Does Molecular Profiling of KRAS-Mutant Non-Squamous Non-Small Cell Lung Cancer (NSCLC) Help in Treatment Strategy Planning?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction from Paraffin Tissue Blocks
2.3. KRAS Mutation at Codons 12 and 13
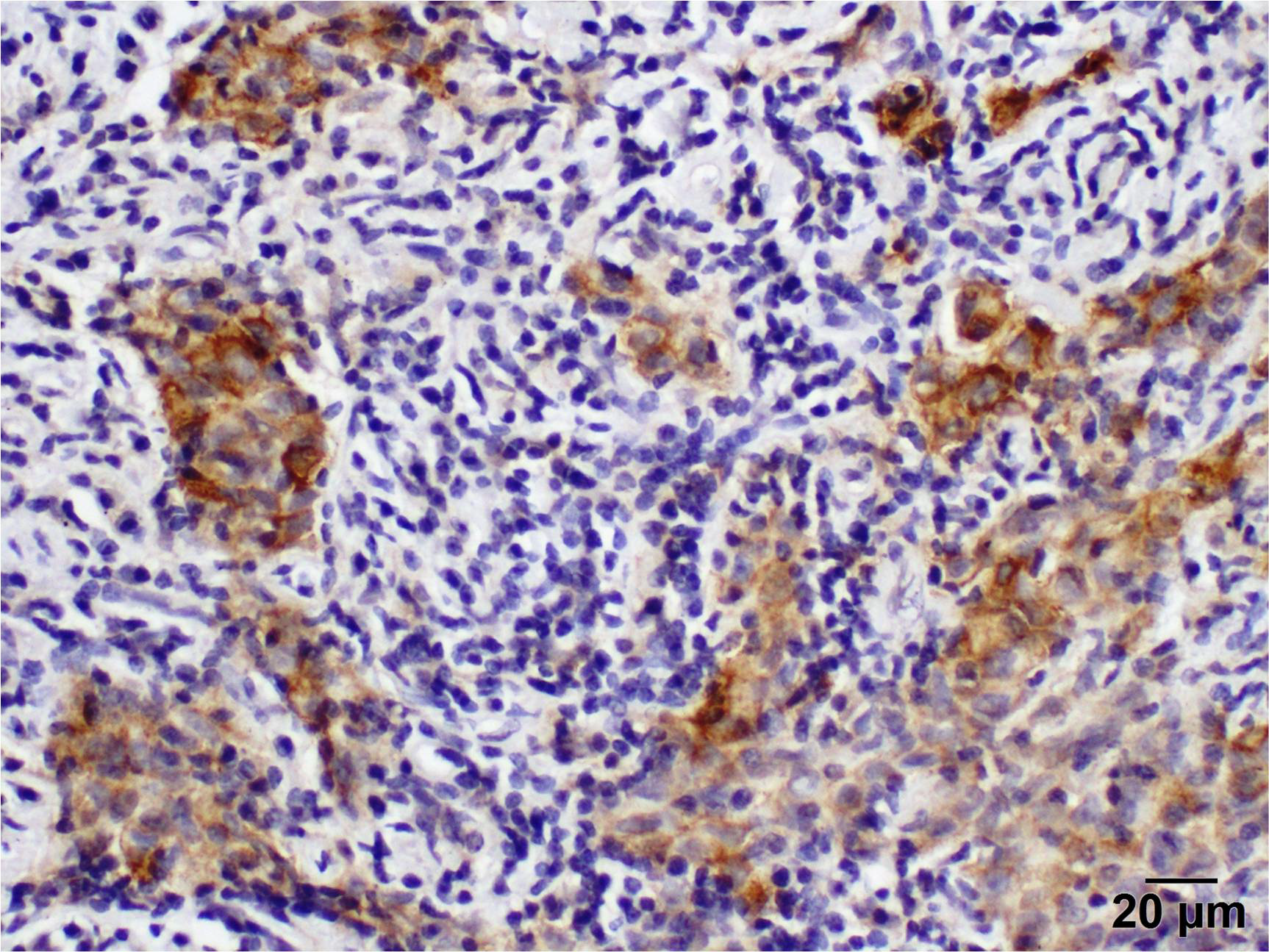
2.4. Immunohistochemistry
2.5. To Evaluate the Level of PD-L1 Expression in the Stroma, the Percentage of Positively
2.6. RNA Extraction from Tissue Blocks
2.7. Quantitative RT-PCR Analysis
2.8. Statistical Analysis
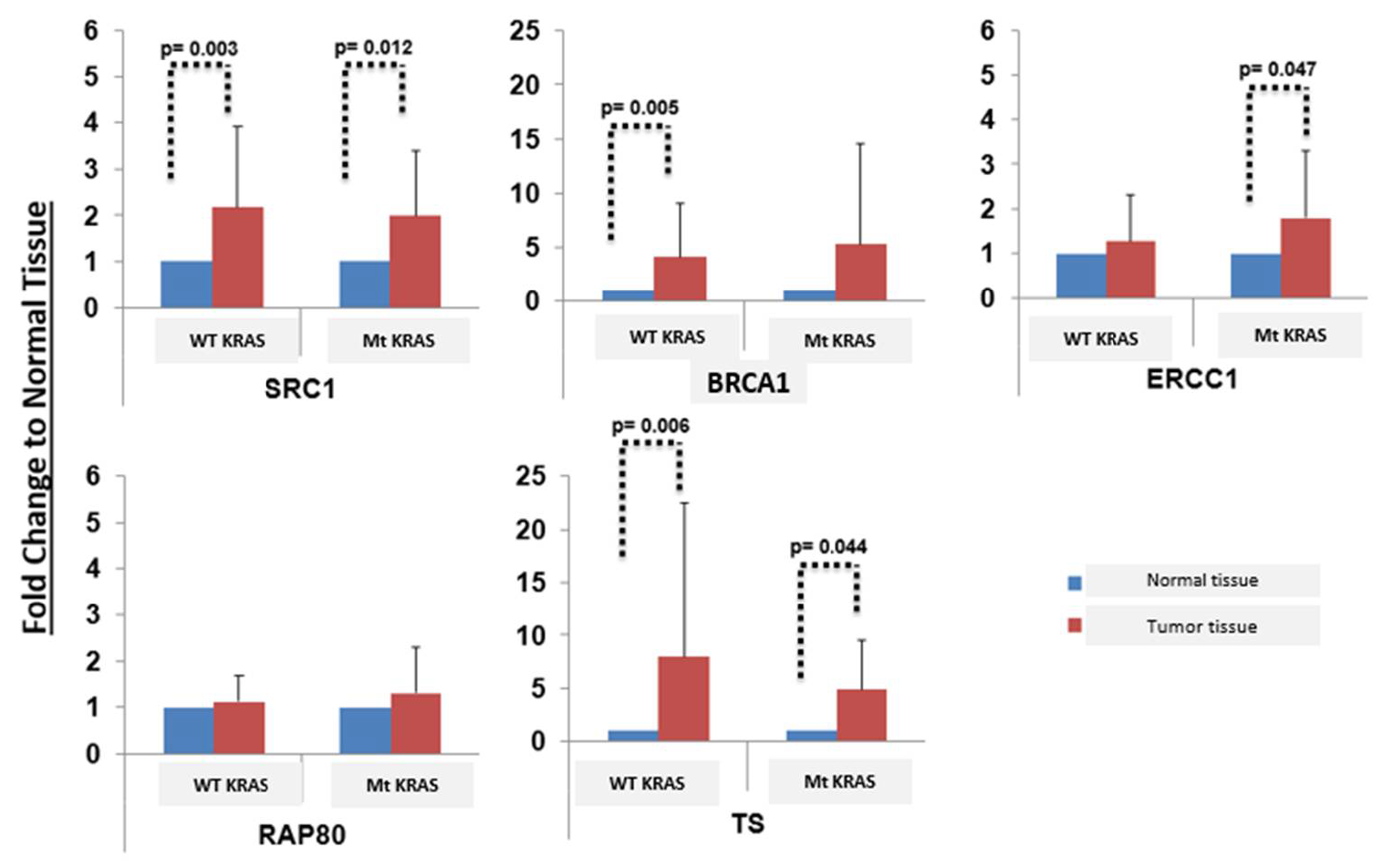
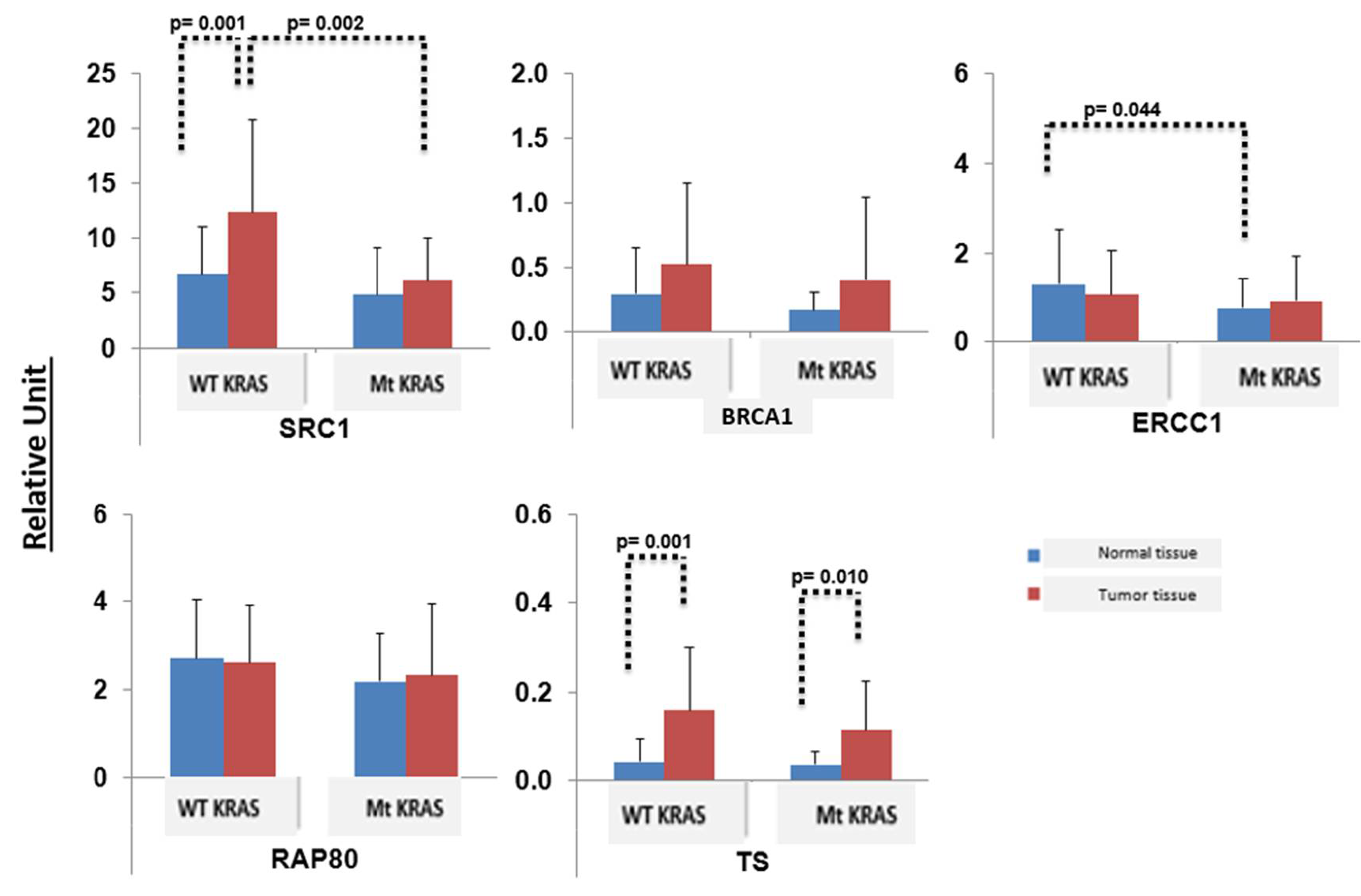
3. Results
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santos, E.; Martin-Zanca, D.; Reddy, E.P.; Pierotti, M.A.; Porta, G.; Barbacid, M. Malignant activation of a K-ras oncogenein ling carcinoma but not in normal tissue of the same patient. Science 1983, 223, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Zugazagoitia, J.; Herbertz, S.; John, W.; Paz-Ares, L.; Schmid-Bindert, G. KRAS-Mutant non-small cell lung cancer: From biology to therapy. Lung Cancer 2018, 124, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Karim, N.A.; Starnes, S.; Morris, J.; Pathrose, P.; Perry, A.; Fathallah, H. Abstract A01: KRAS molecular profiling in non-squamous non-small cell lung cancer (NSCLC). Clin. Cancer Res. 2014, 20 (Suppl. 2), A01. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Socinski, M.A.; Burns, T.F. KRAS mutant lung cancer: Progress thus far on an elusive therapeutic target. Clin. Transl. Med. 2016, 4, 35. [Google Scholar] [CrossRef]
- Zhang, J.; Park, D.; Shin, D.M.; Deng, X. Targeting KRAS-mutant non-small cell lung cancer: Challenges and opportunities. Acta Biochim. Biophys. Sin. 2015, 48, 11–16. [Google Scholar] [CrossRef]
- Riely, G.J.; Marks, J.; Pao, W. KRAS Mutations in Non-Small Cell Lung Cancer. Proc. Am. Thorac. Soc. 2009, 6, 201–205. [Google Scholar] [CrossRef]
- Downward, J. New exchange, new target. Nature 1998, 396, 416–417. [Google Scholar] [CrossRef]
- Shields, J.M.; Pruitt, K.; McFall, A.; Shaub, A.; Der, C.J. Understanding Ras: ‘It ain’t over ’til it’s over’. Trends Cell Biol. 2000, 10, 147–154. [Google Scholar] [CrossRef]
- Govindan, R.; Fakih, M.; Price, T.; Falchook, G.; Desai, J.; Kuo, J.; Strickler, J.; Krauss, J.; Li, B.; Denlinger, C.; et al. Phase I study of safety, tolerability, PK and efficacy of AMG 510, a novel KRAS G12C inhibitor, evaluated in NSCLC. In Proceedings of the IASLC 2019 World Conference on Lung Cancer hosted by the International Association for the Study of Lung Cancer, Barcelona, Spain, 7–10 September 2019. Abstract OA02.02. [Google Scholar]
- de la Fuente, E.C.; Garcia, M.E.O.; Rueda, A.G.; Lage, Y.; Garrido, P. Targeting KRAS in Non-Small Cell Lung Cancer. Front. Oncol. 2022, 11, 792635. [Google Scholar] [CrossRef]
- Friedlaender, A.; Drilon, A.; Weiss, G.J.; Banna, G.L.; Addeo, A. KRAS as a druggable target in NSCLC: Rising like a phoenix after decades of development failures. Cancer Treat. Rev. 2020, 85, 101978. [Google Scholar] [CrossRef]
- Reck, M.; Carbone, D.; Garassino, M.; Barlesi, F. Targeting KRAS in non-small-cell lung cancer: Recent progress and new approaches. Ann. Oncol. 2021, 32, 1101–1110. [Google Scholar] [CrossRef]
- Scheffler, M.; Ihle, M.A.; Hein, R.; Merkelbach-Bruse, S.; Scheel, A.H.; Siemanowski, J.; Brägelmann, J.; Kron, A.; Abedpour, N.; Ueckeroth, F.; et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J. Thorac. Oncol. 2019, 14, 606–616. [Google Scholar] [CrossRef]
- Oh, M.; McBride, A.; Yun, S.; Bhattacharjee, S.; Slack, M.; Martin, J.R.; Jeter, J.; Abraham, I. BRCA1 and BRCA2 Gene Mutations and Colorectal Cancer Risk: Systematic Review and Meta-analysis. JNCI J. Natl. Cancer Inst. 2018, 110, 1178–1189. [Google Scholar] [CrossRef]
- Lee, Y.C.; Lee, Y.C.; Li, C.Y.; Lee, Y.L.; Chen, B.L. BRCA1 and BRCA2 gene mutations and lung cancer risk: A meta-analysis. Medicina 2020, 56, 212. [Google Scholar] [CrossRef]
- Li, S.; Tao, L.; Dai, H.; Gong, X.; Zhuo, Y.; Xiang, H.; Zhao, Y.; Gao, Q.; Deng, L. BRCA1 Versus BRCA2 and PARP Inhibitors Efficacy in Solid Tumors: A Meta-Analysis of Randomized Controlled Trials. Front. Oncol. 2021, 11, 718871. [Google Scholar] [CrossRef]
- Coleman, R.L.; Fleming, G.F.; Brady, M.F.; Swisher, E.M.; Steffensen, K.D.; Friedlander, M.; Okamoto, A.; Moore, K.N.; Efrat Ben-Baruch, N.; Werner, T.L.; et al. Veliparib with First-Line Chemotherapy and as Maintenance Therapy in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2403–2415. [Google Scholar] [CrossRef]
- Han, H.S.; Diéras, V.; Robson, M.; Palácová, M.; Marcom, P.K.; Jager, A.; Puhalla, S. Veliparib with temozolomide or carboplatin/paclitaxel versus placebo with carboplatin/paclitaxel in patients with BRCA1/2 locally recurrent/metastatic breast cancer: Randomized phase II study. Ann. Oncol. 2018, 29, 154–161. [Google Scholar] [CrossRef]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Diossy, M.; Sztupinszki, Z.; Borcsok, J.; Krzystanek, M.; Tisza, V.; Spisak, S.; Rusz, O.; Timar, J.; Csabai, I.; Fillinger, J.; et al. A subset of lung cancer cases shows robust signs of homologous recombination deficiency associated genomic mutational signatures. NPJ Precis. Oncol. 2021, 5, 1–8. [Google Scholar] [CrossRef]
- Rao, D.; Mallick, A.B.; Augustine, T.; Daroqui, C.; Jiffry, J.; Merla, A.; Chaudhary, I.; Seetharam, R.; Sood, A.; Gajavelli, S.; et al. Excision repair cross-complementing group-1 (ERCC1) induction kinetics and polymorphism are markers of inferior outcome in patients with colorectal cancer treated with oxaliplatin. Oncotarget 2019, 10, 5510–5522. [Google Scholar] [CrossRef]
- Chen, L.-H.; Shen, T.-C.; Li, C.-H.; Chiu, K.-L.; Hsiau, Y.-C.; Wang, Y.-C.; Gong, C.-L.; Wang, Z.-H.; Chang, W.-S.; Tsai, C.-W.; et al. The Significant Interaction of Excision Repair Cross-complementing Group 1 Genotypes and Smoking to Lung Cancer Risk. Cancer Genom. Proteom. 2020, 17, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Wang, D.-P.; Lu, G.-Q.; Liu, K.-L.; Zhang, T.-J.; Li, S.; O Mohamed, K.; Xue, W.-H.; Qian, X.-H.; Meng, F.-H. Development of a novel thymidylate synthase (TS) inhibitor capable of up-regulating P53 expression and inhibiting angiogenesis in NSCLC. J. Adv. Res. 2020, 26, 95–110. [Google Scholar] [CrossRef]
- Kulda, V.; Hrda, K.; Houdek, Z.; Dobra, J.K.; Vrzakova, R.; Svaton, M.; Safranek, J.; Dolezal, J.; Babuska, V.; Pesek, M.; et al. Predictive Significance of Thymidylate Synthase Expression in Non-small Cell Lung Cancer. Anticancer Res. 2017, 37, 6953–6958. [Google Scholar] [CrossRef] [PubMed]
- Karim, N.A.; Schuster, J.; Eldessouki, I.; Gaber, O.; Namad, T.; Wang, J.; Xie, C.; Morris, J.C. Pulmonary sarcomatoid carcinoma: University of Cincinnati experience. Oncotarget 2017, 9, 4102–4108. [Google Scholar] [CrossRef]
- Hassan, R.; Gulati, S.; Mahender, Y.; Eldessouki, I.; Siddiqi, N.I.; Xie, C.; Pruemer, J.; Karim, N.A. Impact of Low Molecular Weight Heparin on Overall Survival in Patients with Advanced Lung Cancer: A Retrospective Study. Am. J. Clin. Exp. Med. 2017, 5, 173. [Google Scholar] [CrossRef][Green Version]
- Hotta, K.; Matsuo, K.; Ueoka, H.; Kiura, K.; Tabata, M.; Tanimoto, M. Addition of platinum compounds to a new agent in patients with advanced non-small-cell lung cancer: A literature based meta-analysis of randomised trials. Ann. Oncol. 2004, 15, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Fossella, F.; Pereira, J.R.; Von Pawel, J.; Pluzanska, A.; Gorbounova, V.; Kaukel, E.; Mattson, K.V.; Ramlau, R.; Szczęsna, A.; Fidias, P.; et al. Randomized, Multinational, Phase III Study of Docetaxel Plus Platinum Combinations Versus Vinorelbine Plus Cisplatin for Advanced Non–Small-Cell Lung Cancer: The TAX 326 Study Group. J. Clin. Oncol. 2003, 21, 3016–3024. [Google Scholar] [CrossRef]
- D’Addario, G.; Pintilie, M.; Leighl, N.B.; Feld, R.; Cerny, T.; Shepherd, F.A. Platinum-Based Versus Non-Platinum-Based Chemotherapy in Advanced Non-Small-Cell Lung Cancer: A Meta-Analysis of the Published Literature. J. Clin. Oncol. 2005, 23, 2926–2936. [Google Scholar] [CrossRef]
- Monica, V.; Iacono, M.L.; Bracco, E.; Busso, S.; di Blasio, L.; Primo, L.; Peracino, B.; Papotti, M.; Scagliotti, G. Dasatinib modulates sensitivity to pemetrexed in malignant pleural mesothelioma cell lines. Oncotarget 2016, 7, 76577–76589. [Google Scholar] [CrossRef]
- Qian, X.-L.; Zhang, J.; Li, P.-Z.; Lang, R.-G.; Li, W.-D.; Sun, H.; Liu, F.-F.; Guo, X.-J.; Gu, F.; Fu, L. Dasatinib inhibits c-src phosphorylation and prevents the proliferation of Triple-Negative Breast Cancer (TNBC) cells which overexpress Syndecan-Binding Protein (SDCBP). PLoS ONE 2017, 12, e0171169. [Google Scholar] [CrossRef]
- Bonanno, L.; Costa, C.; Majem, M.; Favaretto, A.; Rugge, M.; Rosell, R. The predictive value of BRCA1 and RAP80 mRNA expression in advanced non-small-cell lung cancer patients treated with platinum-based chemotherapy. Ann. Oncol. 2013, 24, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Perez-Roca, L.; Sánchez, J.J.; Cobo, M.; Moran, T.; Chaib, I.; Provencio, M.; Dómine, M.; Sala, M.; Jimenez, U.; et al. Customized Treatment in Non-Small-Cell Lung Cancer Based on EGFR Mutations and BRCA1 mRNA Expression. PLoS ONE 2009, 4, e5133. [Google Scholar] [CrossRef] [PubMed]
- Friboulet, L.; Olaussen, K.A.; Pignon, J.-P.; Shepherd, F.A.; Tsao, M.-S.; Graziano, S.; Kratzke, R.; Douillard, J.-Y.; Seymour, L.; Pirker, R.; et al. ERCC1 Isoform Expression and DNA Repair in Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2013, 368, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Mack, P.C.; Gandara, D.R.; Bowen, C.; Edelman, M.; Paglieroni, T.; Schnier, J.B.; Gelmann, E.P.; Gumerlock, P.H. RB status as a determinant of response to UCN-01 in non-small cell lung carcinoma. In Clin. Cancer Res.; 1999; 5, pp. 2596–2604. Available online: http://www.ncbi.nlm.nih.gov/pubmed/10499638 (accessed on 4 March 2022).
- Kelber, J.A.; Reno, T.; Kaushal, S.; Metildi, C.; Wright, T.; Stoletov, K.; Weems, J.M.; Park, F.D.; Mose, E.; Wang, Y.; et al. KRas Induces a Src/PEAK1/ErbB2 Kinase Amplification Loop That Drives Metastatic Growth and Therapy Resistance in Pancreatic Cancer. Cancer Res. 2012, 72, 2554–2564. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.A.; Tran, T.; Vilain, R.E.; Madore, J.; Selinger, C.I.; Kohonen-Corish, M.; Yip, P.; Yu, B.; O’Toole, S.A.; McCaughan, B.C.; et al. PD-L1 expression is a favorable prognostic factor in early stage non-small cell carcinoma. Lung Cancer 2015, 89, 181–188. [Google Scholar] [CrossRef]
- Yu, H.; Boyle, T.A.; Zhou, C.; Rimm, D.L.; Hirsch, F.R. PD-L1 expression in Lung Cancer. J. Thorac. Oncol. 2016, 11, 964–975. [Google Scholar] [CrossRef]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for lung cancers with KRAS p. G12C mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 2078–2092. [Google Scholar] [CrossRef]
- Jänne, P.A.; Van Den Heuvel, M.M.; Barlesi, F.; Cobo, M.; Mazieres, J.; Crinò, L.; Orlov, S.; Blackhall, F.; Wolf, J.; Garrido, P.; et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with kras-mutant advanced non–small cell lung cancer: The select-1 randomized clinical trial. JAMA 2017, 317, 1844–1853. [Google Scholar] [CrossRef]
- Garon, E.B.; Ciuleanu, T.-E.; Arrieta, O.; Prabhash, K.; Syrigos, K.N.; Goksel, T.; Park, K.; Gorbunova, V.; Kowalyszyn, R.D.; Pikiel, J.; et al. Ramucirumab plus docetaxel versus placebo plus docetaxel for second-line treatment of stage IV non-small-cell lung cancer after disease progression on platinum-based therapy (REVEL): A multicentre, double-blind, randomised phase 3 trial. Lancet 2014, 384, 665–673. [Google Scholar] [CrossRef]
- Awad, M.M.; Liu, S.; Rybkin, I.I.; Arbour, K.C.; Dilly, J.; Zhu, V.W.; Johnson, M.L.; Heist, R.S.; Patil, T.; Riely, G.J.; et al. Acquired resistance to KRASG12C inhibition in cancer. N. Engl. J. Med. 2021, 384, 2382–2393. [Google Scholar] [CrossRef] [PubMed]


| Oncogene. | ERCC1 | BRCA1 | TS | SRC | PD-L1 |
|---|---|---|---|---|---|
| mt KRAS | + | − | + | − | 25% |
| wtKRAS | − | + | + | + | |
| Normal lung tissue | − | − | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karim, N.A.; Ullah, A.; Pathrose, P.; Fathallah, H.; Perry, A.; Morris, J.C.; Wang, J.; Starnes, S.L. Does Molecular Profiling of KRAS-Mutant Non-Squamous Non-Small Cell Lung Cancer (NSCLC) Help in Treatment Strategy Planning? Curr. Oncol. 2022, 29, 4779-4790. https://doi.org/10.3390/curroncol29070379
Karim NA, Ullah A, Pathrose P, Fathallah H, Perry A, Morris JC, Wang J, Starnes SL. Does Molecular Profiling of KRAS-Mutant Non-Squamous Non-Small Cell Lung Cancer (NSCLC) Help in Treatment Strategy Planning? Current Oncology. 2022; 29(7):4779-4790. https://doi.org/10.3390/curroncol29070379
Chicago/Turabian StyleKarim, Nagla Abdel, Asad Ullah, Peterson Pathrose, Hassana Fathallah, Ashley Perry, John C. Morris, Jiang Wang, and Sandra L. Starnes. 2022. "Does Molecular Profiling of KRAS-Mutant Non-Squamous Non-Small Cell Lung Cancer (NSCLC) Help in Treatment Strategy Planning?" Current Oncology 29, no. 7: 4779-4790. https://doi.org/10.3390/curroncol29070379
APA StyleKarim, N. A., Ullah, A., Pathrose, P., Fathallah, H., Perry, A., Morris, J. C., Wang, J., & Starnes, S. L. (2022). Does Molecular Profiling of KRAS-Mutant Non-Squamous Non-Small Cell Lung Cancer (NSCLC) Help in Treatment Strategy Planning? Current Oncology, 29(7), 4779-4790. https://doi.org/10.3390/curroncol29070379






