Molecular Differences between Squamous Cell Carcinoma and Adenocarcinoma Cervical Cancer Subtypes: Potential Prognostic Biomarkers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of Differential Expression
2.2. Cervical Samples
2.3. RNA Isolation and qPCR
2.4. CC Transcriptome and Pathway Analyses
2.5. OS Analysis
3. Results
3.1. Determination of the Molecular Profile Distinguishing ADC and SCC among CC Patients from the TCGA Database
3.2. Validation of the Molecular Profile Obtained from the TCGA Database in a Mexican-Mestizo Independent Cohort
3.3. Signaling Pathway Enrichment Analysis in DEGs between ADC and SCC Subtypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020, GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Retana, J.; Lasa-Gonsebatt, F.; Lopez-Urrutia, E.; Coronel-Martínez, J.; Cantu De Leon, D.; Jacobo-Herrera, N.; Peralta-Zaragoza, O.; Perez-Montiel, D.; Reynoso-Noveron, N.; Vazquez-Romo, R.; et al. Transcript profiling distinguishes complete treatment responders with locally advanced cervical cancer. Transl. Oncol. 2015, 8, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.B.; Lee, J.M.; Park, C.Y.; Lee, K.B.; Cho, H.Y.; Ha, S.Y. What is the difference between squamous cell carcinoma and adenocarcinoma of the cervix? A matched case-control study. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2006, 16, 1569–1573. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, N.; Bosch, F.X.; de Sanjosé, S.; Herrero, R.; Castellsagué, X.; Shah, K.V.; Snijders, P.J.F.; Meijer, C.J.L.M.; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamzi Abdul Raub, S.; Isa, N.M.; Zailani, H.A.; Omar, B.; Abdullah, M.F.; Mohd Amin, W.A.; Noor, R.M.; Ayub, M.C.; Abidin, Z.; Kassim, F.; et al. Distribution of HPV genotypes in cervical cancer in multi- ethnic Malaysia. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 651–656. [Google Scholar] [CrossRef] [Green Version]
- Tjalma, W.A.A.; Trinh, X.B.; Rosenlund, M.; Makar, A.P.; Kridelka, F.; Rosillon, D.; Van Dam, P.A.; Collas De Souza, S.; Holl, K.; Simon, P.; et al. A cross-sectional, multicentre, epidemiological study on human papillomavirus (HPV) type distribution in adult women diagnosed with invasive cervical cancer in Belgium. Facts Views Vis. ObGyn. 2015, 7, 101–108. [Google Scholar]
- Hadzisejdić, I.; Simat, M.; Bosak, A.; Krasević, M.; Grahovac, B. Prevalence of human papillomavirus genotypes in cervical cancer and precursor lesions. Coll. Antropol. 2006, 30, 879–883. [Google Scholar]
- Koh, W.-J.; Abu-Rustum, N.R.; Bean, S.; Bradley, K.; Campos, S.M.; Cho, K.R.; Chon, H.S.; Chu, C.; Clark, R.; Cohn, D.; et al. Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2019, 17, 64–84. [Google Scholar] [CrossRef] [Green Version]
- Jung, E.J.; Byun, J.M.; Kim, Y.N.; Lee, K.B.; Sung, M.S.; Kim, K.T.; Jeong, D.H. Cervical Adenocarcinoma Has a Poorer Prognosis and a Higher Propensity for Distant Recurrence Than Squamous Cell Carcinoma. Int. J. Gynecol. Cancer Off. J. Int. Gynecol. Cancer Soc. 2017, 27, 1228–1236. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, B.; Zhou, Z.-H.; Han, W.-W. Clinicopathological characteristics and prognostic factors of cervical adenocarcinoma. Sci. Rep. 2021, 11, 7506. [Google Scholar] [CrossRef]
- Yamauchi, M.; Fukuda, T.; Wada, T.; Kawanishi, M.; Imai, K.; Hashiguchi, Y.; Ichimura, T.; Yasui, T.; Sumi, T. Comparison of Outcomes between Squamous Cell Carcinoma and Adenocarcinoma in Patients with Surgically Treated Stage I-II Cervical Cancer. Mol. Clin. Oncol. 2014, 2, 518–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.; Song, K.; Cui, B.; Jiang, J.; Yang, X.; Kong, B. A comparison of the prognosis between adenocarcinoma and squamous cell carcinoma in stage IB-IIA cervical cancer. Int. J. Clin. Oncol. 2018, 23, 522–531. [Google Scholar] [CrossRef] [PubMed]
- PLOS Genetics Staff. Correction, Comparative transcriptomes of adenocarcinomas and squamous cell carcinomas reveal molecular similarities that span classical anatomic boundaries. PLoS Genet. 2017, 13, e1007056. [Google Scholar] [CrossRef] [Green Version]
- Winer, I.; Alvarado-Cabrero, I.; Hassan, O.; Ahmed, Q.F.; Alosh, B.; Bandyopadhyay, S.; Thomas, S.; Albayrak, S.; Talukdar, S.; Al-Wahab, Z.; et al. The Prognostic Significance of Histologic Type in Early Stage Cervical Cancer—A multi-institutional study. Ginecol. Oncol. 2015, 137, 474–478. [Google Scholar] [CrossRef]
- Chao, A.; Wang, T.-H.; Lee, Y.-S.; Hsueh, S.; Chao, A.-S.; Chang, T.-C.; Kung, W.-H.; Huang, S.-L.; Chao, F.-Y.; Wei, M.-L.; et al. Molecular characterization of adenocarcinoma and squamous carcinoma of the uterine cervix using microarray analysis of gene expression. Int. J. Cancer 2006, 119, 91–98. [Google Scholar] [CrossRef]
- Bispo, S.; Farias, T.D.J.; de Araujo-Souza, P.S.; Cintra, R.; Dos Santos, H.G.; Jorge, N.A.N.; Castro, M.A.A.; Wajnberg, G.; de Scherer, M.N.; Genta, M.L.N.D.; et al. Dysregulation of Transcription Factor Networks Unveils Different Pathways in Human Papillomavirus 16-Positive Squamous Cell Carcinoma and Adenocarcinoma of the Uterine Cervix. Front. Oncol. 2021, 11, 626187. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services; Barretos Cancer Hospital; Baylor College of Medicine; Beckman Research Institute of City of Hope; Buck Institute for Research on Aging; Canada’s Michael Smith Genome Sciences Centre; Harvard Medical School; Helen, F.; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks, an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Retana, J.; Zamudio-Meza, H.; Rodriguez-Morales, M.; Pedroza-Torres, A.; Isla-Ortiz, D.; Herrera, L.; Jacobo-Herrera, N.; Peralta-Zaragoza, O.; López-Camarillo, C.; Morales-Gonzalez, F.; et al. Gene signature based on degradome-related genes can predict distal metastasis in cervical cancer patients. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317711895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, S.; Meltzer, P.S. GEOquery, a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinforma Oxf. Engl. 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019, gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrington de González, A.; Sweetland, S.; Green, J. Comparison of risk factors for squamous cell and adenocarcinomas of the cervix, a meta-analysis. Br. J. Cancer 2004, 90, 1787–1791. [Google Scholar] [CrossRef] [Green Version]
- Vidal, A.C.; Skaar, D.; Maguire, R.; Dodor, S.; Musselwhite, L.W.; Bartlett, J.A.; Oneko, O.; Obure, J.; Mlay, P.; Murphy, S.K.; et al. IL-10, IL-15, IL-17, and GMCSF levels in cervical cancer tissue of Tanzanian women infected with HPV16/18 vs. non-HPV16/18 genotypes. Infect. Agent Cancer 2015, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.; Wang, Y.; Chen, C.; Zhu, X.; Zhu, H.; Hu, Y. Effects of Th17 cells and IL-17 in the progression of cervical carcinogenesis with high-risk human papillomavirus infection. Cancer Med. 2018, 7, 297–306. [Google Scholar] [CrossRef]
- Mareti Bonin, C.; Zatorre Almeida-Lugo, L.; Rodrigues Dos Santos, A.; Tezelli Junqueira Padovani, C.; Silva Pina, A.F.; Teixeira Ferreira, A.M.; Dos Santos Fernandes, C.E.; Possati Resende, J.C.; Bovo, A.C.; Tozetti, I.A. Interleukin-17 expression in the serum and exfoliated cervical cells of patients infected with high-risk oncogenic human papillomavirus. Cytokine 2019, 120, 92–98. [Google Scholar] [CrossRef]
- Punt, S.; Fleuren, G.J.; Kritikou, E.; Lubberts, E.; Trimbos, J.B.; Jordanova, E.S.; Gorter, A. Angels and demons, Th17 cells represent a beneficial response, while neutrophil IL-17 is associated with poor prognosis in squamous cervical cancer. Oncoimmunology 2015, 4, e984539. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Zhang, H.-L.; Niu, Z.-Y.; Wang, Z.; Kong, Y.; Yang, X.-S.; Yuan, F. The disease stage-associated imbalance of Th1/Th2 and Th17/Treg in uterine cervical cancer patients and their recovery with the reduction of tumor burden. BMC Womens Health 2020, 20, 126. [Google Scholar] [CrossRef]
- Campos-Parra, A.D.; Padua-Bracho, A.; Pedroza-Torres, A.; Figueroa-González, G.; Fernández-Retana, J.; Millan-Catalan, O.; Peralta-Zaragoza, O.; Cantú de León, D.; Herrera, L.A.; Pérez-Plasencia, C. Comprehensive transcriptome analysis identifies pathways with therapeutic potential in locally advanced cervical cancer. Gynecol. Oncol. 2016, 143, 406–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Shea, J.J.; Lahesmaa, R.; Vahedi, G.; Laurence, A.; Kanno, Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat. Rev. Immunol. 2011, 11, 239–250. [Google Scholar] [CrossRef]
- Zou, Y.; Liu, F.-Y.; Wu, J.; Wan, L.; Fang, S.-F.; Zhang, Z.-Y.; Luo, Y.; Chen, M.-H.; Huang, M.-Z.; He, M.; et al. Mutational analysis of the RAS/RAF/MEK/ERK signaling pathway in 260 Han Chinese patients with cervical carcinoma. Oncol. Lett. 2017, 14, 2427–2431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.; Kim, H.-S.; Seo, S.S.; Park, S.-Y.; Sidransky, D.; Dong, S.M. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol. Oncol. 2007, 105, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.A.; Howitt, B.E.; Myers, A.P.; Dahlberg, S.E.; Palescandolo, E.; Van Hummelen, P.; MacConaill, L.E.; Shoni, M.; Wagle, N.; Jones, R.T.; et al. Oncogenic mutations in cervical cancer, genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer 2013, 119, 3776–3783. [Google Scholar] [CrossRef]
- Jin, Y.; Jin, W.; Zheng, Z.; Chen, E.; Wang, Q.; Wang, Y.; Wang, O.; Zhang, X. GABRB2 plays an important role in the lymph node metastasis of papillary thyroid cancer. Biochem. Biophys. Res. Commun. 2017, 492, 323–330. [Google Scholar] [CrossRef]
- Zhu, R.; Gires, O.; Zhu, L.; Liu, J.; Li, J.; Yang, H.; Ju, G.; Huang, J.; Ge, W.; Chen, Y.; et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat. Commun. 2019, 10, 2863. [Google Scholar] [CrossRef]
- Tang, Y.; Xie, J.; Chen, Y.; Zhang, T.; Zhuo, H.; Chen, J.; Yang, P.; Cui, Q.; Zhao, L.; Li, L. Tspan8 Is Highly Expressed in Clear Cell Renal Cell Carcinoma and Indicates Poor Prognosis. Ann. Clin. Lab. Sci. 2020, 50, 638–644. [Google Scholar]
- Heiler, S.; Wang, Z.; Zöller, M. Pancreatic cancer stem cell markers and exosomes—The incentive push. World J. Gastroenterol. 2016, 22, 5971–6007. [Google Scholar] [CrossRef]
- Zhang, Z.-F.; Zhang, H.-R.; Zhang, Q.-Y.; Lai, S.-Y.; Feng, Y.-Z.; Zhou, Y.; Zheng, S.-R.; Shi, R.; Zhou, J.-Y. High expression of TMEM40 is associated with the malignant behavior and tumorigenesis in bladder cancer. J. Transl. Med. 2018, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-Y.; Fu, M.-T.; Zhang, Z.-F.; Feng, Y.-Z.; Wei, M.; Zhou, J.-Y.; Shi, R. Expression of TMEM40 in bladder cancer and its correlation with clinicopathological parameters. Int. J. Clin. Exp. Pathol. 2017, 10, 8050–8057. [Google Scholar] [PubMed]
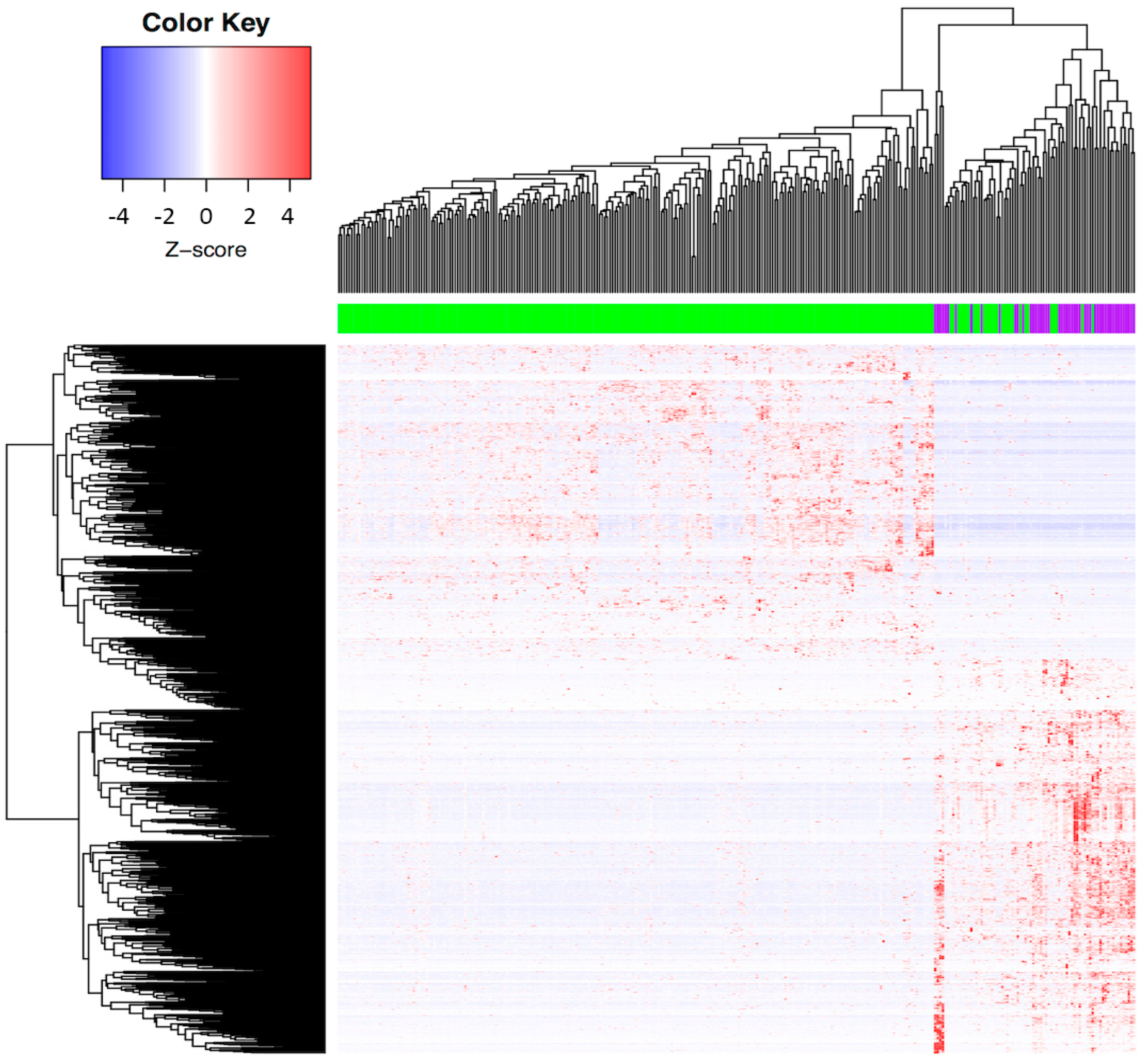
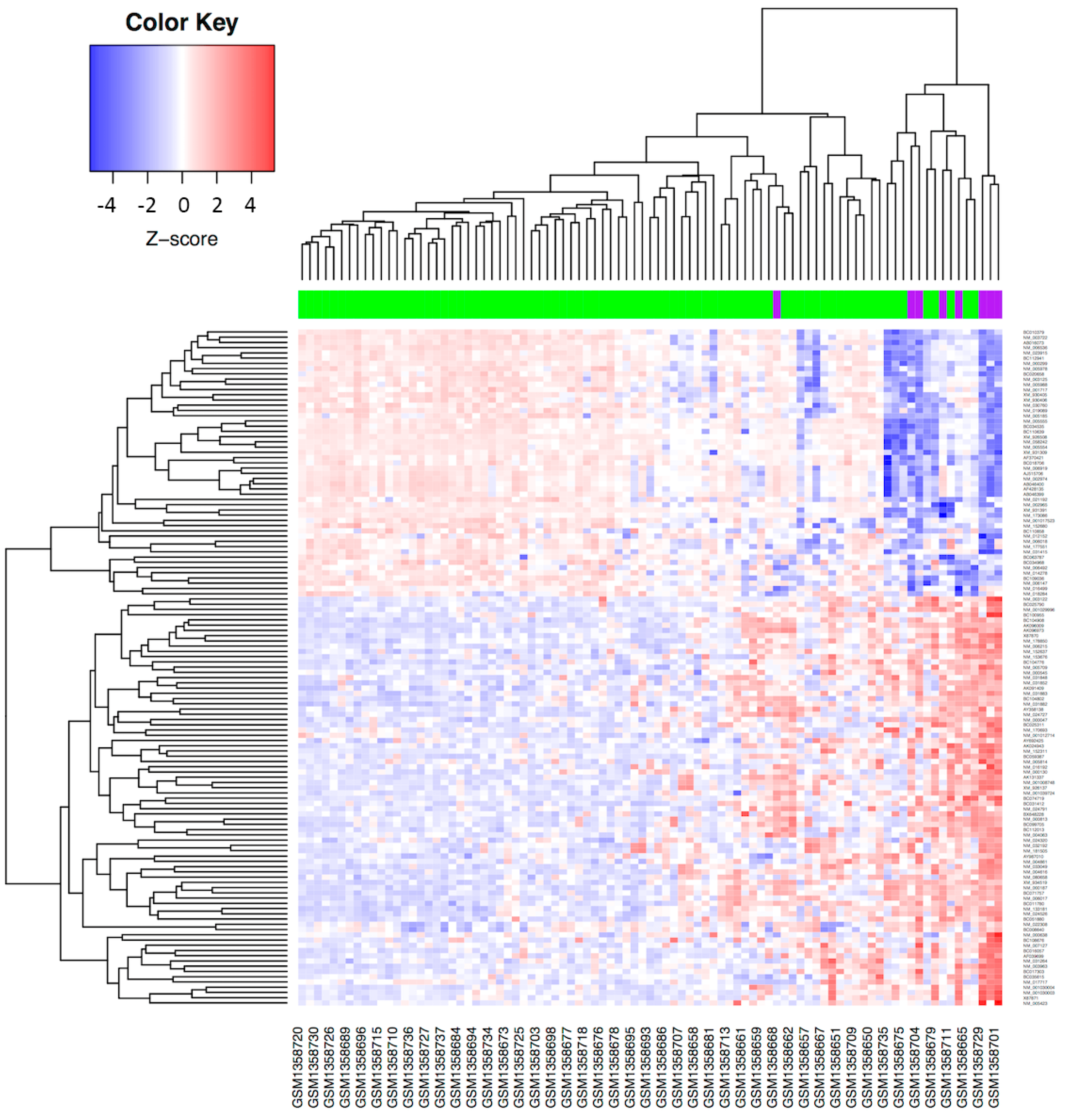
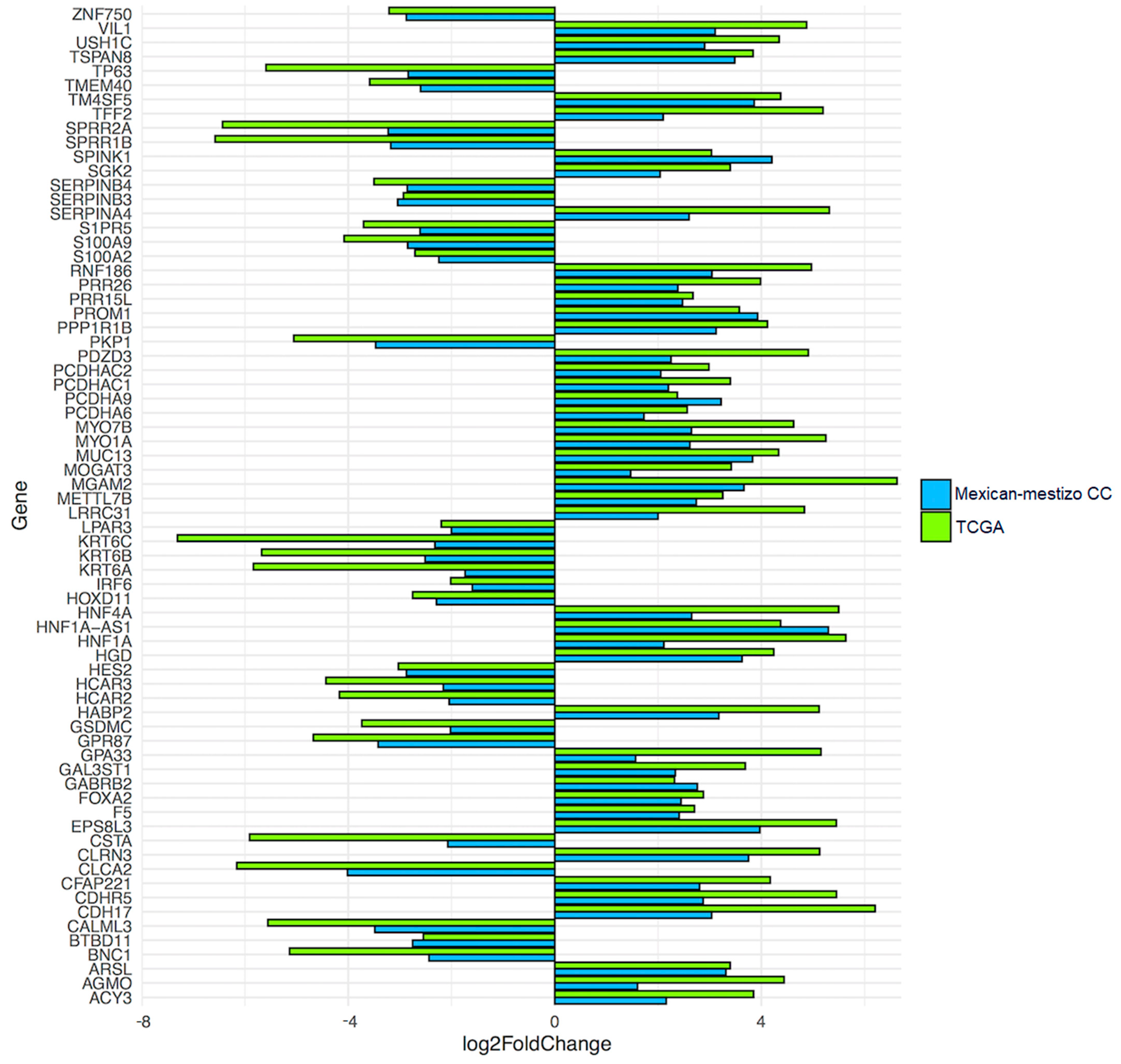
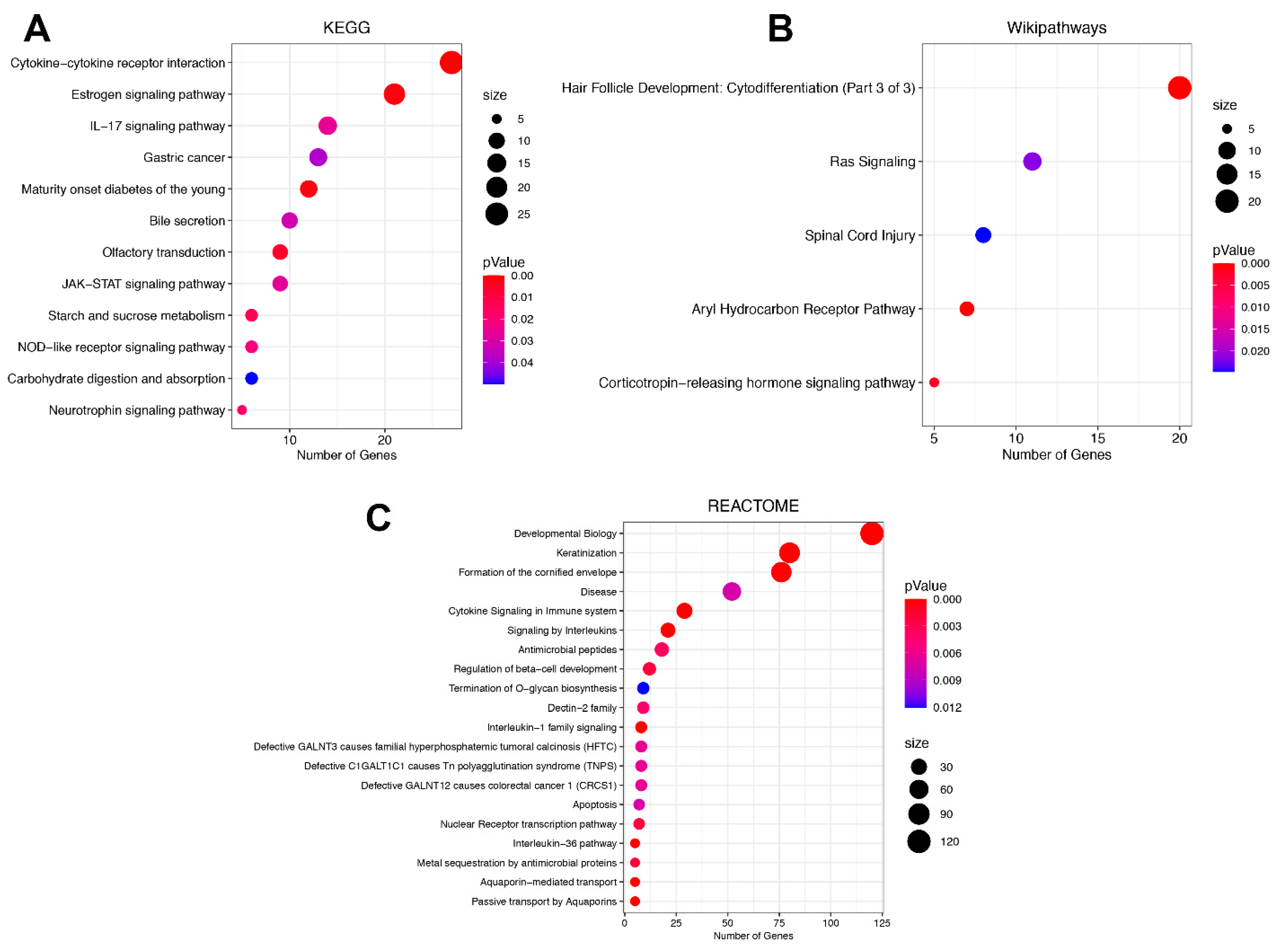
| Characteristics | Total (n = 309) | SCC (n = 260) | ADC (n = 49) | |
|---|---|---|---|---|
| Ethnicity | Hispanic or Latino | 24 (7.76%) | 21 (8.07%) | 3 (6.12%) |
| Not Hispanic or Latino | 171 (55.33%) | 145 (55.76%) | 26 (53.06%) | |
| Not reported | 114 (36.91%) | 94 (36.15%) | 20 (40.81%) | |
| FIGO Stage | ||||
| Stage I | 163 (52.75%) | 129 (49.61%) | 34 (69.38%) | |
| Stage II | 70 (22.65%) | 63 (24.3%) | 7 (14.28%) | |
| Stage III | 46 (14.88%) | 43 (16.53%) | 3 (6.12%) | |
| Stage IV | 21 (6.79%) | 17 (6.53%) | 4 (8.16%) | |
| Not reported | 9 (2.91%) | 8 (3.07%) | 1 (2.04%) |
| Over-Expressed | Under-Expressed | ||
|---|---|---|---|
| Gene | log2FC | Gene | log2FC |
| REG4 | 8.41 | CRNN | −9.38 |
| TM4SF4 | 8.27 | KRT14 | −8.57 |
| TAAR1 | 7.54 | TMPRSS11B | −8.17 |
| FABP1 | 7.52 | CLEC2A | −8.07 |
| SLC17A4 | 7.42 | SPRR2E | −7.84 |
| ANKRD40CL | 7.32 | TMPRSS11BNL | −7.82 |
| REG1A | 6.77 | LCE3E | −7.78 |
| REG1B | 6.65 | SERPINB12 | −7.78 |
| MGAM2 | 6.63 | KPRP | −7.73 |
| LOC105375166 | 6.57 | LCE3D | −7.66 |
| MUC17 | 6.57 | TMPRSS11A | −7.56 |
| CCL15 | 6.33 | SPRR2G | −7.33 |
| SLC39A5 | 6.26 | KRT6C | −7.31 |
| LOC107105282 | 6.23 | LINC02582 | −7.30 |
| CDH17 | 6.20 | KRT1 | −7.26 |
| Overexpressed | Underexpressed | ||
|---|---|---|---|
| Gene | log2FC | Gene | log2FC |
| HNF1A-AS1 | 5.30 | CLCA2 | −4.02 |
| SPINK1 | 4.20 | CALML3 | −3.48 |
| EPS8L3 | 3.97 | PKP1 | −3.47 |
| PROM1 | 3.93 | GPR87 | −3.42 |
| TM4SF5 | 3.86 | TP63 | −3.41 |
| EPS8L3 | 3.86 | SPRR2A | −3.23 |
| MUC13 | 3.83 | SPRR1B | −3.18 |
| USH1C | 3.77 | SERPINB3 | −3.17 |
| CLRN3 | 3.75 | GPR87 | −2.98 |
| HGD | 3.73 | ZNF750 | −2.88 |
| MGAM2 | 3.66 | SERPINB4 | −2.86 |
| HGD | 3.63 | S100A9 | −2.85 |
| TSPAN8 | 3.48 | TP63 | −2.84 |
| ARSL | 3.31 | BTBD11 | −2.75 |
| HNF4A | 3.25 | KRT6C | −2.73 |
| Univariate Analysis | Multivariate Analysis | ||||
| Overall Survival | HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
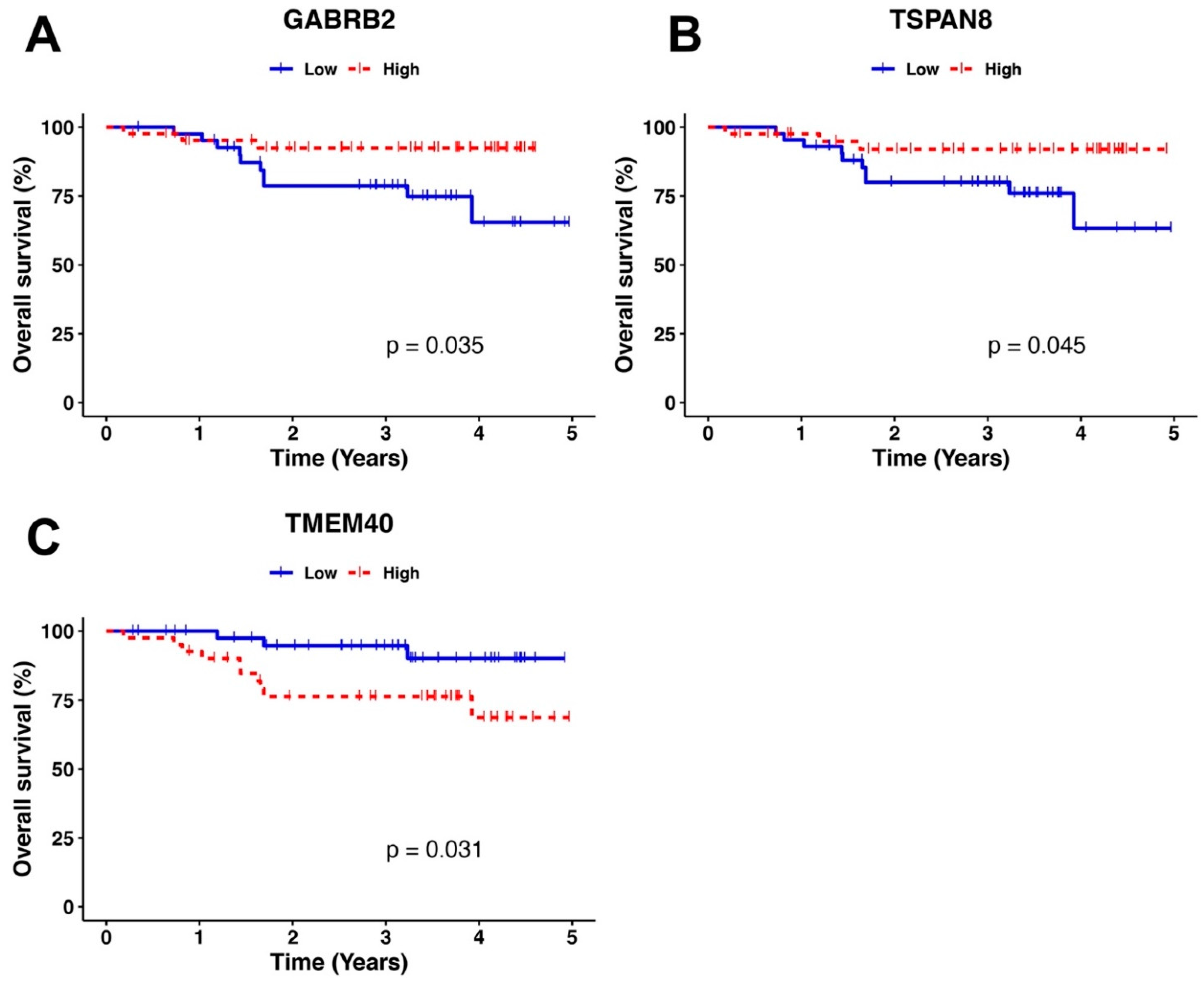
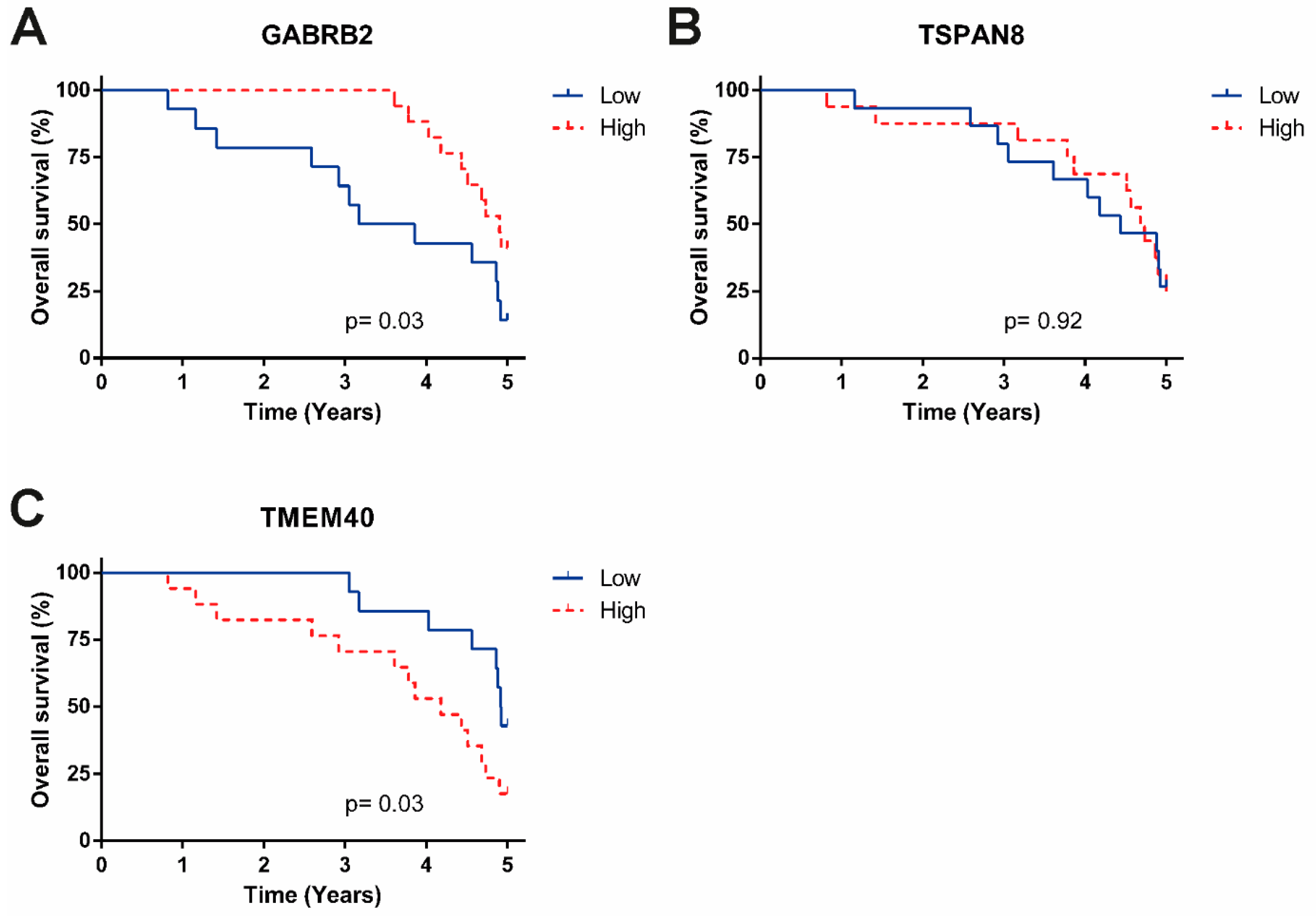
| TMEM40 | High vs. low expression | 0.27 (0.073–0.97) | 0.045 | 0.32 (0.08–1.22) | 0.096 |
| GABRB2 | 3.7 (1–13) | 0.049 | 2.93 (0.78–10.9) | 0.108 | |
| TSPAN8 | 3.5 (0.95–13) | 0.059 | |||
| HPV | Negative vs. positive | 1.1 (0.31–4.1) | 0.86 | 1.05 (0.28–3.84) | 0.937 |
| Hystology | Adenocarcinoma vs. epidermoid | 1.2 (0.15–9) | 0.88 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Parra, A.D.; Pérez-Quintanilla, M.; Martínez-Gutierrez, A.D.; Pérez-Montiel, D.; Coronel-Martínez, J.; Millan-Catalan, O.; De León, D.C.; Pérez-Plasencia, C. Molecular Differences between Squamous Cell Carcinoma and Adenocarcinoma Cervical Cancer Subtypes: Potential Prognostic Biomarkers. Curr. Oncol. 2022, 29, 4689-4702. https://doi.org/10.3390/curroncol29070372
Campos-Parra AD, Pérez-Quintanilla M, Martínez-Gutierrez AD, Pérez-Montiel D, Coronel-Martínez J, Millan-Catalan O, De León DC, Pérez-Plasencia C. Molecular Differences between Squamous Cell Carcinoma and Adenocarcinoma Cervical Cancer Subtypes: Potential Prognostic Biomarkers. Current Oncology. 2022; 29(7):4689-4702. https://doi.org/10.3390/curroncol29070372
Chicago/Turabian StyleCampos-Parra, Alma D., Milagros Pérez-Quintanilla, Antonio Daniel Martínez-Gutierrez, Delia Pérez-Montiel, Jaime Coronel-Martínez, Oliver Millan-Catalan, David Cantú De León, and Carlos Pérez-Plasencia. 2022. "Molecular Differences between Squamous Cell Carcinoma and Adenocarcinoma Cervical Cancer Subtypes: Potential Prognostic Biomarkers" Current Oncology 29, no. 7: 4689-4702. https://doi.org/10.3390/curroncol29070372
APA StyleCampos-Parra, A. D., Pérez-Quintanilla, M., Martínez-Gutierrez, A. D., Pérez-Montiel, D., Coronel-Martínez, J., Millan-Catalan, O., De León, D. C., & Pérez-Plasencia, C. (2022). Molecular Differences between Squamous Cell Carcinoma and Adenocarcinoma Cervical Cancer Subtypes: Potential Prognostic Biomarkers. Current Oncology, 29(7), 4689-4702. https://doi.org/10.3390/curroncol29070372







