Immune-Related Long Non-Coding RNA Signatures for Tongue Squamous Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
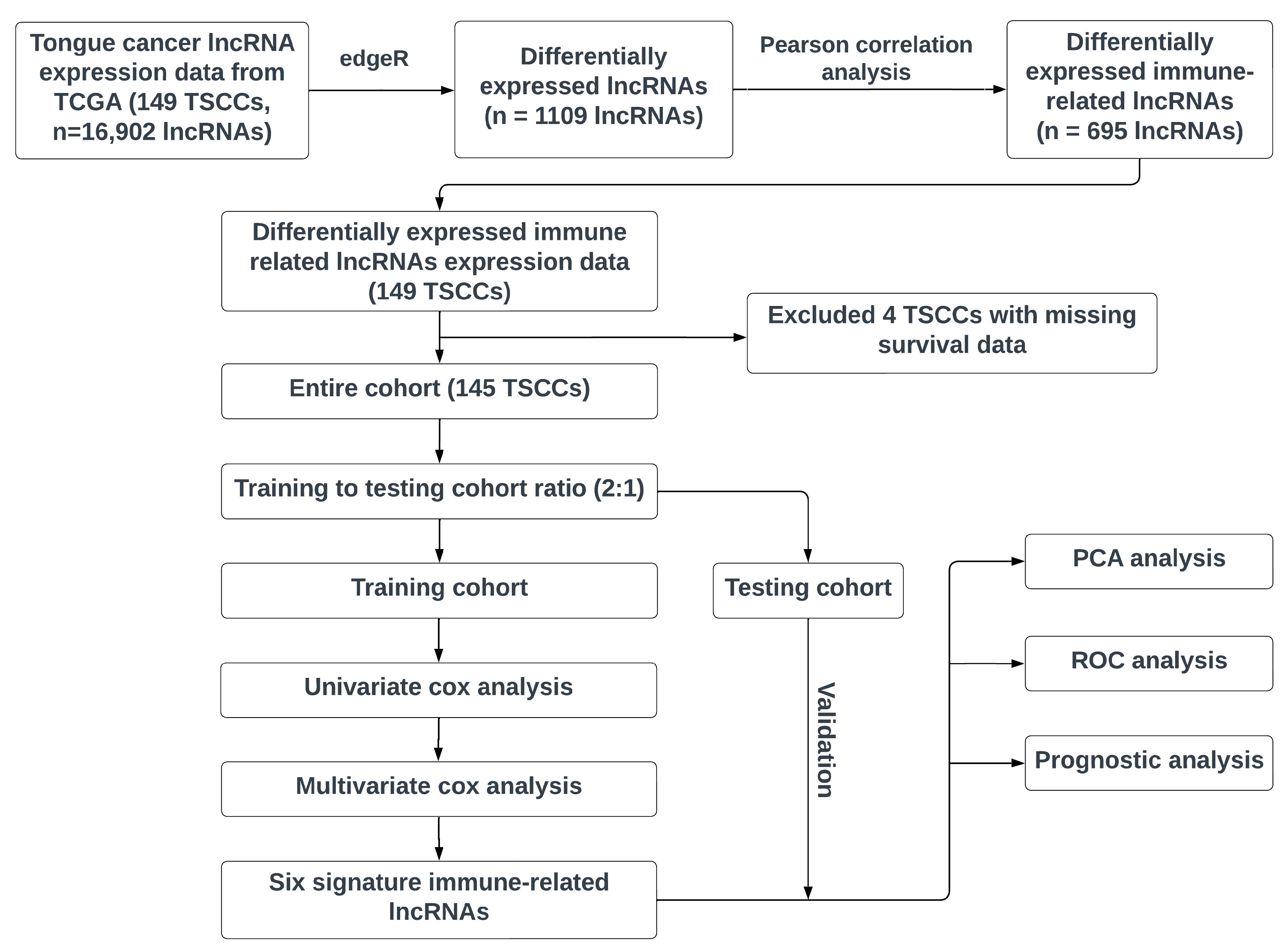
2.1. Workflow
2.2. Data Resource
2.3. Immune-Related lncRNAs
2.4. Construction of the Immune-Related lncRNA Prognostic Model
2.5. Application and Validation of the Risk Score Model
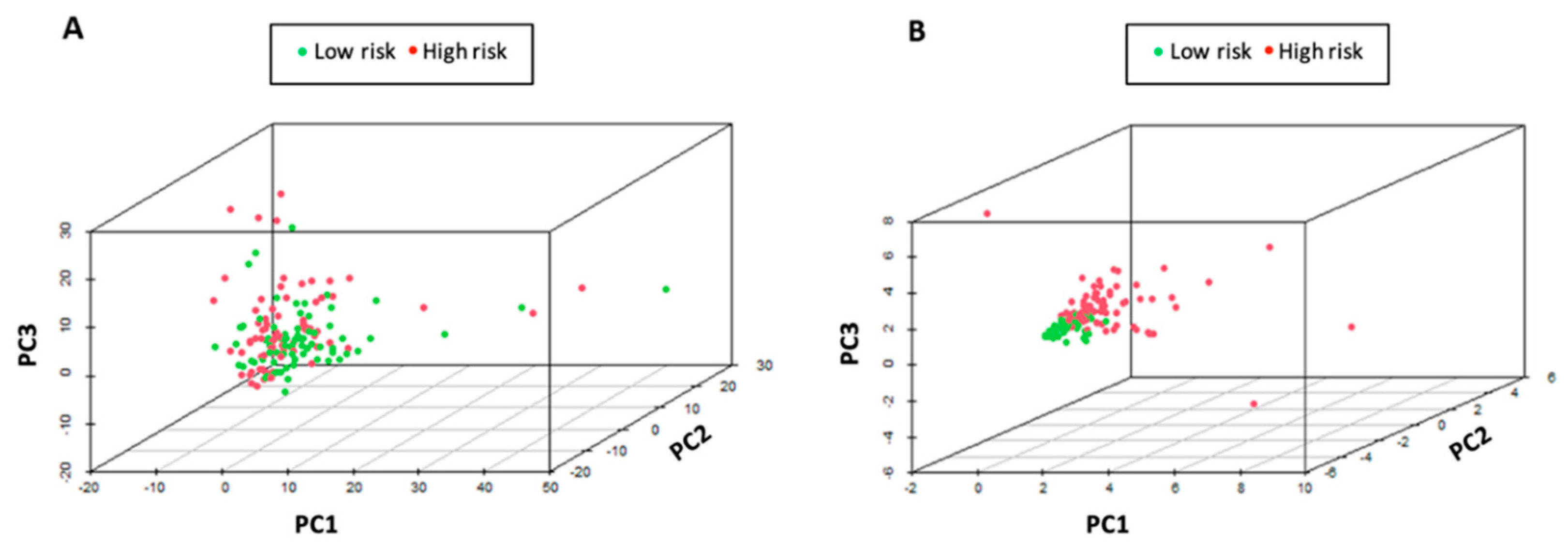
2.6. Principal Components Analysis (PCA)
3. Results
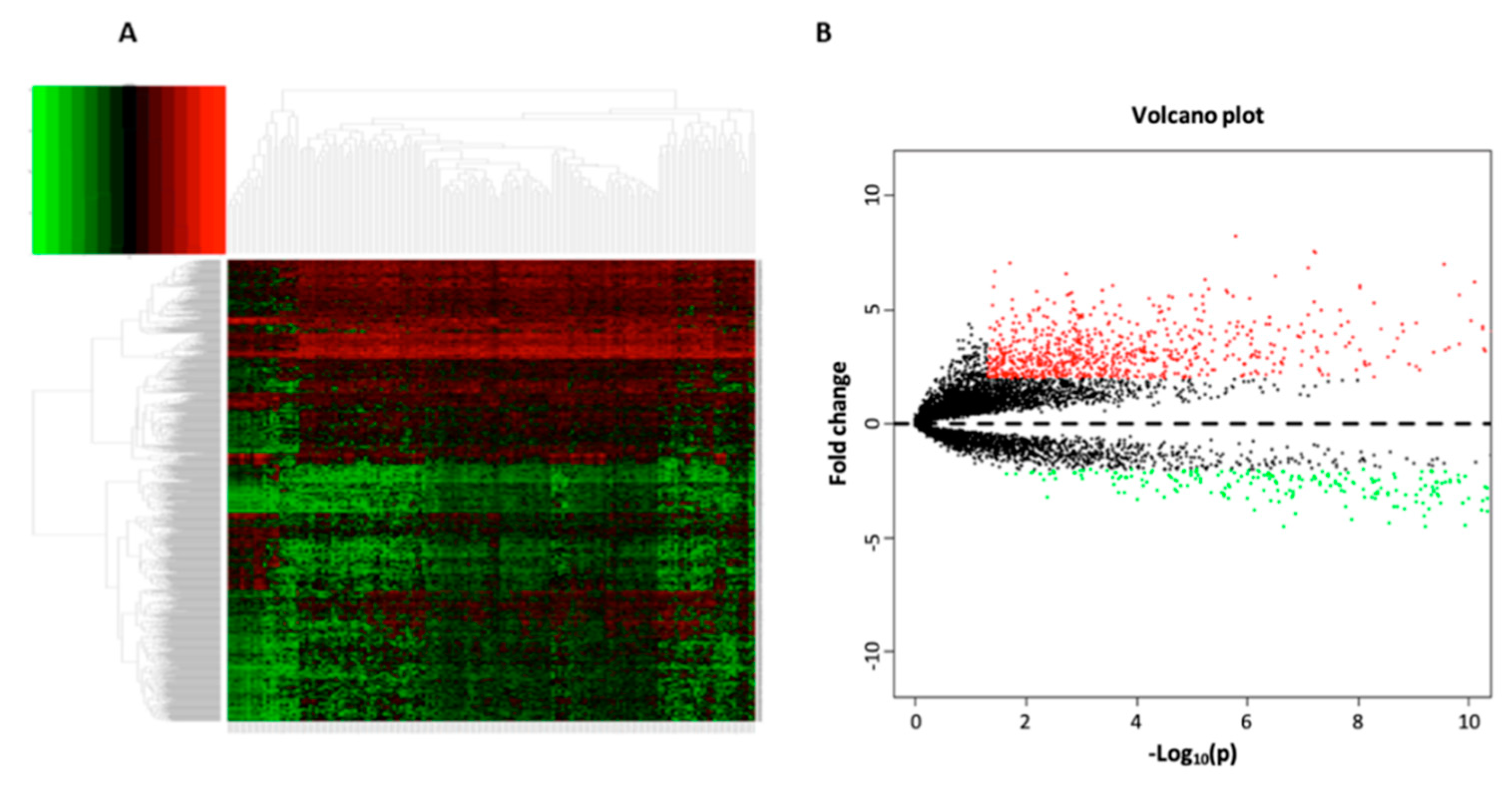
3.1. Data Processing and Differential Expression Analysis
3.2. Identification of Immune-Related Signature lncRNAs and Construction of Six-lncRNA Prognostic Model
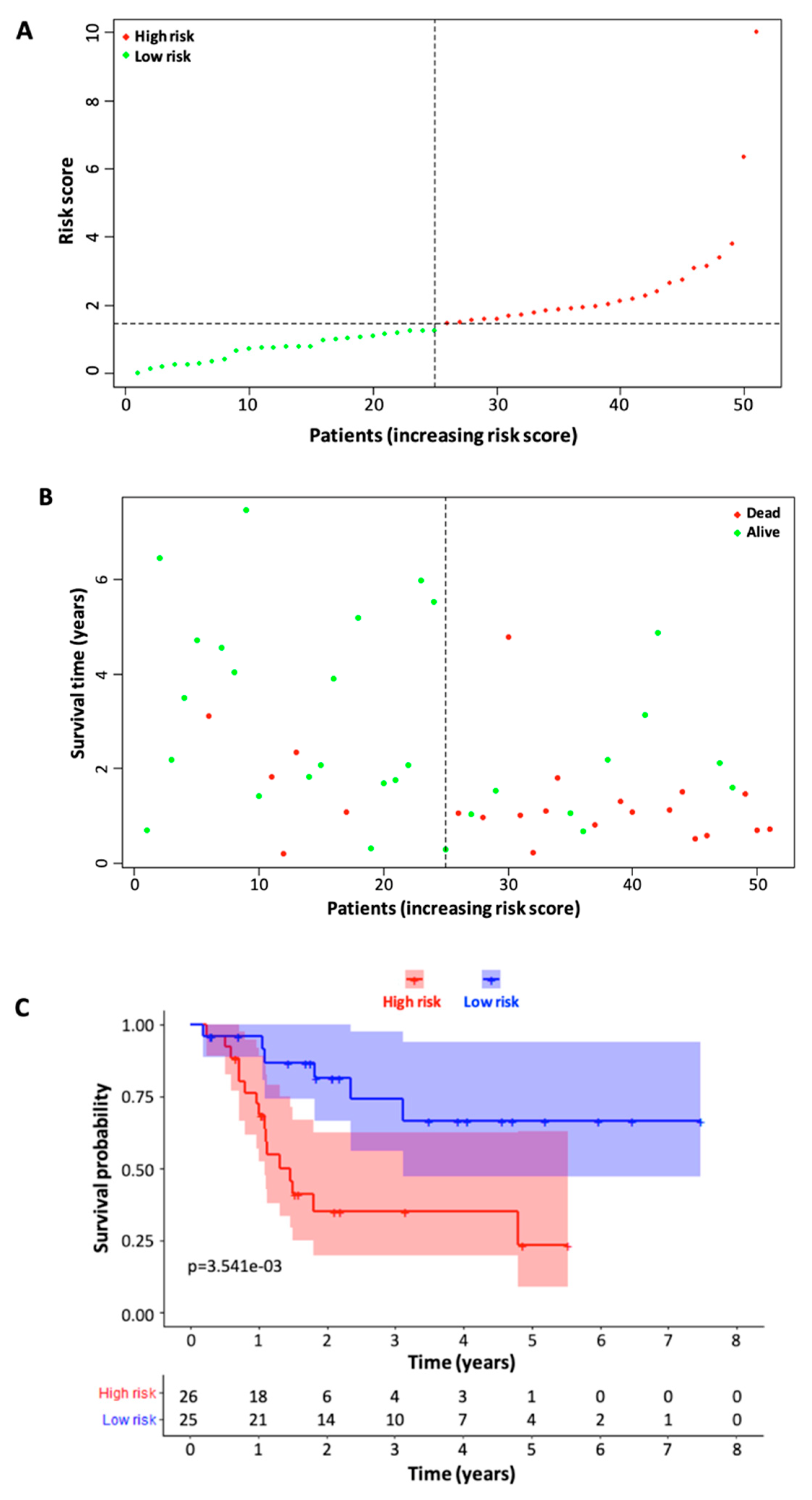
3.3. Validation of Six Immune-Related lncRNA Prognostic Model
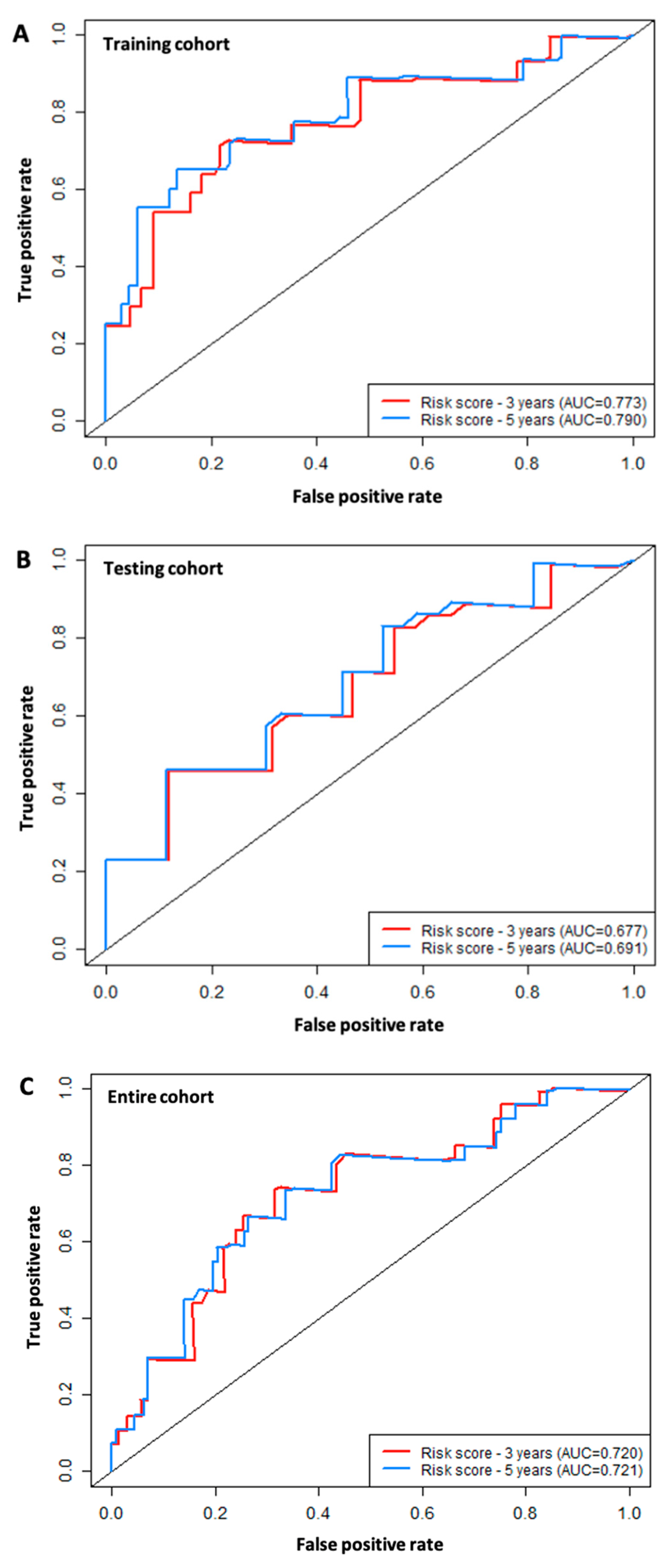
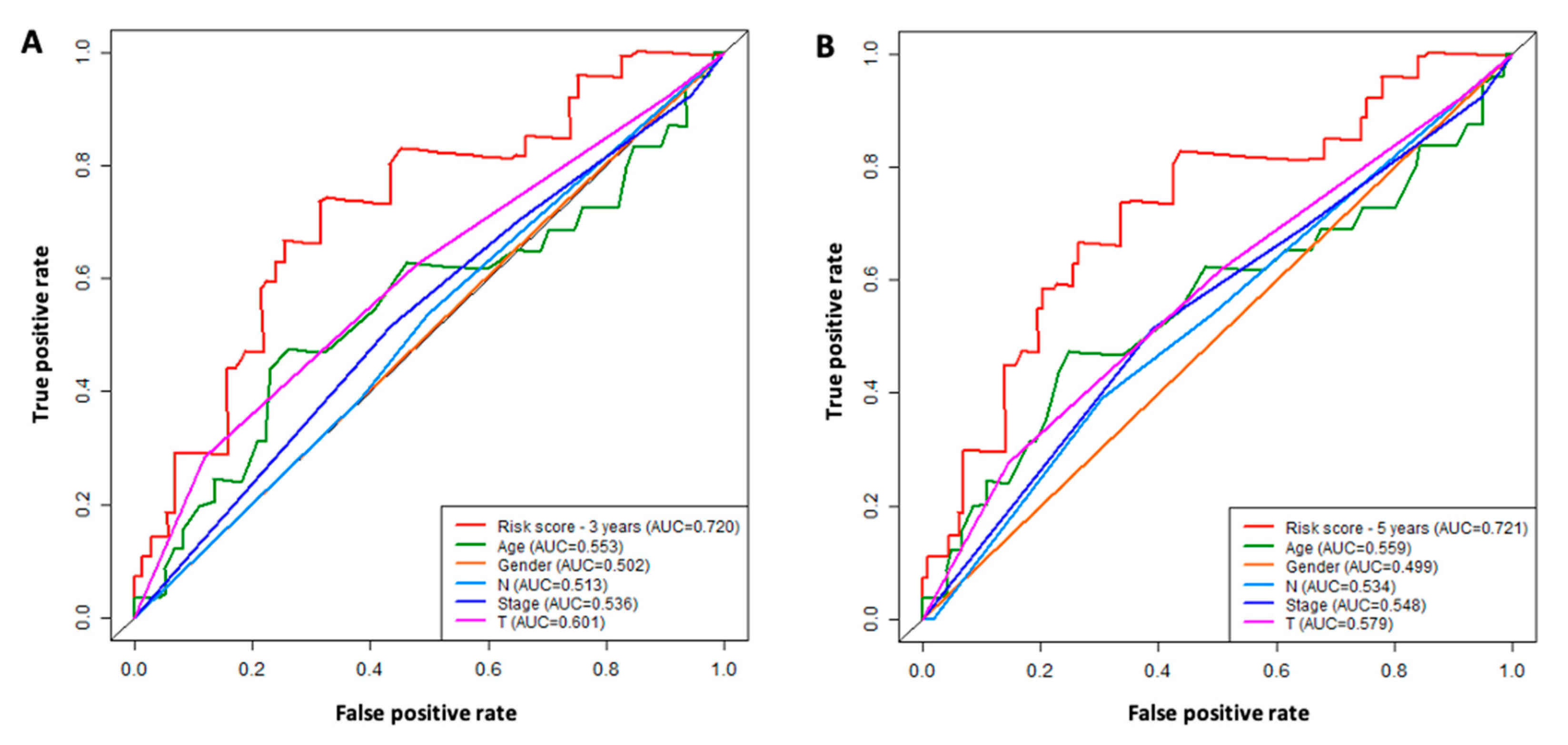
3.4. ROC Analysis
3.5. Comparison of the Six-lncRNA Model with Other Clinicopathological Parameters in TSCC
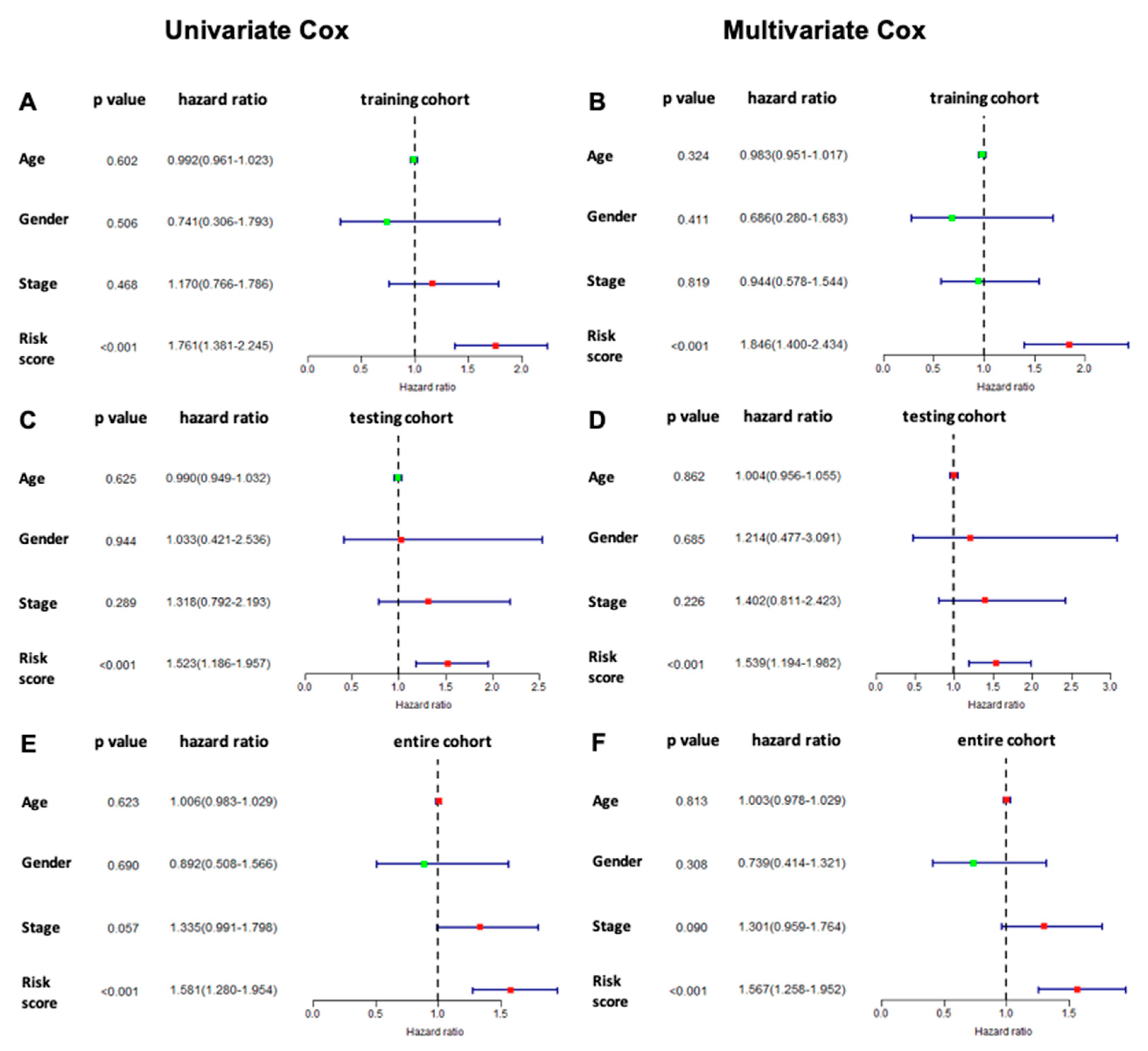
3.6. Prognostic Value of the Six Immune-Related lncRNA Model
3.7. Immune Status Associated with the Six Signature lncRNAs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Almangush, A.; Heikkinen, I.; Mäkitie, A.A.; Coletta, R.D.; Läärä, E.; Leivo, I.; Salo, T. Prognostic biomarkers for oral tongue squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2017, 117, 856–866. [Google Scholar] [CrossRef]
- Bello, I.O.; Soini, Y.; Salo, T. Prognostic evaluation of oral tongue cancer: Means, markers and perspectives (I). Oral Oncol. 2010, 46, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.-H.; Kim, M.G.; Park, J.Y.; Lee, J.H.; Kim, M.J.; Myoung, H.; Choi, S.W. Analysis of the outcome of young age tongue squamous cell carcinoma. Maxillofac. Plast. Reconstr. Surg. 2017, 39, 41. [Google Scholar] [CrossRef] [PubMed]
- Garavello, W.; Spreafico, R.; Gaini, R.M. Oral tongue cancer in young patients: A matched analysis. Oral Oncol. 2007, 43, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Vered, M.; Dayan, D.; Dobriyan, A.; Yahalom, R.; Shalmon, B.; Barshack, I.; Bedrin, L.; Talmi, Y.P.; Taicher, S. Oral tongue squamous cell carcinoma: Recurrent disease is associated with histopathologic risk score and young age. J. Cancer Res. Clin. Oncol. 2010, 136, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Moon, E.K.; Park, J.Y.; Jung, K.W.; Oh, C.M.; Kong, H.J.; Won, Y.J. Trends in the incidence of and survival rates for oral cavity cancer in the Korean population. Oral Dis. 2014, 20, 773–779. [Google Scholar] [CrossRef]
- Poling, J.S.; Ma, X.J.; Bui, S.; Luo, Y.; Li, R.; Koch, W.M.; Westra, W.H. Human papillomavirus (HPV) status of non-tobacco related squamous cell carcinomas of the lateral tongue. Oral Oncol. 2014, 50, 306–310. [Google Scholar] [CrossRef]
- National Cancer Institute. Cancer Stat Facts: Tongue Cancer. Available online: https://seer.cancer.gov/statfacts/html/tongue.html (accessed on 7 January 2023).
- Almangush, A.; Bello, I.O.; Keski–Säntti, H.; Mäkinen, L.K.; Kauppila, J.H.; Pukkila, M.; Hagström, J.; Laranne, J.; Tommola, S.; Nieminen, O.; et al. Depth of invasion, tumor budding, and worst pattern of invasion: Prognostic indicators in early-stage oral tongue cancer. Head Neck 2014, 36, 811–818. [Google Scholar] [CrossRef]
- Li, G.; Li, X.; Yang, M.; Xu, L.; Deng, S.; Ran, L. Prediction of biomarkers of oral squamous cell carcinoma using microarray technology. Sci. Rep. 2017, 7, 42105. [Google Scholar] [CrossRef]
- Yang, M.; Xiong, X.; Chen, L.; Yang, L.; Li, X. Identification and validation long non-coding RNAs of oral squamous cell carcinoma by bioinformatics method. Oncotarget 2017, 8, 107469–107476. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Duncan, L.; Shay, C.; Teng, Y. Multifaceted Roles of Long Non-Coding RNAs in Head and Neck Cancer. Adv. Exp. Med. Biol. 2021, 1286, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gong, Z.; Ma, L.; Wang, Q. lncRNA RPSAP52 induced the development of tongue squamous cell carcinomas via miR-423-5p/MYBL2. J. Cell. Mol. Med. 2021, 25, 4744–4752. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Li, H.; An, J.; Wu, M.; Zheng, Q.; Gui, X.; Li, T.; Pu, H.; Lu, D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015, 6, 27847–27864. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, F.; Zhao, M.; Yang, Z.; Li, J.; Zhang, S.; Zhang, W.; Ye, L.; Zhang, X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J. Biol. Chem. 2017, 292, 15395–15407. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef]
- De Martino, M.; Esposito, F.; Pallante, P. Long non-coding RNAs regulating multiple proliferative pathways in cancer cell. Transl. Cancer Res. 2021, 10, 3140–3157. [Google Scholar] [CrossRef]
- Xie, S.-J.; Diao, L.-T.; Cai, N.; Zhang, L.-T.; Xiang, S.; Jia, C.-C.; Qiu, D.-B.; Liu, C.; Sun, Y.-J.; Lei, H.; et al. mascRNA and its parent lncRNA MALAT1 promote proliferation and metastasis of hepatocellular carcinoma cells by activating ERK/MAPK signaling pathway. Cell Death Discov. 2021, 7, 110. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef]
- Özeş, A.R.; Wang, Y.; Zong, X.; Fang, F.; Pilrose, J.; Nephew, K.P. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 2017, 7, 894. [Google Scholar] [CrossRef]
- Xu, F.; Zhan, X.; Zheng, X.; Xu, H.; Li, Y.; Huang, X.; Lin, L.; Chen, Y. A signature of immune-related gene pairs predicts oncologic outcomes and response to immunotherapy in lung adenocarcinoma. Genomics 2020, 112, 4675–4683. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Qiang, W.; Lin, K.; Gui, Y.; Lan, X.; Wang, D. An immune-related gene signature for predicting survival and immunotherapy efficacy in hepatocellular carcinoma. Cancer Immunol. Immunother. 2021, 70, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Peng, X.; Shen, C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics 2020, 112, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, Y.; Huang, W. Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int. Immunopharmacol. 2021, 92, 107333. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Ren, F.; Chu, Y.; Cui, B. Identification and Validation of a Four-Long Non-Coding RNA Signature Associated with Immune Infiltration and Prognosis in Colon Cancer. Front. Genet. 2021, 12, 671128. [Google Scholar] [CrossRef]
- Tang, C.; Qu, G.; Xu, Y.; Yang, G.; Wang, J.; Xiang, M. An immune-related lncRNA risk coefficient model to predict the outcomes in clear cell renal cell carcinoma. Aging 2021, 13, 26046–26062. [Google Scholar] [CrossRef]
- Miao, T.; Si, Q.; Wei, Y.; Fan, R.; Wang, J.; An, X. Identification and validation of seven prognostic long non-coding RNAs in oral squamous cell carcinoma. Oncol. Lett. 2020, 20, 939–946. [Google Scholar] [CrossRef]
- Jia, B.; Zheng, X.; Qiu, X.; Jiang, X.; Liu, J.; Huang, Z.; Xiang, S.; Chen, G.; Zhao, J. Long non-coding RNA MIR4713HG aggravates malignant behaviors in oral tongue squamous cell carcinoma via binding with microRNA let-7c-5p. Int. J. Mol. Med. 2021, 47, 84. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, Y.; Li, X.; Lou, M.; Gao, Z.; Tong, J.; Yuan, K. Long Non-Coding RNA AL513318.2 as ceRNA Binding to hsa-miR-26a-5p Upregulates SLC6A8 Expression and Predicts Poor Prognosis in Non-Small Lung Cancer. Front. Oncol. 2022, 12, 781903. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, X.; Jiao, D.; Zhu, J.; Xiao, H.; Yang, Y.; Zhao, S.; Zhang, J.; Jiao, F.; Liu, Z. Analysis of the expression profile of serum exosomal lncRNA in breast cancer patients. Ann. Transl. Med. 2021, 9, 1382. [Google Scholar] [CrossRef]
- Hu, J.; Xu, L.; Shou, T.; Chen, Q. Systematic analysis identifies three-lncRNA signature as a potentially prognostic biomarker for lung squamous cell carcinoma using bioinformatics strategy. Transl. Lung Cancer Res. 2019, 8, 614–635. [Google Scholar] [CrossRef]
- Fan, C.N.; Ma, L.; Liu, N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J. Transl. Med. 2018, 16, 264. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, T.; Chen, Q. Competitive endogenous RNA network identifies four long non-coding RNA signature as a candidate prognostic biomarker for lung adenocarcinoma. Transl Cancer Res 2019, 8, 1046–1064. [Google Scholar] [CrossRef] [PubMed]
- Boerrigter, E.; Benoist, G.E.; van Oort, I.M.; Verhaegh, G.W.; de Haan, A.F.J.; van Hooij, O.; Groen, L.; Smit, F.; Oving, I.M.; de Mol, P.; et al. RNA Biomarkers as a Response Measure for Survival in Patients with Metastatic Castration-Resistant Prostate Cancer. Cancers 2021, 13, 6279. [Google Scholar] [CrossRef]
- Groen, L.; Yurevych, V.; Ramu, H.; Chen, J.; Steenge, L.; Boer, S.; Kuiper, R.; Smit, F.P.; Verhaegh, G.W.; Mehra, N.; et al. The Androgen Regulated lncRNA NAALADL2-AS2 Promotes Tumor Cell Survival in Prostate Cancer. Non-Coding RNA 2022, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.; Parameswaran, S.; Donmez, O.A.; Miller, D.; Forney, C.; Lape, M.; Saint Just Ribeiro, M.; Liang, J.; Edsall, L.E.; Magnusen, A.F.; et al. Epstein-Barr virus nuclear antigen 2 extensively rewires the human chromatin landscape at autoimmune risk loci. Genome Res. 2021, 31, 2185–2198. [Google Scholar] [CrossRef] [PubMed]

| Variable | Case (n) |
|---|---|
| Age | |
| ≤53 | 48 |
| >53 | 100 |
| N/A | 1 |
| Gender | |
| Male | 104 |
| Female | 45 |
| T stage | |
| T1 + T2 | 68 |
| T3 + T4 | 74 |
| TX or N/A | 7 |
| Lymph Node Status | |
| N0 | 71 |
| N1–3 | 70 |
| NX or N/A | 8 |
| Metastasis | |
| M0 | 136 |
| M1 | 1 |
| MX or N/A | 12 |
| Stage | |
| Stage I + Stage II | 49 |
| Stage III + Stage IV | 93 |
| N/A | 7 |
| ID | Hazard Ratio | HR.95L | HR.95H | p Value |
|---|---|---|---|---|
| AC008147.2 | 1.618197807 | 1.159476115 | 2.258402832 | 0.0047 |
| MIR4713HG | 1.465332135 | 1.096913289 | 1.957491341 | 0.0097 |
| AC009226.1 | 1.502288429 | 1.160577329 | 1.944610211 | 0.0020 |
| AC079160.1 | 1.212107231 | 1.058402392 | 1.388133615 | 0.0054 |
| AC104088.1 | 1.361078975 | 1.098591619 | 1.686282639 | 0.0048 |
| LINC00534 | 1.322660403 | 1.108148844 | 1.578696356 | 0.0020 |
| NAALADL2-AS2 | 1.243322929 | 1.075414330 | 1.437447748 | 0.0033 |
| AC003092.1 | 1.336654339 | 1.078292842 | 1.656919858 | 0.0081 |
| AC083967.1 | 1.650994013 | 1.349623551 | 2.019660393 | <0.0001 |
| FNDC1-IT1 | 1.662942115 | 1.171056335 | 2.361437613 | 0.0045 |
| ID | Coefficient | Hazard Ratio | HR.95L | HR.95H | p Value |
|---|---|---|---|---|---|
| MIR4713HG | 0.4766 | 1.610596 | 1.128103 | 2.299451 | 0.0087 |
| AC104088.1 | 0.3380 | 1.402013 | 1.104656 | 1.779414 | 0.0055 |
| LINC00534 | 0.1640 | 1.178198 | 0.979253 | 1.417559 | 0.0082 |
| NAALADL2-AS2 | 0.1566 | 1.169497 | 0.987941 | 1.384418 | 0.0069 |
| AC083967.1 | 0.5811 | 1.787962 | 1.418558 | 2.253560 | <0.0001 |
| FNDC1-IT1 | 0.4401 | 1.552811 | 1.066420 | 2.261043 | 0.0217 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, D.; Messadi, D.V. Immune-Related Long Non-Coding RNA Signatures for Tongue Squamous Cell Carcinoma. Curr. Oncol. 2023, 30, 4817-4832. https://doi.org/10.3390/curroncol30050363
Hu D, Messadi DV. Immune-Related Long Non-Coding RNA Signatures for Tongue Squamous Cell Carcinoma. Current Oncology. 2023; 30(5):4817-4832. https://doi.org/10.3390/curroncol30050363
Chicago/Turabian StyleHu, Daniel, and Diana V. Messadi. 2023. "Immune-Related Long Non-Coding RNA Signatures for Tongue Squamous Cell Carcinoma" Current Oncology 30, no. 5: 4817-4832. https://doi.org/10.3390/curroncol30050363
APA StyleHu, D., & Messadi, D. V. (2023). Immune-Related Long Non-Coding RNA Signatures for Tongue Squamous Cell Carcinoma. Current Oncology, 30(5), 4817-4832. https://doi.org/10.3390/curroncol30050363




