Spatially Resolved Microglia/Macrophages in Recurrent Glioblastomas Overexpress Fatty Acid Metabolism and Phagocytic Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Digital Spatial Profiling—Selection and Segmenting of Regions of Interest
2.3. Data Analysis
2.3.1. Nanostring Data Normalization
2.3.2. Unsupervised Clustering
2.3.3. Differential Expression and Pathway Analyses
2.3.4. Survival Analyses
3. Results
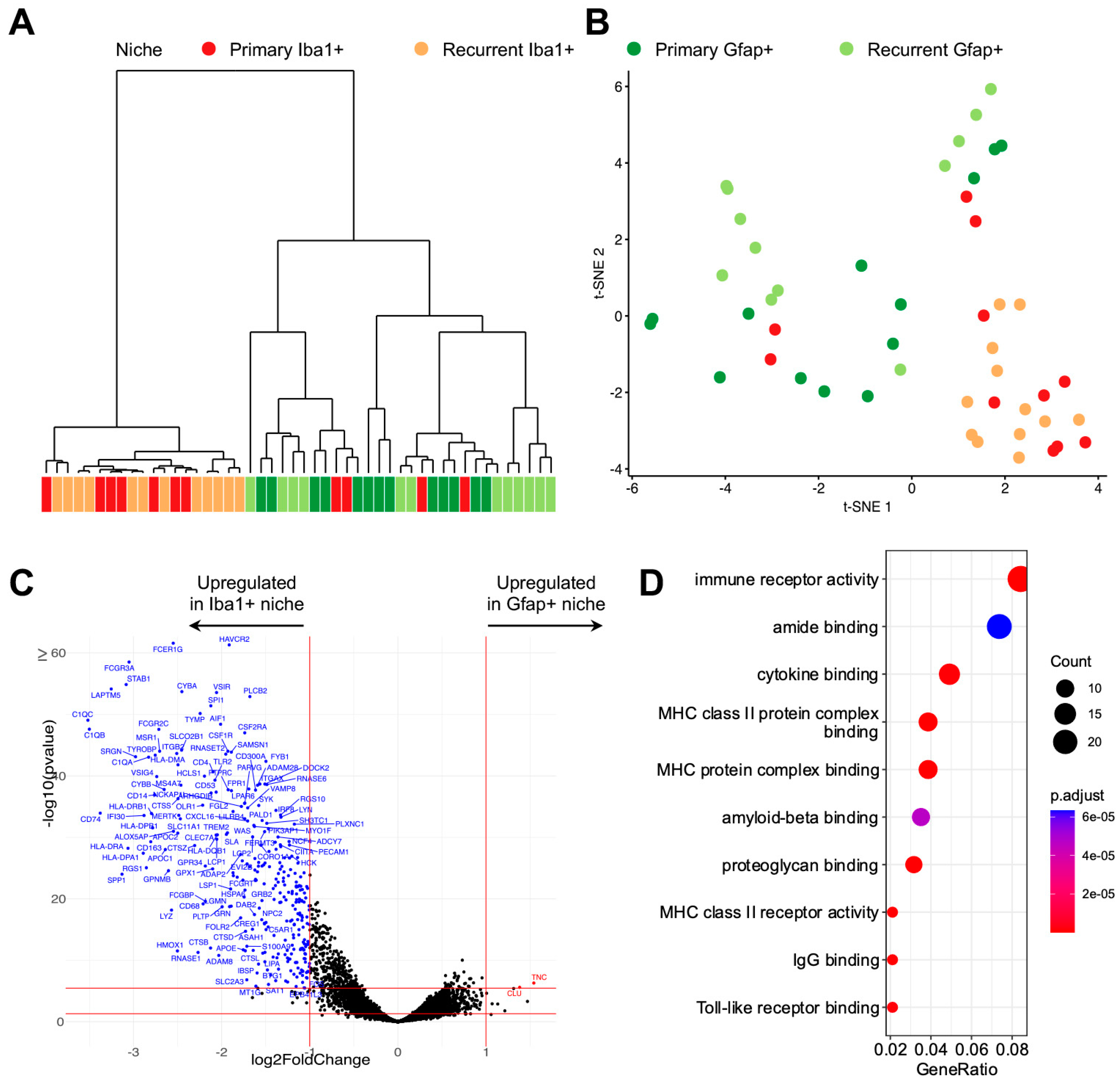
3.1. The Presence of a Tumor-Associated Microglia/Macrophage Niche in GBM
3.2. Digital Spatial Transcriptomic Profiling of Tumor-Associated Microglia/Macrophages
3.3. Differentially Expressed Microglial/Macrophage Genes in Recurrent GBM
3.4. Lipid Metabolism Gene PLD3 Correlates with Survival in Several GBM Cohorts
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro Oncol. 2022, 24, v1–v95. [Google Scholar] [CrossRef]
- Nabors, L.B.; Portnow, J.; Ahluwalia, M.; Baehring, J.; Brem, H.; Brem, S.; Butowski, N.; Campian, J.L.; Clark, S.W.; Fabiano, A.J.; et al. Central Nervous System Cancers, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 1537–1570. [Google Scholar] [CrossRef]
- Bagley, S.J.; Kothari, S.; Rahman, R.; Lee, E.Q.; Dunn, G.P.; Galanis, E.; Chang, S.M.; Nabors, L.B.; Ahluwalia, M.S.; Stupp, R.; et al. Glioblastoma Clinical Trials: Current Landscape and Opportunities for Improvement. Clin. Cancer Res. 2022, 28, 594–602. [Google Scholar] [CrossRef]
- Gorlia, T.; Stupp, R.; Brandes, A.A.; Rampling, R.R.; Fumoleau, P.; Dittrich, C.; Campone, M.M.; Twelves, C.C.; Raymond, E.; Hegi, M.E.; et al. New prognostic factors and calculators for outcome prediction in patients with recurrent glioblastoma: A pooled analysis of EORTC Brain Tumour Group phase I and II clinical trials. Eur. J. Cancer 2012, 48, 1176–1184. [Google Scholar] [CrossRef]
- Vaz-Salgado, M.A.; Villamayor, M.; Albarran, V.; Alia, V.; Sotoca, P.; Chamorro, J.; Rosero, D.; Barrill, A.M.; Martin, M.; Fernandez, E.; et al. Recurrent Glioblastoma: A Review of the Treatment Options. Cancers 2023, 15, 4279. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Reardon, D.A.; Brandes, A.A.; Omuro, A.; Mulholland, P.; Lim, M.; Wick, A.; Baehring, J.; Ahluwalia, M.S.; Roth, P.; Bahr, O.; et al. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1003–1010. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; Brandes, A.A.; Carpentier, A.F.; Idbaih, A.; Reardon, D.A.; Cloughesy, T.; Sumrall, A.; Baehring, J.; van den Bent, M.; Bahr, O.; et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: An international randomized phase III trial. Neuro Oncol. 2023, 25, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Hersh, D.S.; Wadajkar, A.S.; Roberts, N.; Perez, J.G.; Connolly, N.P.; Frenkel, V.; Winkles, J.A.; Woodworth, G.F.; Kim, A.J. Evolving Drug Delivery Strategies to Overcome the Blood Brain Barrier. Curr. Pharm. Des. 2016, 22, 1177–1193. [Google Scholar] [CrossRef]
- Omuro, A.; Vlahovic, G.; Lim, M.; Sahebjam, S.; Baehring, J.; Cloughesy, T.; Voloschin, A.; Ramkissoon, S.H.; Ligon, K.L.; Latek, R.; et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: Results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 2018, 20, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, W.; Sanchez-Perez, L.; Gajewski, T.F.; Sampson, J.H. Brain Tumor Microenvironment and Host State: Implications for Immunotherapy. Clin. Cancer Res. 2019, 25, 4202–4210. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Weller, M.; Idbaih, A.; Steinbach, J.; Finocchiaro, G.; Raval, R.R.; Ansstas, G.; Baehring, J.; Taylor, J.W.; Honnorat, J.; et al. Phase III trial of chemoradiotherapy with temozolomide plus nivolumab or placebo for newly diagnosed glioblastoma with methylated MGMT promoter. Neuro Oncol. 2022, 24, 1935–1949. [Google Scholar] [CrossRef]
- Baxter, M.E.; Miller, H.A.; Chen, J.; Williams, B.J.; Frieboes, H.B. Metabolomic differentiation of tumor core versus edge in glioma. Neurosurg. Focus 2023, 54, E4. [Google Scholar] [CrossRef]
- Barber, H.; Tofias, A.; Lander, B.; Daniels, A.; Gong, J.; Ren, Y.; Ren, X.; Liang, Y.; White, P.; Kurian, K.M. Advanced Molecular Characterization Using Digital Spatial Profiling Technology on Immunooncology Targets in Methylated Compared with Unmethylated IDH-Wildtype Glioblastoma. J. Oncol. 2021, 2021, 8819702. [Google Scholar] [CrossRef]
- Hernandez, S.; Lazcano, R.; Serrano, A.; Powell, S.; Kostousov, L.; Mehta, J.; Khan, K.; Lu, W.; Solis, L.M. Challenges and Opportunities for Immunoprofiling Using a Spatial High-Plex Technology: The NanoString GeoMx((R)) Digital Spatial Profiler. Front Oncol. 2022, 12, 890410. [Google Scholar] [CrossRef]
- Unanue, E.R.; Turk, V.; Neefjes, J. Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu. Rev. Immunol. 2016, 34, 265–297. [Google Scholar] [CrossRef]
- Miller, D.M.; Yadanapudi, K.; Rai, V.; Rai, S.N.; Chen, J.; Frieboes, H.B.; Masters, A.; McCallum, A.; Williams, B.J. Untangling the web of glioblastoma treatment resistance using a multi-omic and multidisciplinary approach. Am. J. Med. Sci. 2023, 366, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.P.; Sells, B.E.; Haque, S.J.; Chakravarti, A. Tumor Heterogeneity in Glioblastomas: From Light Microscopy to Molecular Pathology. Cancers 2021, 13, 761. [Google Scholar] [CrossRef]
- Rajaratnam, V.; Islam, M.M.; Yang, M.; Slaby, R.; Ramirez, H.M.; Mirza, S.P. Glioblastoma: Pathogenesis and Current Status of Chemotherapy and Other Novel Treatments. Cancers 2020, 12, 937. [Google Scholar] [CrossRef]
- Goldmann, T.; Wieghofer, P.; Jordao, M.J.; Prutek, F.; Hagemeyer, N.; Frenzel, K.; Amann, L.; Staszewski, O.; Kierdorf, K.; Krueger, M.; et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 2016, 17, 797–805. [Google Scholar] [CrossRef]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; de Bruijn, M.F.; Geissmann, F.; et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef]
- Crotti, A.; Ransohoff, R.M. Microglial Physiology and Pathophysiology: Insights from Genome-wide Transcriptional Profiling. Immunity 2016, 44, 505–515. [Google Scholar] [CrossRef]
- Sousa, C.; Biber, K.; Michelucci, A. Cellular and Molecular Characterization of Microglia: A Unique Immune Cell Population. Front. Immunol. 2017, 8, 198. [Google Scholar] [CrossRef]
- Korin, B.; Ben-Shaanan, T.L.; Schiller, M.; Dubovik, T.; Azulay-Debby, H.; Boshnak, N.T.; Koren, T.; Rolls, A. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat. Neurosci. 2017, 20, 1300–1309. [Google Scholar] [CrossRef]
- Chinnery, H.R.; Ruitenberg, M.J.; McMenamin, P.G. Novel characterization of monocyte-derived cell populations in the meninges and choroid plexus and their rates of replenishment in bone marrow chimeric mice. J. Neuropathol. Exp. Neurol. 2010, 69, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Bowman, R.L.; Klemm, F.; Akkari, L.; Pyonteck, S.M.; Sevenich, L.; Quail, D.F.; Dhara, S.; Simpson, K.; Gardner, E.E.; Iacobuzio-Donahue, C.A.; et al. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell Rep. 2016, 17, 2445–2459. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Gabrusiewicz, K.; Ellert-Miklaszewska, A.; Lipko, M.; Sielska, M.; Frankowska, M.; Kaminska, B. Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS ONE 2011, 6, e23902. [Google Scholar] [CrossRef]
- Brandenburg, S.; Muller, A.; Turkowski, K.; Radev, Y.T.; Rot, S.; Schmidt, C.; Bungert, A.D.; Acker, G.; Schorr, A.; Hippe, A.; et al. Resident microglia rather than peripheral macrophages promote vascularization in brain tumors and are source of alternative pro-angiogenic factors. Acta Neuropathol. 2016, 131, 365–378. [Google Scholar] [CrossRef]
- Li, W.; Graeber, M.B. The molecular profile of microglia under the influence of glioma. Neuro. Oncol. 2012, 14, 958–978. [Google Scholar] [CrossRef]
- Sorensen, M.D.; Dahlrot, R.H.; Boldt, H.B.; Hansen, S.; Kristensen, B.W. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol. Appl. Neurobiol. 2018, 44, 185–206. [Google Scholar] [CrossRef]
- Geribaldi-Doldan, N.; Fernandez-Ponce, C.; Quiroz, R.N.; Sanchez-Gomar, I.; Escorcia, L.G.; Velasquez, E.P.; Quiroz, E.N. The Role of Microglia in Glioblastoma. Front. Oncol. 2020, 10, 603495. [Google Scholar] [CrossRef] [PubMed]
- Park, J.B.; Lee, C.S.; Jang, J.H.; Ghim, J.; Kim, Y.J.; You, S.; Hwang, D.; Suh, P.G.; Ryu, S.H. Phospholipase signalling networks in cancer. Nat. Rev. Cancer 2012, 12, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef]
- Park, M.H.; Ahn, B.H.; Hong, Y.K.; Min, D.S. Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-kappaB/Sp1-mediated signaling pathways. Carcinogenesis 2009, 30, 356–365. [Google Scholar] [CrossRef]
- Parik, S.; Fernandez-Garcia, J.; Lodi, F.; De Vlaminck, K.; Derweduwe, M.; De Vleeschouwer, S.; Sciot, R.; Geens, W.; Weng, L.; Bosisio, F.M.; et al. GBM tumors are heterogeneous in their fatty acid metabolism and modulating fatty acid metabolism sensitizes cancer cells derived from recurring GBM tumors to temozolomide. Front. Oncol. 2022, 12, 988872. [Google Scholar] [CrossRef] [PubMed]
- Eyme, K.M.; Sammarco, A.; Jha, R.; Mnatsakanyan, H.; Pechdimaljian, C.; Carvalho, L.; Neustadt, R.; Moses, C.; Alnasser, A.; Tardiff, D.F.; et al. Targeting de novo lipid synthesis induces lipotoxicity and impairs DNA damage repair in glioblastoma mouse models. Sci. Transl. Med. 2023, 15, eabq6288. [Google Scholar] [CrossRef]
- Gimple, R.C.; Kidwell, R.L.; Kim, L.J.Y.; Sun, T.; Gromovsky, A.D.; Wu, Q.; Wolf, M.; Lv, D.; Bhargava, S.; Jiang, L.; et al. Glioma Stem Cell-Specific Superenhancer Promotes Polyunsaturated Fatty-Acid Synthesis to Support EGFR Signaling. Cancer Discov. 2019, 9, 1248–1267. [Google Scholar] [CrossRef] [PubMed]
- Miska, J.; Chandel, N.S. Targeting fatty acid metabolism in glioblastoma. J. Clin. Investig. 2023, 133. [Google Scholar] [CrossRef] [PubMed]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell 2021, 56, 1363–1393. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Bell, E.H.; Chakravarti, A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013, 2, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.; Al-Khami, A.A.; Wyczechowska, D.; Hernandez, C.; Zheng, L.; Reiss, K.; Valle, L.D.; Trillo-Tinoco, J.; Maj, T.; Zou, W.; et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol. Res. 2015, 3, 1236–1247. [Google Scholar] [CrossRef] [PubMed]
- Miska, J.; Lee-Chang, C.; Rashidi, A.; Muroski, M.E.; Chang, A.L.; Lopez-Rosas, A.; Zhang, P.; Panek, W.K.; Cordero, A.; Han, Y.; et al. HIF-1alpha Is a Metabolic Switch between Glycolytic-Driven Migration and Oxidative Phosphorylation-Driven Immunosuppression of Tregs in Glioblastoma. Cell Rep. 2019, 27, 226–237.e4. [Google Scholar] [CrossRef]
- Wang, H.; Franco, F.; Tsui, Y.C.; Xie, X.; Trefny, M.P.; Zappasodi, R.; Mohmood, S.R.; Fernandez-Garcia, J.; Tsai, C.H.; Schulze, I.; et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020, 21, 298–308. [Google Scholar] [CrossRef]
- Jakovlevs, A.; Vanags, A.; Gardovskis, J.; Strumfa, I. Molecular classification of diffuse gliomas. Pol. J. Pathol. 2019, 70, 246–258. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; de Carvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e6. [Google Scholar] [CrossRef]
- Cosenza-Contreras, M.; Schafer, A.; Sing, J.; Cook, L.; Stillger, M.N.; Chen, C.Y.; Hidalgo, J.V.; Pinter, N.; Meyer, L.; Werner, T.; et al. Proteometabolomics of initial and recurrent glioblastoma highlights an increased immune cell signature with altered lipid metabolism. Neuro Oncol. 2023, noad208. [Google Scholar] [CrossRef]
- Wang, L.B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 2021, 39, 509–528.e520. [Google Scholar] [CrossRef]
- Kesarwani, P.; Prabhu, A.; Kant, S.; Chinnaiyan, P. Metabolic remodeling contributes towards an immune-suppressive phenotype in glioblastoma. Cancer Immunol. Immunother. 2019, 68, 1107–1120. [Google Scholar] [CrossRef] [PubMed]



| Patient Study ID | MGMT Promoter Status | Gender | Age at Diagnosis | Collection Date (Days Relevant to Diagnosis) | Disease Status at Collection | Treatment | RT End Date (Days Relevant to Collection) | Overall Survival (Days) |
|---|---|---|---|---|---|---|---|---|
| CDSR-01276-CNS | Unmethylated | Male | 43 | 0 | De Novo | Resection/RT/ Temozolomide | 336 | |
| CDSR-01276-CNS | Male | 206 | Progression | Reresection | 151 | |||
| CDSR-01396-CNS | Methylated | Female | 62 | 0 | De Novo | Resection/RT/ Temozolomide | 564 | |
| CDSR-01396-CNS | Female | 358 | Progression | Needle Biopsy | 36 | |||
| CDSR-01595-CNS | Unmethylated | Male | 68 | 0 | De Novo | Resection/ Lomustine | 148 | |
| CDSR-01595-CNS | Male | 77 | Progression | Reresection | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mistry, A.M.; Daneshmand, J.; Seo, S.J.; Lehman, N.L.; Miller, D.M.; Goodin, D.A.; Frieboes, H.B.; Chen, J.; Masters, A.; Williams, B.J.; et al. Spatially Resolved Microglia/Macrophages in Recurrent Glioblastomas Overexpress Fatty Acid Metabolism and Phagocytic Genes. Curr. Oncol. 2024, 31, 1183-1194. https://doi.org/10.3390/curroncol31030088
Mistry AM, Daneshmand J, Seo SJ, Lehman NL, Miller DM, Goodin DA, Frieboes HB, Chen J, Masters A, Williams BJ, et al. Spatially Resolved Microglia/Macrophages in Recurrent Glioblastomas Overexpress Fatty Acid Metabolism and Phagocytic Genes. Current Oncology. 2024; 31(3):1183-1194. https://doi.org/10.3390/curroncol31030088
Chicago/Turabian StyleMistry, Akshitkumar M., Jonah Daneshmand, SeonYeong Jamie Seo, Norman L. Lehman, Donald M. Miller, Dylan A. Goodin, Hermann B. Frieboes, Joseph Chen, Adrianna Masters, Brian J. Williams, and et al. 2024. "Spatially Resolved Microglia/Macrophages in Recurrent Glioblastomas Overexpress Fatty Acid Metabolism and Phagocytic Genes" Current Oncology 31, no. 3: 1183-1194. https://doi.org/10.3390/curroncol31030088
APA StyleMistry, A. M., Daneshmand, J., Seo, S. J., Lehman, N. L., Miller, D. M., Goodin, D. A., Frieboes, H. B., Chen, J., Masters, A., Williams, B. J., & Yaddanapudi, K. (2024). Spatially Resolved Microglia/Macrophages in Recurrent Glioblastomas Overexpress Fatty Acid Metabolism and Phagocytic Genes. Current Oncology, 31(3), 1183-1194. https://doi.org/10.3390/curroncol31030088






