Retrospective Assessment of Complementary Liquid Biopsy on Tissue Single-Gene Testing for Tumor Genotyping in Advanced NSCLC
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Mol. Diagn. 2018, 20, 129–159. [Google Scholar]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2022, 20, 497–530. [Google Scholar] [CrossRef] [PubMed]
- Smeltzer, M.P.; Wynes, M.W.; Lantuejoul, S.; Soo, R.; Ramalingam, S.S.; Varella-Garcia, M.; Meadows Taylor, M.; Richeimer, K.; Wood, K.; Howell, K.E.; et al. The International Association for the Study of Lung Cancer Global Survey on Molecular Testing in Lung Cancer. J. Thorac. Oncol. 2020, 15, 1434–1448. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Olsen, S.; Ku, B.M.; Lee, M.S.; Jung, H.A.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; Choi, Y.L.; et al. High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program. Cancer 2021, 127, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.H.; Allison, D.H.R.; Feng, Y.; Jour, G.; Park, K.; Zhou, F.; Moreira, A.L.; Shen, G.; Feng, X.; Sabari, J.; et al. Comparison of solid tissue sequencing and liquid biopsy accuracy in identification of clinically relevant gene mutations and rearrangements in lung adenocarcinomas. Mod. Pathol. 2021, 34, 2168–2174. [Google Scholar] [CrossRef] [PubMed]
- Mack, P.C.; Banks, K.C.; Espenschied, C.R.; Burich, R.A.; Zill, O.A.; Lee, C.E.; Riess, J.W.; Mortimer, S.A.; Talasaz, A.; Lanman, R.B.; et al. Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: Analysis of over 8000 cases. Cancer 2020, 126, 3219–3228. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Schwartzberg, L.S.; Li, G.; Tolba, K.; Bourla, A.B.; Schulze, K.; Gadgil, R.; Fine, A.; Lofgren, K.T.; Graf, R.P.; Oxnard, G.R.; et al. Complementary Roles for Tissue- and Blood-Based Comprehensive Genomic Profiling for Detection of Actionable Driver Alterations in Advanced NSCLC. JTO Clin. Res. Rep. 2022, 3, 100386. [Google Scholar] [CrossRef]
- Desmeules, P.; Boudreau, D.K.; Bastien, N.; Boulanger, M.C.; Bosse, Y.; Joubert, P.; Couture, C. Performance of an RNA-Based Next-Generation Sequencing Assay for Combined Detection of Clinically Actionable Fusions and Hotspot Mutations in NSCLC. JTO Clin. Res. Rep. 2022, 3, 100276. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Sabari, J.K.; Offin, M.; Stephens, D.; Ni, A.; Lee, A.; Pavlakis, N.; Clarke, S.; Diakos, C.I.; Datta, S.; Tandon, N.; et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J. Natl. Cancer Inst. 2019, 111, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.C.; Aggarwal, C.; Wong, J.; Nimgaonkar, V.; Hwang, W.T.; Andronov, M.; Dibardino, D.M.; Hutchinson, C.T.; Ma, K.C.; Lanfranco, A.; et al. Plasma Genotyping at the Time of Diagnostic Tissue Biopsy Decreases Time-to-Treatment in Patients With Advanced NSCLC-Results From a Prospective Pilot Study. JTO Clin. Res. Rep. 2022, 3, 100301. [Google Scholar] [CrossRef] [PubMed]
- Francaviglia, I.; Magliacane, G.; Lazzari, C.; Grassini, G.; Brunetto, E.; Dal Cin, E.; Girlando, S.; Medicina, D.; Smart, C.E.; Bulotta, A.; et al. Identification and monitoring of somatic mutations in circulating cell-free tumor DNA in lung cancer patients. Lung Cancer 2019, 134, 225–232. [Google Scholar] [CrossRef]
- Garcia-Pardo, M.; Czarnecka, K.; Law, J.H.; Salvarrey, A.; Fernandes, R.; Fan, J.; Corke, L.; Waddell, T.K.; Yasufuku, K.; Donahoe, L.L.; et al. Plasma-first: Accelerating lung cancer diagnosis and molecular profiling through liquid biopsy. Ther. Adv. Med. Oncol. 2022, 14, 17588359221126151. [Google Scholar] [CrossRef]
- Low, S.K.; Ariyasu, R.; Uchibori, K.; Hayashi, R.; Chan, H.T.; Chin, Y.M.; Akita, T.; Harutani, Y.; Kiritani, A.; Tsugitomi, R.; et al. Rapid genomic profiling of circulating tumor DNA in non-small cell lung cancer using Oncomine Precision Assay with Genexus integrated sequencer. Transl. Lung Cancer Res. 2022, 11, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Mondaca, S.; Lebow, E.S.; Namakydoust, A.; Razavi, P.; Reis-Filho, J.S.; Shen, R.; Offin, M.; Tu, H.Y.; Murciano-Goroff, Y.; Xu, C.; et al. Clinical utility of next-generation sequencing-based ctDNA testing for common and novel ALK fusions. Lung Cancer 2021, 159, 66–73. [Google Scholar] [CrossRef]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef] [PubMed]
- Moroz, I.; Monika, S.D. A Blueprint for Optimizing Lung Cancer Care for Better Patient Outcomes; The Conference Board of Canada: Ottawa, ON, Canada, 2021. [Google Scholar]
- Cheema, P.K.; Menjak, I.B.; Winterton-Perks, Z.; Raphael, S.; Cheng, S.Y.; Verma, S.; Muinuddin, A.; Freedman, R.; Toor, N.; Perera, J.; et al. Impact of Reflex EGFR/ ALK Testing on Time to Treatment of Patients With Advanced Nonsquamous Non-Small-Cell Lung Cancer. J. Oncol. Pract. 2017, 13, e130–e138. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Tsao, M.S.; Le, L.W.; Shepherd, F.A.; Feld, R.; Burkes, R.L.; Liu, G.; Kamel-Reid, S.; Hwang, D.; Tanguay, J.; et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 2015, 26, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Chan, J.M.; Kubota, D.; Sato, H.; Rizvi, H.; Daneshbod, Y.; Chang, J.C.; Paik, P.K.; Offin, M.; Arcila, M.E.; et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin. Cancer Res. 2020, 26, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- Offin, M.; Somwar, R.; Rekhtman, N.; Benayed, R.; Chang, J.C.; Plodkowski, A.; Lui, A.J.W.; Eng, J.; Rosenblum, M.; Li, B.T.; et al. Acquired ALK and RET Gene Fusions as Mechanisms of Resistance to Osimertinib in EGFR-Mutant Lung Cancers. JCO Precis. Oncol. 2018, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
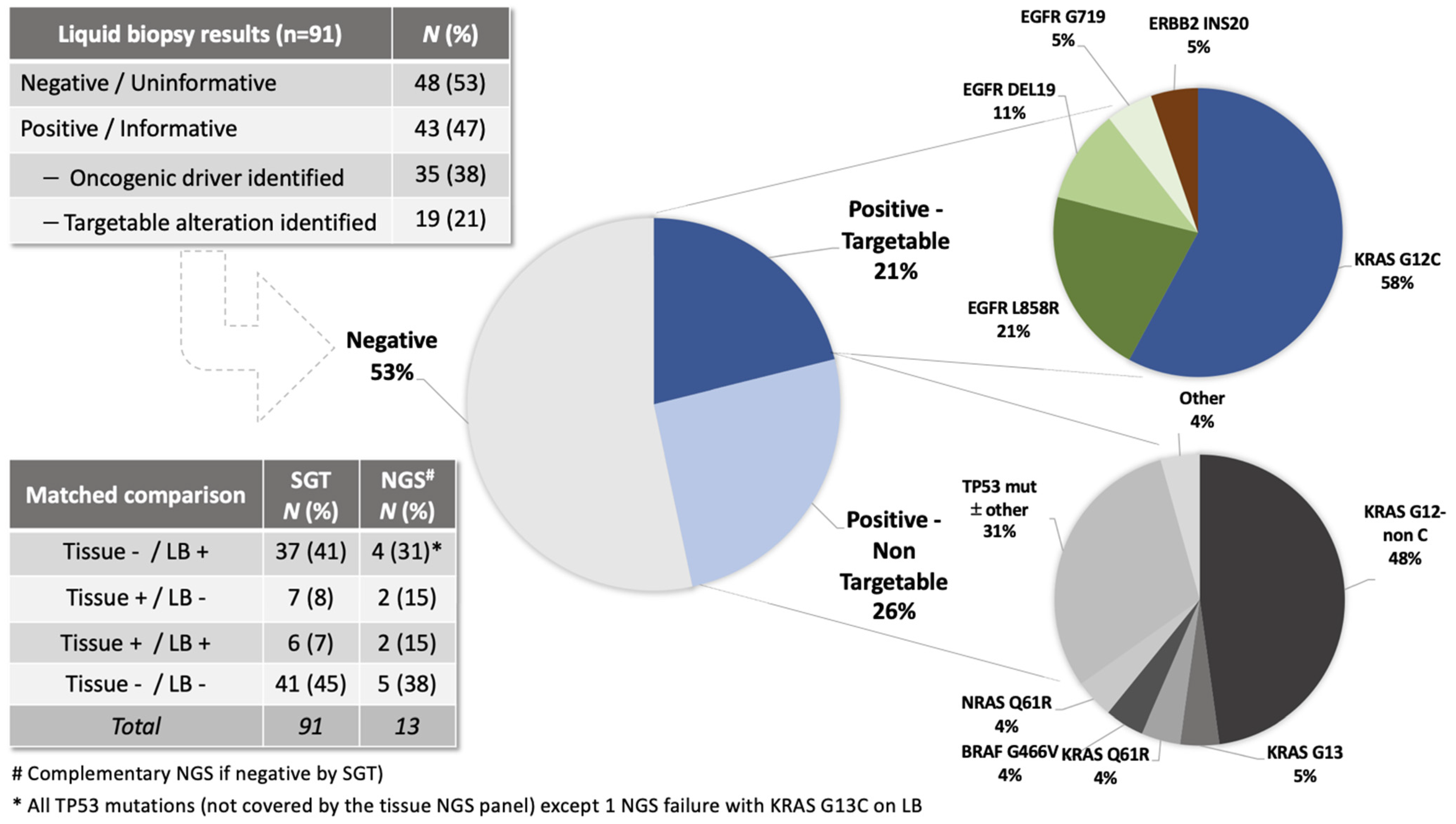
| Clinical and Pathological Characteristics | N (%) |
|---|---|
| Patients with liquid biopsy | 91 |
| Median age (range, y) | 66 (27–83) |
| Sex | |
| Male | 37 (41) |
| Female | 54 (59) |
| Histology | |
| Adenocarcinoma | 60 (66) |
| Non-small cell lung carcinoma, NOS | 29 (32) |
| Squamous cell carcinoma | 2 (2) |
| Stage at blood draw | |
| IIIB/C | 4 (4) |
| IV | 84 (92) |
| N/A | 3 (3) |
| Site of metastatic disease | |
| Intra-thoracic | 35 (38) |
| Extra-thoracic | 51 (56) |
| Not available | 5 (5) |
| Lines of treatment completed at time of liquid biopsy | |
| None | 27 (30) |
| 1 | 37 (41) |
| 2 to 4 | 25 (27) |
| Not available | 2 (2) |
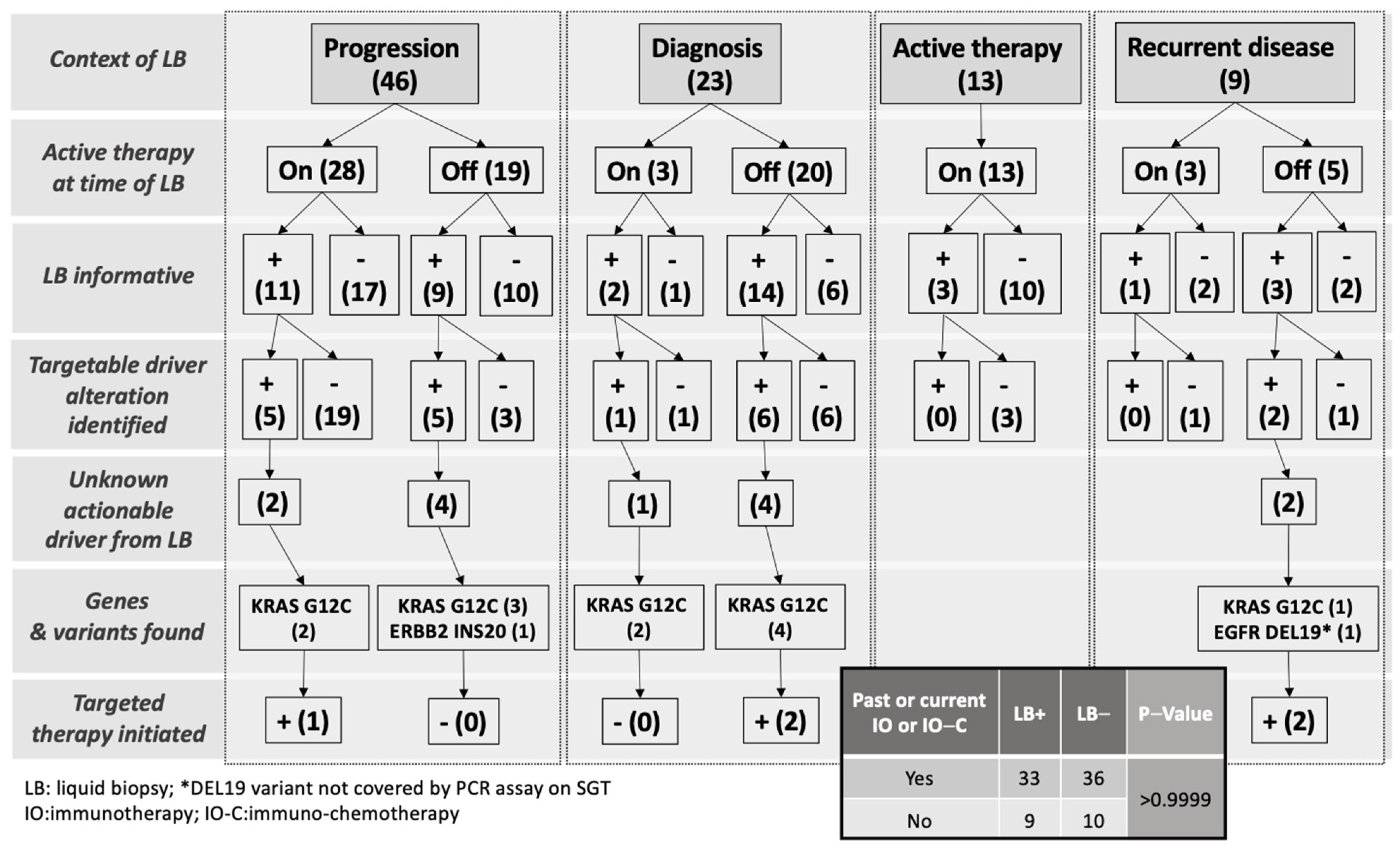
| Clinical context at time of liquid biopsy | |
| Progression of disease | 46 (51) |
| Diagnosis | 23 (25) |
| Active therapy | 13 (14) |
| Recurrent disease | 9 (10) |
| Tissue biomarker testing performed by single-gene testing (SGT) | |
| EGFR/ALK | 91 (100) |
| EGFR/ALK/PD-L1 | 88 (98) |
| EGFR/ALK/ROS1/PD-L1 | 76 (84) |
| EGFR/ALK/ROS1/BRAFV600/PD-L1 | 42 (46) |
| SGT + complementary NGS panel | 13 (14) |
| Tissue biomarker result at blood draw by single-gene testing | |
| Driver Known and actionable | 13 (14) |
| Driver Unknown | 78 (86) |
| Plasma Positive | Plasma Negative | Detection Rate (%) | Total (n) Evaluable | p-Value | |
|---|---|---|---|---|---|
| All patients | 43 | 48 | 47 | 91 | |
| Tissue biopsy positive * | 6 | 7 | 46 | 91 | >0.9999 |
| Tissue biopsy negative * | 37 | 41 | 47 | ||
| Tissue PD-L1 > 50% | 17 | 24 | 41 | 88 | 0.2930 |
| Tissue PD-L1 50% or less | 22 | 25 | 47 | ||
| No extra-thoracic spread | 11 | 24 | 31 | 86 | 0.0278 |
| Extra-thoracic spread | 29 | 22 | 57 | ||
| On treatment | 16 | 30 | 35 | 90 | 0.0340 |
| Off treatment/naïve to treatment | 26 | 18 | 59 |
| Patient | Tissue Genotype | Plasma Finding | Therapy after BL Finding | Clinical Evolution |
|---|---|---|---|---|
| 1 | Negative | EGFR DEL19 * | Osimertinib | SD |
| 2 | Negative | ERBB2 INS20 | Conventional | DOD |
| 3 | Negative | KRAS G12C | Sotorasib 2nd L | SD |
| 4 | Negative | KRAS G12C | Conventional | SD |
| 5 | Negative | KRAS G12C | Sotorasib 2nd L | Active treatment # |
| 6 | Negative | KRAS G12C | NA | NA |
| 7 | Negative | KRAS G12C | Sotorasib 2nd L | PD |
| 8 | Negative | KRAS G12C | Sotorasib 3rd L | Active treatment # |
| 9 | Negative | KRAS G12C | NA | NA |
| 10 | Negative | KRAS G12C | Conventional | SD |
| 11 | Negative | KRAS G12C | Conventional | PD |
| Patient | Tissue Genotype | Plasma Finding | Therapy 1st L | Clinical Evolution |
|---|---|---|---|---|
| 1 | EGFR L861Q | Negative | Osimertinib | PR |
| 2 | EGFR L858R | Negative | Osimertinib | PR |
| 3 | EGFR DEL19 | Negative | Osimertinib | PR |
| 4 | EGFR DEL19 | Negative | Osimertinib | PD |
| 5 | EGFR DEL19 | Negative | Osimertinib | PR |
| 6 | ALK fusion | Negative | Alectinib | SD |
| 7 | ALK fusion | Negative | Alectinib | PR |
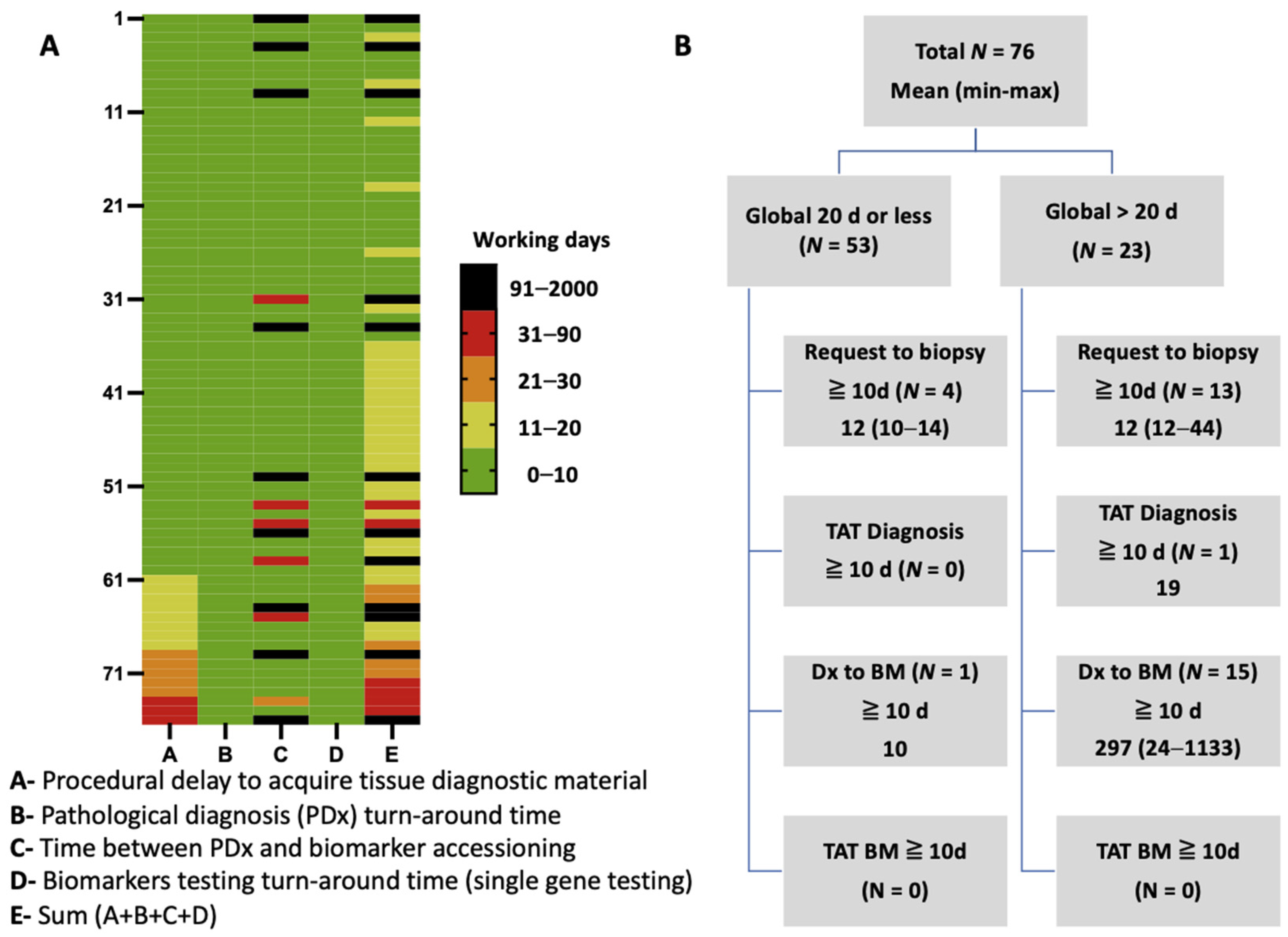
| Delay Category; Days (Median); Range | Entire Cohort (n = 76) | Exclusion of Excessive Delays * (n = 61) | Liquid Biopsy (n = 91) |
|---|---|---|---|
| Procedures to acquire diagnostic material | 7.9 (4); 1–44 | 6.8 (4); 1–35 | – |
| Pathological diagnosis TAT | 2.3 (2); 1–7 | 2.1 (2); 1–5 | – |
| Pathological diagnosis to biomarker request | 60.8 (2); 0–1133 | 3.0 (2); 2–24 | – |
| Biomarker results TAT (single-gene testing) | 2.8 (2); 2–7 | 2.8 (2); 2–7 | – |
| Total trajectory for tissue testing | 73.7 (13) 7–1137 | 14.7 (12); 7–64– | – |
| Liquid biopsy (blood draw to results) | – | – | 10 (10); 6–25 |
| p-value (tissue vs. liquid biopsy) | 0.002 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desmeules, P.; Dusselier, M.; Bouffard, C.; Bafaro, J.; Fortin, M.; Labbé, C.; Joubert, P. Retrospective Assessment of Complementary Liquid Biopsy on Tissue Single-Gene Testing for Tumor Genotyping in Advanced NSCLC. Curr. Oncol. 2023, 30, 575-585. https://doi.org/10.3390/curroncol30010045
Desmeules P, Dusselier M, Bouffard C, Bafaro J, Fortin M, Labbé C, Joubert P. Retrospective Assessment of Complementary Liquid Biopsy on Tissue Single-Gene Testing for Tumor Genotyping in Advanced NSCLC. Current Oncology. 2023; 30(1):575-585. https://doi.org/10.3390/curroncol30010045
Chicago/Turabian StyleDesmeules, Patrice, Matthieu Dusselier, Cédrik Bouffard, Josée Bafaro, Marc Fortin, Catherine Labbé, and Philippe Joubert. 2023. "Retrospective Assessment of Complementary Liquid Biopsy on Tissue Single-Gene Testing for Tumor Genotyping in Advanced NSCLC" Current Oncology 30, no. 1: 575-585. https://doi.org/10.3390/curroncol30010045
APA StyleDesmeules, P., Dusselier, M., Bouffard, C., Bafaro, J., Fortin, M., Labbé, C., & Joubert, P. (2023). Retrospective Assessment of Complementary Liquid Biopsy on Tissue Single-Gene Testing for Tumor Genotyping in Advanced NSCLC. Current Oncology, 30(1), 575-585. https://doi.org/10.3390/curroncol30010045







