim, H.J.; Yeom, J.; Yoo, C.; Shin, J.; Yoo, J.; Kang, C.S.; Lee, C. Lowered expression of galectin-2 is associated with lymph node metastasis in gastric cancer. J. Gastroenterol. 2012, 47, 37–48. [Google Scholar] [CrossRef]
- Negedu, M.N.; Duckworth, C.A.; Yu, L.G. Galectin-2 in Health and Diseases. Int. J. Mol. Sci. 2022, 24, 341. [Google Scholar] [CrossRef]
- Zhang, M.H.; Xu, X.H.; Wang, Y.; Linq, Q.X.; Bi, Y.T.; Miao, X.J.; Ye, C.F.; Gao, S.X.; Gong, C.Y.; Xiang, H.; et al. A prognostic biomarker for gastric cancer with lymph node metastases. Anat. Rec. 2013, 296, 590–594. [Google Scholar] [CrossRef]
- Ma, Y.; Li, Y.F.; Wang, T.; Pang, R.; Xue, Y.W.; Zhao, S.P. Identification of proteins associated with lymph node metastasis of gastric cancer. J. Cancer Res. Clin. Oncol. 2014, 140, 1739–1749. [Google Scholar] [CrossRef]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef]
- Saitta, C.; Raffa, G.; Alibrandi, A.; Brancatelli, S.; Lombardo, D.; Tripodi, G.; Raimondo, G.; Pollicino, T. PIVKA-II is a useful tool for diagnostic characterization of ultrasound-detected liver nodules in cirrhotic patients. Medicine 2017, 96, e7266. [Google Scholar] [CrossRef]
- Benson, A.B.; D’Angelica, M.I.; Abbott, D.E.; Anaya, D.A.; Anders, R.; Are, C.; Bachini, M.; Borad, M.; Brown, D.; Burgoyne, A.; et al. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 541–565. [Google Scholar] [CrossRef]
- Xiaohong, S.; Huikai, L.; Feng, W.; Ti, Z.; Yunlong, C.; Qiang, L. Clinical significance of lymph node metastasis in patients undergoing partial hepatectomy for hepatocellular carcinoma. World J. Surg. 2010, 34, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.; Roshandel, G.; McCormack, V.; Malekzadeh, R. Current Status and Future Prospects for Esophageal Cancer. Cancers 2023, 15, 765. [Google Scholar] [CrossRef] [PubMed]
- Kakeji, Y.; Oshikiri, T.; Takiguchi, G.; Kanaji, S.; Matsuda, T.; Nakamura, T.; Suzuki, S. Multimodality approaches to control esophageal cancer: Development of chemoradiotherapy, chemotherapy, and immunotherapy. Esophagus 2021, 18, 25–32. [Google Scholar] [CrossRef]
- Yang, Y.M.; Hong, P.; Xu, W.W.; He, Q.Y.; Li, B. Advances in targeted therapy for esophageal cancer. Signal Transduct. Target. Ther. 2020, 5, 229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Chen, L.Q.; Liu, R.L.; Shi, Y.T.; He, M.; Meng, X.L.; Bai, S.X.; Ping, Y.M. The number of lymph node metastases influences survival and International Union Against Cancer tumor-node-metastasis classification for esophageal squamous cell carcinoma. Dis. Esophagus 2010, 23, 53–58. [Google Scholar] [CrossRef]
- Hu, J.X.; Zhao, C.F.; Chen, W.B.; Liu, Q.C.; Li, Q.W.; Lin, Y.Y.; Gao, F. Pancreatic cancer: A review of epidemiology, trend, and risk factors. World J. Gastroenterol. 2021, 27, 4298–4321. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.C.; Goggins, M.G.; Fishman, E.K. Diagnosis and Detection of Pancreatic Cancer. Cancer J. 2017, 23, 333–342. [Google Scholar] [CrossRef]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Baldwin, S.; Kukar, M.; Gabriel, E.; Attwood, K.; Wilkinson, N.; Hochwald, S.N.; Kuvshinoff, B. Pancreatic cancer metastatic to a limited number of lymph nodes has no impact on outcome. HPB 2016, 18, 523–528. [Google Scholar] [CrossRef]
- Cui, Y.; Wu, J.; Zong, M.; Song, G.; Jia, Q.; Jiang, J.; Han, J. Proteomic profiling in pancreatic cancer with and without lymph node metastasis. Int. J. Cancer 2009, 124, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, K.; Jones, R.; Dmitrovic, B.; Wijesuriya, N.; Kocher, H.; Hart, I.R.; Crnogorac-Jurcevic, T. Proteome of formalin-fixed paraffin-embedded pancreatic ductal adenocarcinoma and lymph node metastases. J. Pathol. 2012, 226, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Takadate, T.; Mizuma, M.; Shima, H.; Suzuki, T.; Tachibana, T.; Shimura, M.; Hata, T.; Iseki, M.; Kawaguchi, K.; et al. Stromal expression of hemopexin is associated with lymph-node metastasis in pancreatic ductal adenocarcinoma. PLoS ONE 2020, 15, e0235904. [Google Scholar] [CrossRef] [PubMed]
- Randi, G.; Franceschi, S.; La Vecchia, C. Gallbladder cancer worldwide: Geographical distribution and risk factors. Int. J. Cancer 2006, 118, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Moreno, P.; Riquelme, I.; García, P.; Brebi, P.; Roa, J.C. Environmental and Lifestyle Risk Factors in the Carcinogenesis of Gallbladder Cancer. J. Pers. Med. 2022, 12, 234. [Google Scholar] [CrossRef]
- Shirai, Y.; Sakata, J.; Wakai, T.; Ohashi, T.; Ajioka, Y.; Hatakeyama, K. Assessment of lymph node status in gallbladder cancer: Location, number, or ratio of positive nodes. World J. Surg. Oncol. 2012, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.G.; Kim, C.Y.; Cho, S.H.; Kim, H.J.; Koh, Y.S.; Kim, J.C.; Cho, C.K.; Kim, H.J.; Hur, Y.H. Impact of lymph node ratio as a valuable prognostic factor in gallbladder carcinoma, focusing on stage IIIB gallbladder carcinoma. J. Korean Surg. Soc. 2013, 84, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Amini, N.; Kim, Y.; Wilson, A.; Margonis, G.A.; Ethun, C.G.; Poultsides, G.; Tran, T.; Idrees, K.; Isom, C.A.; Fields, R.C.; et al. Prognostic Implications of Lymph Node Status for Patients with Gallbladder Cancer: A Multi-Institutional Study. Ann. Surg. Oncol. 2016, 23, 3016–3023. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.K.; Kalayarasan, R.; Javed, A.; Sakhuja, P. Role of routine 16b1 lymph node biopsy in the management of gallbladder cancer: An analysis. HPB 2014, 16, 229–234. [Google Scholar] [CrossRef][Green Version]
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019, 39, 19–31. [Google Scholar] [CrossRef]
- Rogacka, N.A.; Benkö, T.; Saner, F.H.; Malamutmann, E.; Kaths, M.; Treckmann, J.W.; Hoyer, D.P. Lymph Node Staging in Perihilar Cholangiocarcinoma: The Key to the Big Picture. Curr. Oncol. 2023, 17, 5849–5862. [Google Scholar] [CrossRef]
- Overman, M.J.; Hu, C.Y.; Wolff, R.A.; Chang, G.J. Prognostic value of lymph node evaluation in small bowel adenocarcinoma: Analysis of the surveillance, epidemiology, and end results database. Cancer 2010, 116, 5374–5382. [Google Scholar] [CrossRef]
- Gondal, T.A.; Chaudhary, N.; Bajwa, H.; Rauf, A.; Le, D.; Ahmed, S. Anal Cancer: The Past, Present and Future. Curr. Oncol. 2023, 30, 3232–3250. [Google Scholar] [CrossRef] [PubMed]
- Prognosis for Anal Cancer. Available online: https://www.healthline.com/health/cancer/prognosis-for-anal-cancer (accessed on 13 May 2024).
- Jain, V.; Akhtar, J.; Priya, R.; Sakhuja, P.; Goyal, S.; Agarwal, A.K.; Ghose, V.; Polisetty, R.V.; Sirdeshmukh, R.; Siraj, F.; et al. Tissue proteome analysis for profiling proteins associated with lymph node metastasis in gallbladder cancer. BMC Cancer 2023, 23, 402. [Google Scholar] [CrossRef] [PubMed]
- Assinder, S.J.; Stanton, J.A.; Prasad, P.D. Transgelin: An actin-binding protein and tumour suppressor. Int. J. Biochem. Cell Biol. 2009, 41, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Han, G.Y.; Park, H.W.; Song, Y.J.; Kim, C.W. Transgelin promotes migration and invasion of cancer stem cells. J. Proteome Res. 2010, 9, 5108–5117. [Google Scholar] [CrossRef]
- Zhao, Z.; Lu, L.; Li, W. TAGLN2 promotes the proliferation, invasion, migration and epithelial-mesenchymal transition of colorectal cancer cells by activating STAT3 signaling through ANXA2. Oncol. Lett. 2021, 22, 737. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, P.; Xu, X.L.; Cai, L.; Song, Z.S.; Cao, D.Y.; Tao, K.S.; Zhou, W.P.; Chen, Z.N.; Dou, K.F. Annexin A2 promotes the migration and invasion of human hepatocellular carcinoma cells in vitro by regulating the shedding of CD147-harboring microvesicles from tumor cells. PLoS ONE 2023, 8, e67268. [Google Scholar] [CrossRef]
- Han, L.; Jiang, Y.; Han, D.; Tan, W. Hsp27 regulates epithelial mesenchymal transition, metastasis and proliferation in colorectal carcinoma. Oncol. Lett. 2018, 16, 5309–5316. [Google Scholar] [CrossRef]
- Sato, Y.; Kumamoto, K.; Saito, K.; Okayama, H.; Hayase, S.; Kofunato, Y.; Miyamoto, K.; Nakamura, I.; Ohki, S.; Koyama, Y.; et al. Up-regulated Annexin A1 expression in gastrointestinal cancer is associated with cancer invasion and lymph node metastasis. Exp. Ther. Med. 2011, 2, 239–243. [Google Scholar] [CrossRef]
- Kirana, C.; Shi, H.; Laing, E.; Hood, K.; Miller, R.; Bethwaite, P.; Keating, J.; Jordan, T.W.; Hayes, M.; Stubbs, R. Cathepsin D Expression in Colorectal Cancer: From Proteomic Discovery through Validation Using Western Blotting, Immunohistochemistry, and Tissue Microarrays. Int. J. Proteom. 2012, 2012, 245819. [Google Scholar] [CrossRef]
- Jin, M.; Zhang, H.; Yang, J.; Zheng, Z.; Liu, K. Expression mode and prognostic value of FXYD family members in colon cancer. Aging 2021, 13, 18404–18422. [Google Scholar] [CrossRef]
- Niu, Y.; Shao, Z.; Wang, H.; Yang, J.; Zhang, F.; Luo, Y.; Xu, L.; Ding, Y.; Zhao, L. LASP1-S100A11 axis promotes colorectal cancer aggressiveness by modulating TGFβ/Smad signaling. Sci. Rep. 2016, 6, 26112. [Google Scholar] [CrossRef]
- Yi, N.; Xiao, M.B.; Ni, W.K.; Jiang, F.; Lu, C.H.; Ni, R.Z. High expression of peroxiredoxin 4 affects the survival time of colorectal cancer patients, but is not an independent unfavorable prognostic factor. Mol. Clin. Oncol. 2014, 2, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Wallin, U.; Glimelius, B.; Jirström, K.; Darmanis, S.; Nong, R.Y.; Pontén, F.; Johansson, C.; Påhlman, L.; Birgisson, H. Growth differentiation factor 15: A prognostic marker for recurrence in colorectal cancer. Br. J. Cancer 2011, 104, 1619–1627. [Google Scholar] [CrossRef]
- Yusufu, A.; Shayimu, P.; Tuerdi, R.; Fang, C.; Wang, F.; Wang, H. TFF3 and TFF1 expression levels are elevated in colorectal cancer and promote the malignant behavior of colon cancer by activating the EMT process. Int. J. Oncol. 2019, 55, 789–804. [Google Scholar] [CrossRef]
- Torer, N.; Kayaselcuk, F.; Nursal, T.Z.; Yildirim, S.; Tarim, A.; Nòyan, T.; Karakayali, H. Adhesion molecules as prognostic markers in pancreatic adenocarcinoma. J. Surg. Oncol. 2007, 96, 419–423. [Google Scholar] [CrossRef]
- Mogal, M.R.; Junayed, A.; Mahmod, M.R.; Sompa, S.A.; Lima, S.A.; Kar, N.; TasminaTarin Khatun, M.; Zubair, M.A.; Sikder, M.A. A Computational Approach to Justifying Stratifin as a Candidate Diagnostic and Prognostic Biomarker for Pancreatic Cancer. Biomed Res. Int. 2022, 2022, 1617989. [Google Scholar] [CrossRef]
- Wang, C.; Chu, M. Advances in Drugs Targeting Lymphangiogenesis for Preventing Tumor Progression and Metastasis. Front. Oncol. 2022, 11, 783309. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, H.; Xu, Y.; Xu, C.; Sun, H.; Li, Z.; Ge, Y.; Wang, H.; Zhao, T.; Gao, S.; et al. BICC1 drives pancreatic cancer progression by inducing VEGF-independent angiogenesis. Signal Transduct. Target. Ther. 2023, 8, 271. [Google Scholar] [CrossRef]
- Qin, X.; Ruan, H.; Yuan, L.; Lin, L. Colorectal cancer tumor stem cells mediate bevacizumab resistance through the signal IL-22-STAT3 signaling pathway. 3 Biotech 2023, 13, 327. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, M. Laser Capture Microdissection: Insights into Methods and Applications. Methods Mol. Biol. 2018, 1723, 1–17. [Google Scholar] [PubMed]
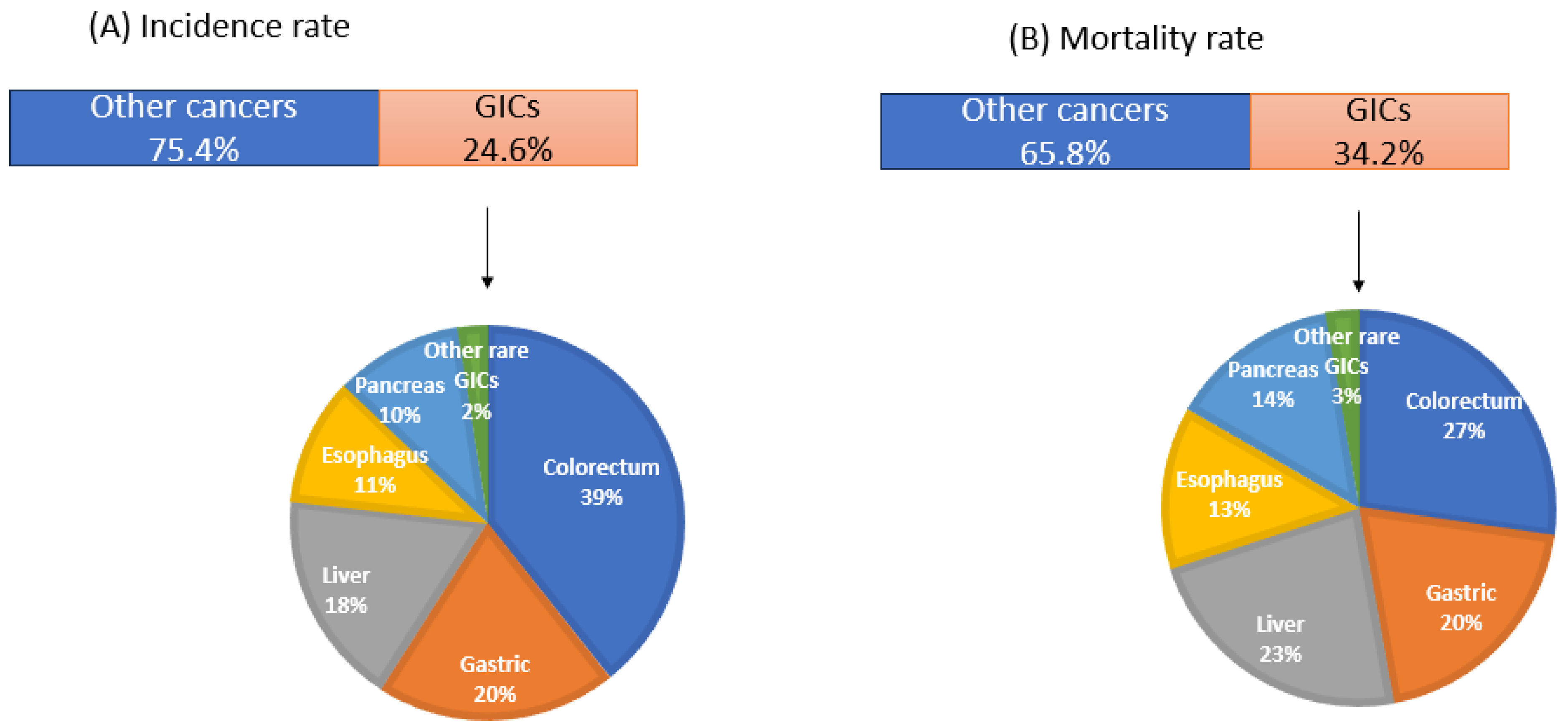
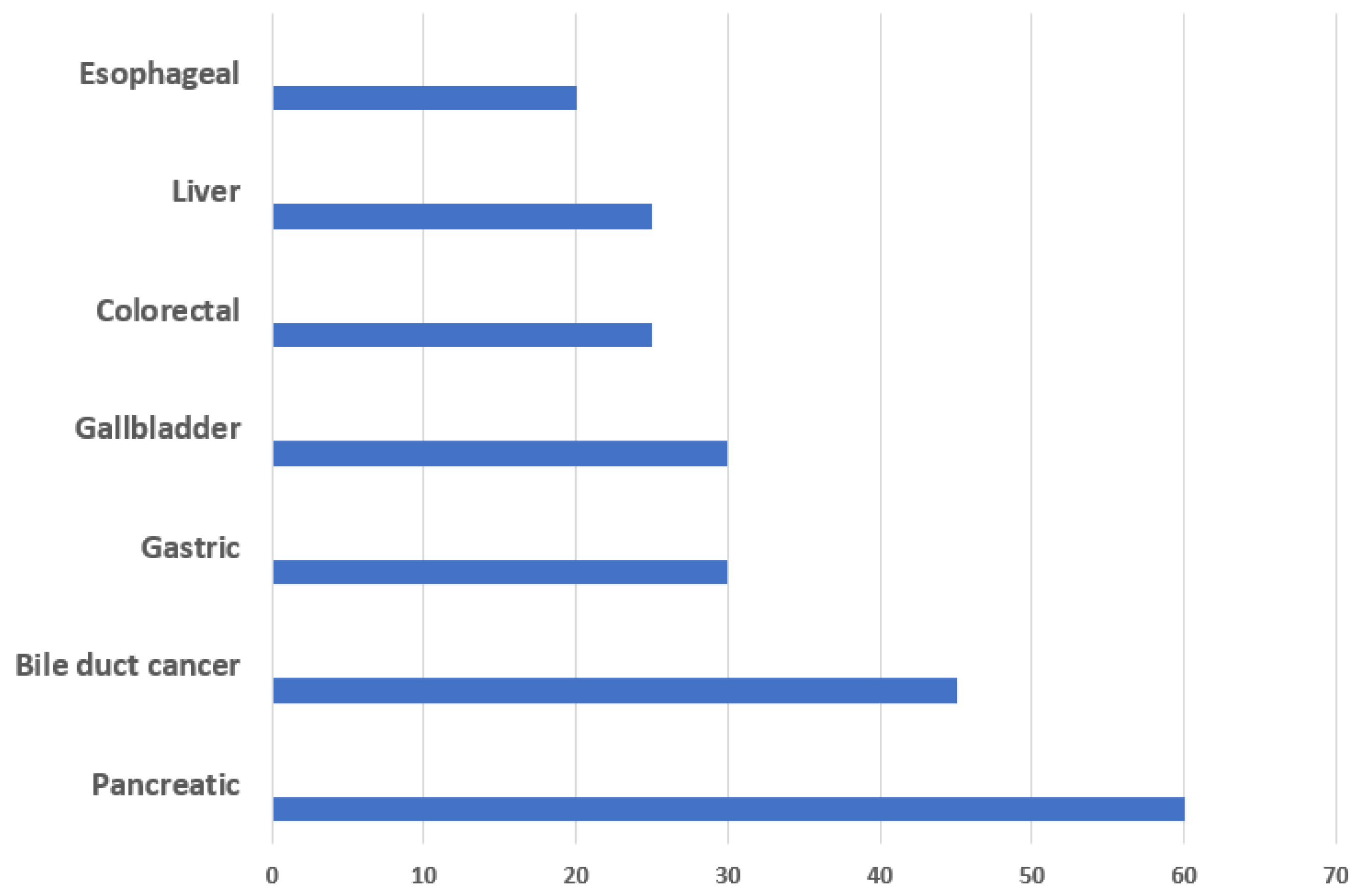
| S. No. | Cancer | Description |
|---|---|---|
| 1. | Colorectal Carcinoma | N1a—metastasis in 1 regional LN N1b—metastasis in 2–3 regional LNs N1c—metastasis in the regional lymph nodes does not contain cancer, but the cancer cells are in the tissue near the tumor N2a—metastasis in 4–6 regional LNs N2b—metastasis in ≥ 7 regional LNs |
| 2. | Gastric Carcinoma | N1—metastasis in 1–2 regional LNs N2—metastasis in 3–6 regional LNs N3a—metastasis in 7–15 regional LNs N3b—metastasis in ≥15 regional LNs |
| 3. | Hepatocellular Carcinoma | N1—metastasis in ≥1 regional LN |
| 4. | Esophageal Carcinoma | N1—metastasis in 1–2 regional LNs N2—metastasis in 3–6 regional LNs N3—≥7 regional LNs |
| 5. | Pancreatic Carcinoma | N1—metastasis in 1–3 regional LNs N2—metastasis in ≥4 regional LNs |
| 6. | Gallbladder Carcinoma | N1—metastasis in 1–3 regional LNs N2—metastasis in ≥4 regional LNs |
| 7. | Bile Duct Cancer | N1—metastasis in 1–3 regional LNs N2—metastasis in ≥4 regional LNs |
| 8. | Small Intestine Cancer | N1—metastasis in 1–2 regional LNs N2—metastasis in ≥3 regional LNs |
| 9. | Anal Cancer | N1—metastasis in nearby lymph nodes |
| Reference | Proteomics Approach/Samples | DEPs | Validation Method/Samples | Validated Proteins | Result | Functional Characterization |
|---|---|---|---|---|---|---|
| [66] | ICAT labeling LC-MS/MS/ FFr tissue (primary tumor) 4 LN positive 3 LN negative | 151 (FC > 1.5, p < 0.05) | Western blot and IHC/120 FFPE primary tumor | Galectin -2 | Downregulated expression of Galectin-2 in LNM | NA |
| [68] | 2DE, LFQ MS/MS/ FFr tissue (primary tumor) 12 LN positive 12 LN negative | 26 (Mascot score > 60) | Validations by IHC/FFPE 43 LN negative and 85 LN positive | 14-3-3β and profilin-1 | 14-3-3β protein upregulated in LNM Profilin-1 downregulated in LNM | NA |
| [69] | LC-MS/MS/ Serum 33 LN-negative patients and 157 LN-positive patients | NA | Not undertaken | Fibrinopeptide A (FPA) with alanine truncation at N terminal | 85.4% of patients with LNM had FPA with alanine truncation at the N-terminal | NA |
| Reference | Proteomics Approach/Samples | DEPs | Validation Method/Samples | Validated Proteins | Result | Functional Characterization |
|---|---|---|---|---|---|---|
| [82] | DIGE, MS/FFr tissue (primary tumor) 8 LN positive and 7 LN negative | 33 (FC > 2, p < 0.05) | Western blot and IHC/63 pancreatic cancer tissues (37 LN positive and 26 LN negative) and 11 normal pancreas tissues | Radixin, moesin, ezrin and c14orf166 | Radixin, moesin and c14orf166 upregulated in LNM Ezrin—no significant results | NA |
| [83] | LFQ LC-MS/MS/ FFPE tissue (primary tumor) 7 LN negative 7 LN positive | 115 g test (g-value ≥ 3.8) | IHCs/55 FFPE tissues of matched primary PDAC and LN metastases (LNs) | S100P, moesin, stratifin | S100P and stratifin significantly upregulated in LNM Moesin—no significant results | NA |
| [84] | LFQ LC-MS/MS/FFPE tissue (primary tumor) 5 LN positive and 5 LN negative | 9 (log2 ratio, >1 or <0.05) | IHCs/FFPE tissue (primary tumor) 5 LN positive and 5 LN negative | Hemopexin FTL | Hemopexin expression was associated with LN metastasis, whereas FTL expression was not associated | In vitro Stimulant hemopexin given It promotes invasion and migration of pancreatic cancer cells |
| Reference | Proteomics Approach/Samples | DEPs | Validation Method/Samples | Validated Proteins | Results | Functional Characterization |
|---|---|---|---|---|---|---|
| [96] | iTRAQ labeling LC-MS/MS/ FFr tissue (primary tumor) 4 gallstone disease cases, 3 LN negative and 4 LN positive | 58 (FC ≥ 2, p < 0.05) | Western blotting and IHCs/FFPE primary tumor of 15 GSD, 15 LN-negative GBC and 15 LN-positive GBC | KRT7, KRT19, sorcin and NPM1 | KRT7 and sorcin upregulated in LNM KRT19 and NPM1—no significant results | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, V.; Sakhuja, P.; Agarwal, A.K.; Sirdeshmukh, R.; Siraj, F.; Gautam, P. Lymph Node Metastasis in Gastrointestinal Carcinomas: A View from a Proteomics Perspective. Curr. Oncol. 2024, 31, 4455-4475. https://doi.org/10.3390/curroncol31080333
Jain V, Sakhuja P, Agarwal AK, Sirdeshmukh R, Siraj F, Gautam P. Lymph Node Metastasis in Gastrointestinal Carcinomas: A View from a Proteomics Perspective. Current Oncology. 2024; 31(8):4455-4475. https://doi.org/10.3390/curroncol31080333
Chicago/Turabian StyleJain, Vaishali, Puja Sakhuja, Anil Kumar Agarwal, Ravi Sirdeshmukh, Fouzia Siraj, and Poonam Gautam. 2024. "Lymph Node Metastasis in Gastrointestinal Carcinomas: A View from a Proteomics Perspective" Current Oncology 31, no. 8: 4455-4475. https://doi.org/10.3390/curroncol31080333
APA StyleJain, V., Sakhuja, P., Agarwal, A. K., Sirdeshmukh, R., Siraj, F., & Gautam, P. (2024). Lymph Node Metastasis in Gastrointestinal Carcinomas: A View from a Proteomics Perspective. Current Oncology, 31(8), 4455-4475. https://doi.org/10.3390/curroncol31080333





