Upfront Next Generation Sequencing in Non-Small Cell Lung Cancer
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Comprehensive Cancer Network. Non-Small Cell Lung Cancer. Version 3.2022. 16 March 2022. Available online: https://www.nccn.org/login?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 13 May 2022).
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular Testing Guideline for the Selection of Patients with Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar] [CrossRef] [Green Version]
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29 (Suppl. 4), iv192–iv237. [Google Scholar] [CrossRef] [PubMed]
- Sukhai, M.A.; Craddock, K.J.; Thomas, M.; Hansen, A.R.; Zhang, T.; Siu, L.; Bedard, P.; Stockley, T.L.; Kamel-Reid, S. A classification system for clinical relevance of somatic variants identified in molecular profiling of cancer. Genet. Med. 2016, 18, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Fiset, P.O.; Labbé, C.; Young, K.; Craddock, K.J.; Smith, A.C.; Tanguay, J.; Pintilie, M.; Wang, R.; Torlakovic, E.; Cheung, C.; et al. Anaplastic lymphoma kinase 5A4 immunohistochemistry as a diagnostic assay in lung cancer: A Canadian reference testing center’s results in population-based reflex testing. Cancer 2019, 125, 4043–4051. [Google Scholar] [CrossRef]
- Hwang, D.M.; Albaqer, T.; Santiago, R.C.; Weiss, J.; Tanguay, J.; Cabanero, M.; Leung, Y.; Pal, P.; Khan, Z.; Lau, S.C.M.; et al. Prevalence and Heterogeneity of PD-L1 Expression by 22C3 Assay in Routine Population-Based and Reflexive Clinical Testing in Lung Cancer. J. Thorac. Oncol. 2021, 16, 1490–1500. [Google Scholar] [CrossRef]
- Cheung, C.C.; Smith, A.C.; Albadine, R.; Bigras, G.; Bojarski, A.; Couture, C.; Cutz, J.C.; Huang, W.Y.; Ionescu, D.; Itani, D.; et al. Canadian ROS proto-oncogene 1 study (CROS) for multi-institutional implementation of ROS1 testing in non-small cell lung cancer. Lung Cancer 2021, 160, 127–135. [Google Scholar] [CrossRef]
- Gauthier, M.; Law, J.; Le, L.; Li, J.J.N.; Zahir, S.; Nirmalakumar, S.; Sung, M.; Pettengell, C.; Aviv, S.; Chu, R.; et al. Automating Access to Real-World Evidence. J. Thorac. Oncol. 2022, 3, 100340. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal. Transduct. Target Ther. 2019, 4, 61. [Google Scholar] [CrossRef] [Green Version]
- Rivera, G.A.; Wakelee, H. Lung Cancer in Never Smokers. Adv. Exp. Med. Biol. 2016, 893, 43–57. [Google Scholar] [CrossRef]
- Kempf, E.; Rousseau, B.; Besse, B.; Paz-Ares, L. KRAS oncogene in lung cancer: Focus on molecularly driven clinical trials. Eur. Respir. Rev. 2016, 25, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Illumina, I. Multi-Site Analytical Validation of TruSight® Tumor 15 (TST15) Determining Robustness and Concordance. 2022. Available online: https://www.illumina.com/products/by-type/clinical-research-products/trusight-tumor-15-gene.html (accessed on 8 April 2022).
- Catania, C.; Botteri, E.; Barberis, M.; Conforti, F.; Toffalorio, F.; De Marinis, F.; Boselli, S.; Noberasco, C.; Delmonte, A.; Spitaleri, G.; et al. Molecular features and clinical outcome of lung malignancies in very young people. Future Oncol. 2015, 11, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Gerstung, M.; Jolly, C.; Leshchiner, I.; Dentro, S.C.; Gonzalez, S.; Rosebrock, D.; Mitchell, T.J.; Rubanova, Y.; Anur, P.; Yu, K.; et al. The evolutionary history of 2,658 cancers. Nature 2020, 578, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.S.; Squire, J.; et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 2013, 8, 823–859. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.; Tsao, M.S.; Le, L.W.; Shepherd, F.A.; Feld, R.; Burkes, R.L.; Liu, G.; Kamel-Reid, S.; Hwang, D.; Tanguay, J.; et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 2015, 26, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Arbour, K.C.; Rizvi, H.; Iqbal, A.N.; Gadgeel, S.M.; Girshman, J.; Kris, M.G.; Riely, G.J.; Yu, H.A.; Hellmann, M.D. Severe immune-related adverse events are common with sequential PD-(L)1 blockade and osimertinib. Ann. Oncol. 2019, 30, 839–844. [Google Scholar] [CrossRef]
- Shiau, C.J.; Babwah, J.P.; da Cunha Santos, G.; Sykes, J.R.; Boerner, S.L.; Geddie, W.R.; Leighl, N.B.; Wei, C.; Kamel-Reid, S.; Hwang, D.M.; et al. Sample features associated with success rates in population-based EGFR mutation testing. J. Thorac. Oncol. 2014, 9, 947–956. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.; Sekhon, H.S.; Cutz, J.C.; Hwang, D.M.; Kamel-Reid, S.; Carter, R.F.; Santos, G.D.C.; Waddell, T.; Binnie, M.; Patel, M.; et al. Improving molecular testing and personalized medicine in non-small-cell lung cancer in Ontario. Curr. Oncol. 2017, 24, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Zer, A.; Cutz, J.C.; Sekhon, H.; Hwang, D.M.; Sit, C.; Maganti, M.; Sung, M.; Binnie, M.; Brade, A.; Chung, T.B.; et al. Translation of Knowledge to Practice-Improving Awareness in NSCLC Molecular Testing. J. Thorac. Oncol. 2018, 13, 1004–1011. [Google Scholar] [CrossRef] [Green Version]
- Makarem, M.; Ezeife, D.A.; Smith, A.C.; Li, J.J.N.; Law, J.H.; Tsao, M.S.; Leighl, N.B. Reflex ROS1 IHC Screening with FISH Confirmation for Advanced Non-Small Cell Lung Cancer-A Cost-Efficient Strategy in a Public Healthcare System. Curr. Oncol. 2021, 28, 3268–3279. [Google Scholar] [CrossRef]
- Tsao, M.S.; Yatabe, Y. Old Soldiers Never Die: Is There Still a Role for Immunohistochemistry in the Era of Next-Generation Sequencing Panel Testing? J. Thorac. Oncol. 2019, 14, 2035–2038. [Google Scholar] [CrossRef] [PubMed]
- Leighl, N.B.; Nirmalakumar, S.; Ezeife, D.A.; Gyawali, B. An Arm and a Leg: The Rising Cost of Cancer Drugs and Impact on Access. Am. Soc. Clin. Oncol. Educ. Book 2021, 41, e1–e12. [Google Scholar] [CrossRef] [PubMed]
- Perdrizet, K.; Stockley, T.; Law, J.; Smith, A.; Zhang, T.; Fernandes, R.; Shabir, M.; Sabatini, P.; Youssef, N.; Ishu, C.; et al. Integrating comprehensive genomic sequencing of non-small cell lung cancer into a public healthcare system. Cancer Treat. Res. Commun. 2022, 31, 100534. [Google Scholar] [CrossRef] [PubMed]
| Number (%) | |
|---|---|
| N = 1395 | |
| Age at diagnosis, median (range) | 68.6 years (18.8–97.2) |
| Sex | |
| Female | 730 (52.3%) |
| Male | 665 (47.7%) |
| Smoking status | |
| Never | 444 (33.1%) |
| Former Smoker | 492 (36.7%) |
| Current Smoker | 405 (30.2%) |
| Unknown | 54 |
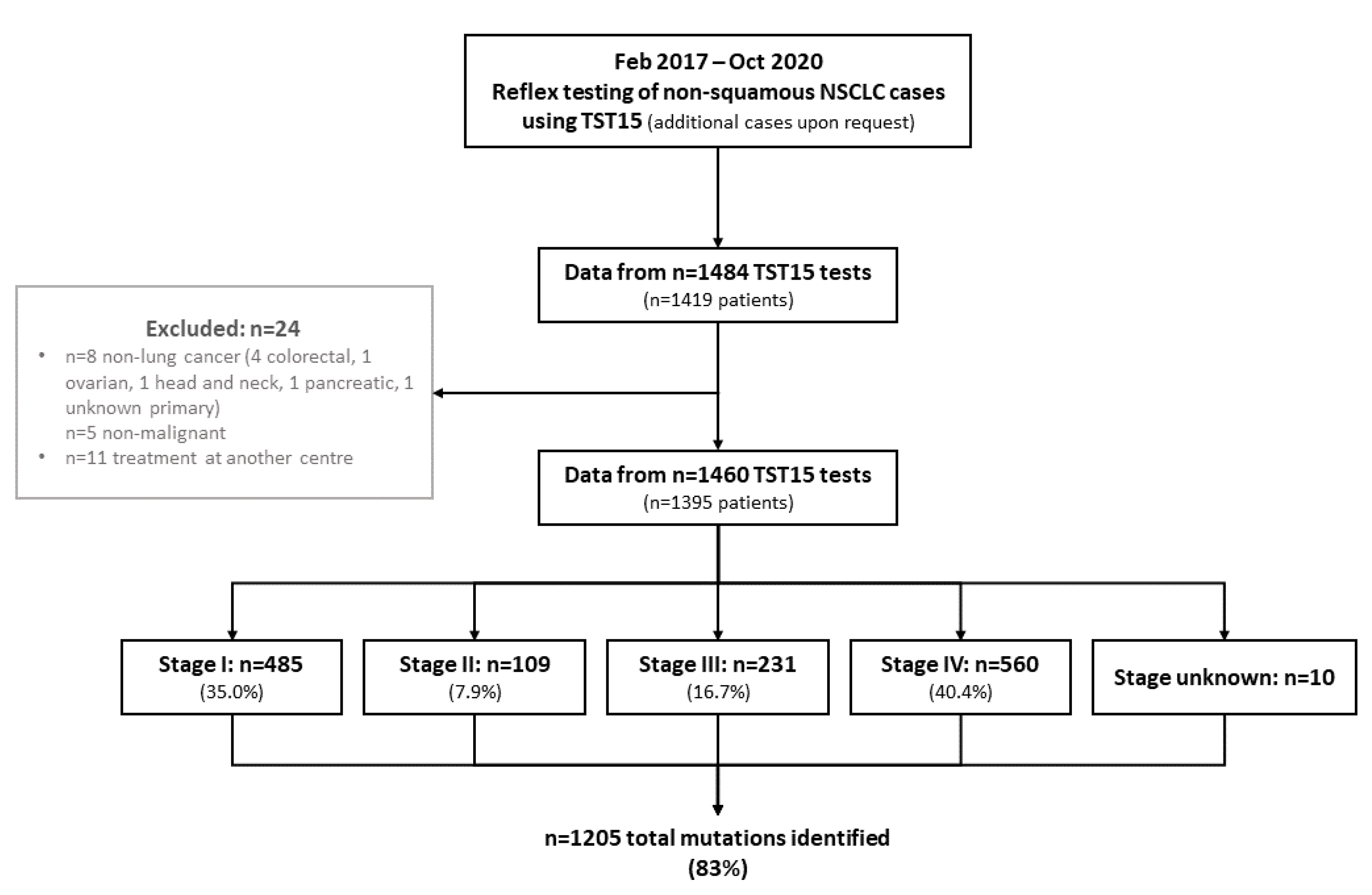
| Stage at diagnosis | |
| I | 485 (35.0%) |
| II | 109 (7.9%) |
| III | 231 (16.7%) |
| IV | 560 (40.4%) |
| Unknown | 10 |
| Histology | |
| Adenocarcinoma | 1198 (85.9%) |
| Large Cell | 40 (2.9%) |
| Squamous | 34 (2.4%) |
| Pleomorphic/Sarcomatoid | 14 (1.0%) |
| Small Cell | 12 (0.9%) |
| Mixed histology | 6 (1.2%) |
| Not otherwise specified | 91 (6.5%) |
| Number (%) | |
|---|---|
| N = 1460 | |
| Samples tested per patient * | |
| 1 | 1335 (95.7%) |
| 2 | 55 (3.9%) |
| 3 | 5 (0.4%) |
| Sample type | |
| Core biopsy | 665 (45.5%) |
| Surgical specimen | 379 (26.0%) |
| FNA cytology | 353 (24.2%) |
| Exfoliative cytology | 62 (4.2%) |
| Unknown | 1 |
| Sample site | |
| Primary (lung) | 997 (68.3%) |
| Non-bone visceral or soft tissue metastasis | 357 (24.5%) |
| Pleural fluid | 57 (3.9%) |
| Bone metastasis | 33 (2.3%) |
| Other | 16 (1.1%) |
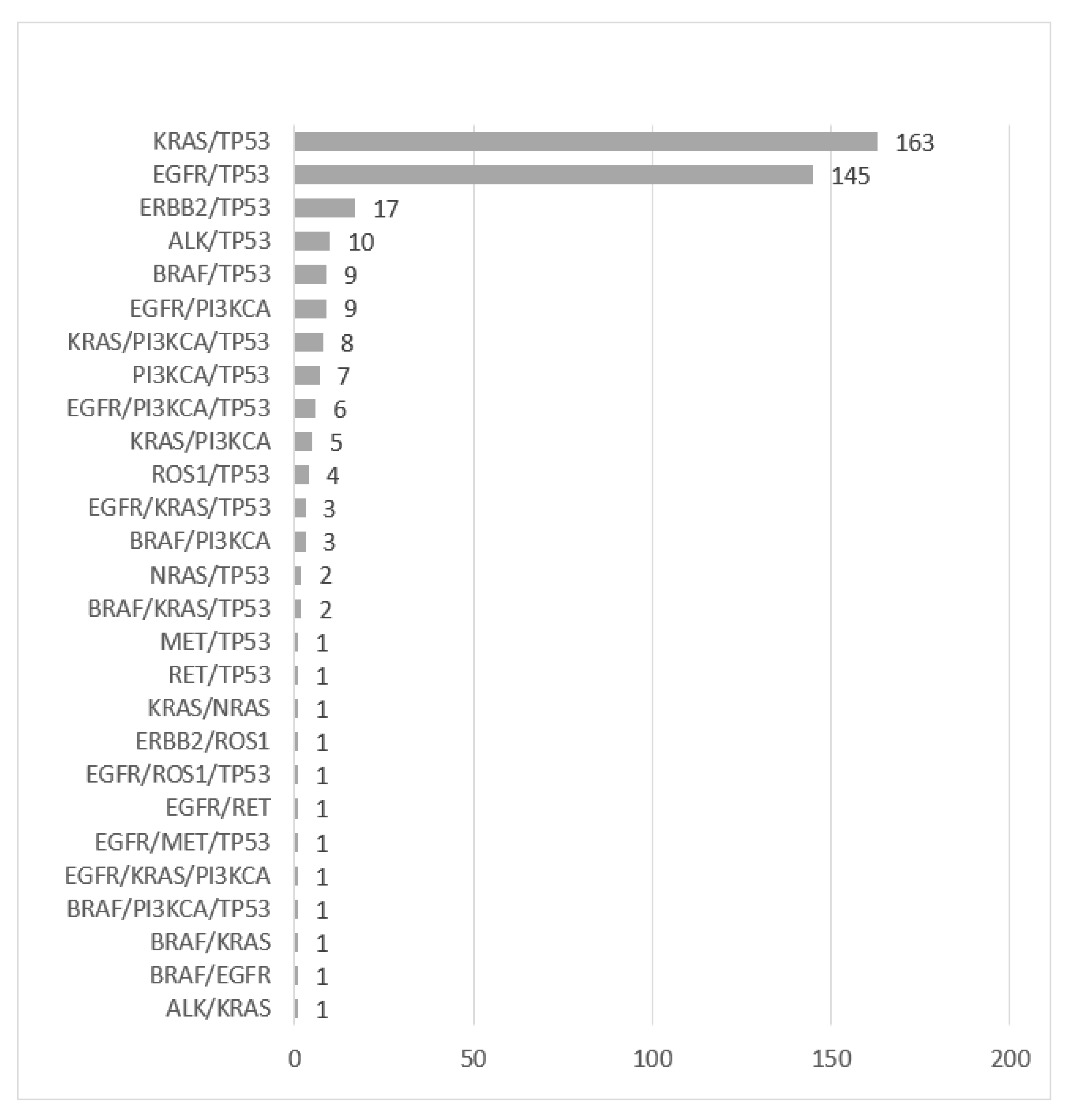
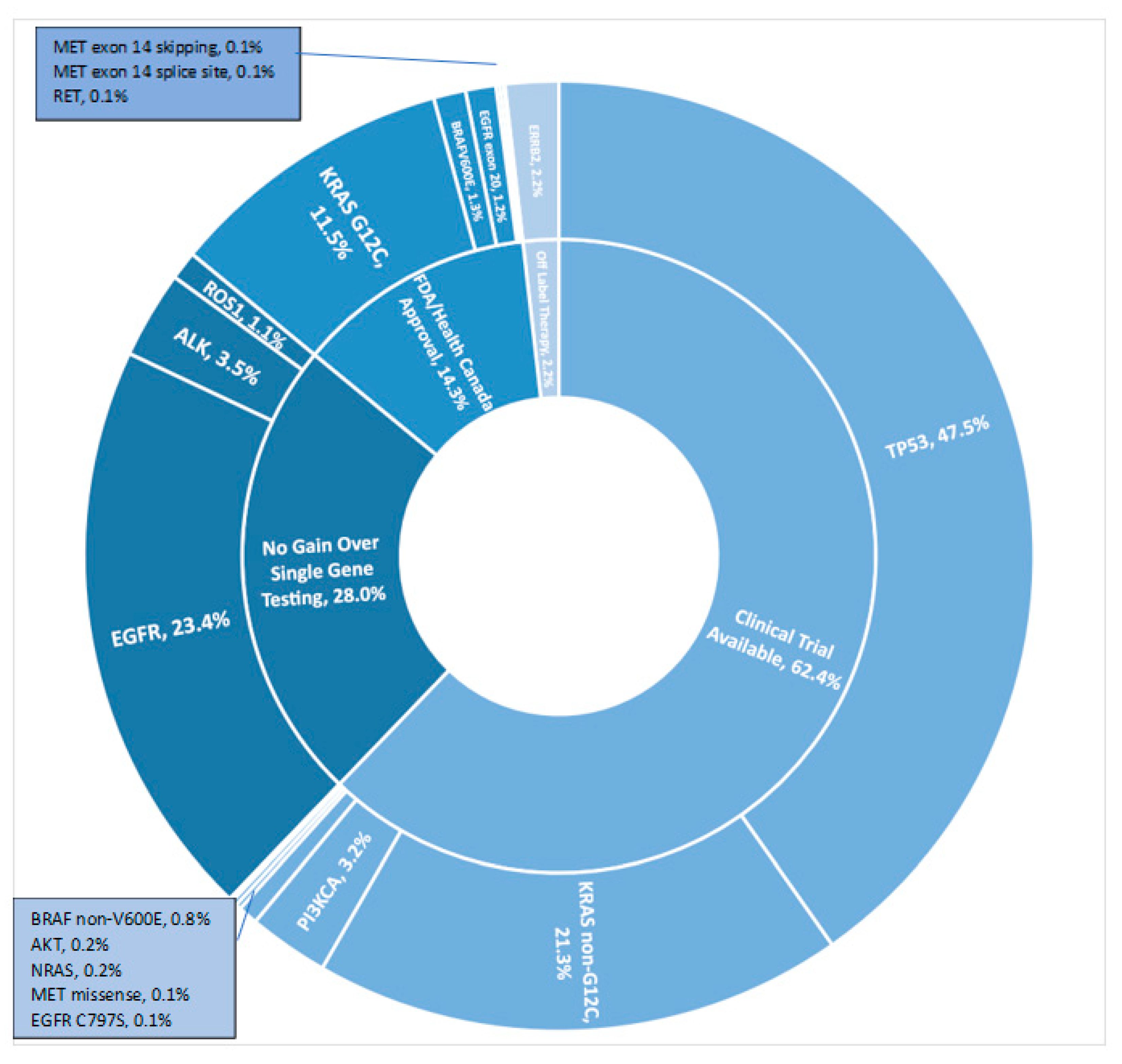
| Gene Variant | Never Smoker (N = 444) | Former/Current Smoker (N = 897) | All Patients (N = 1395) |
|---|---|---|---|
| AKT | 0 | 3 (0.3%) | 3 (0.2%) |
| BRAF | 7 (1.6%) | 21 (2.3%) | 29 (2.1%) |
| V600E * | 6 | 11 | 18 |
| Non-V600E | 1 | 10 | 11 |
| EGFR | 228 (51.4%) | 97 (10.8%) | 337 (24.2%) |
| Exon 19 deletion * | 102 | 41 | 147 |
| L858R * | 98 | 34 | 137 |
| Other Exon 19/20/21 | 12 | 12 | 27 |
| Exon 18 * | 13 | 10 | 24 |
| Exon 20 insertion * | 17 | 1 | 18 |
| T790M * | 11 | 15 | 16 |
| L861Q * | 5 | 5 | 10 |
| C797S | 1 | 0 | 1 |
| ≥2 EGFR variants | 28 | 11 | 40 |
| ERBB2 | 20 (4.5%) | 10 (1.1%) | 30 (2.2%) |
| Exon 20 | 17 | 5 | 22 |
| Transmembrane domain | 2 | 2 | 4 |
| Other | 1 | 3 | 4 |
| KRAS | 40 (9.0%) | 393 (43.8%) | 449 (32.2%) |
| G12C * | 4 | 152 | 161 |
| Non-G12C | 36 | 253 | 300 |
| ≥2 KRAS variants | 0 | 12 | 12 |
| MET | 1 (0.1%) | 3 (0.3%) | 4 (0.3%) |
| Exon 14 splice site * | 1 | 1 | 2 |
| Exon 14 skipping * | 0 | 1 | 1 |
| Non-splice site missense | 0 | 1 | 1 |
| NRAS | 1 (0.1%) | 2 (0.2%) | 3 (0.2%) |
| PI3KCA | 15 (3.4%) | 29 (3.2%) | 45 (3.2%) |
| RET * | 0 | 2 (0.2%) | 2 (0.1%) |
| TP53 | 168 (37.8%) | 469 (52.3%) | 662 (47.5%) |
| Gene Variant | <40 years (N = 19) | 40-65 years (N = 506) | ≥65 years (N = 870) |
|---|---|---|---|
| AKT | 0 | 1 (0.2%) | 2 (0.2%) |
| BRAF | 0 | 6 (1.2%) | 23 (2.6%) |
| V600E * | 0 | 5 | 13 |
| Non-V600E | 0 | 1 | 10 |
| EGFR | 2 (10.5%) | 132 (26.1%) | 203 (23.3%) |
| Exon 19 deletion * | 0 | 68 | 79 |
| L858R * | 1 | 48 | 88 |
| Other Exon 19/20/21 | 0 | 10 | 17 |
| Exon 18 * | 1 | 7 | 16 |
| Exon 20 insertion * | 0 | 7 | 11 |
| T790M * | 0 | 8 | 8 |
| L861Q * | 0 | 2 | 8 |
| C797S | 0 | 1 | 0 |
| ≥2 EGFR variants | 0 | 17 | 23 |
| ERBB2 | 1 (5.3%) | 14 (2.8%) | 15 (1.7%) |
| Exon 20 | 1 | 9 | 12 |
| Transmembrane domain | 0 | 2 | 2 |
| Other | 0 | 3 | 1 |
| KRAS | 1 (5.3%) | 153 (30.2%) | 295 (33.9%) |
| G12C * | 0 | 55 | 106 |
| Non-G12C | 1 | 102 | 197 |
| ≥2 KRAS variants | 0 | 4 | 8 |
| MET | 0 | 0 | 4 (0.5%) |
| Exon 14 splice site * | 0 | 0 | 2 |
| Exon 14 skipping * | 0 | 0 | 1 |
| Non-splice site missense | 0 | 0 | 1 |
| NRAS | 0 | 0 | 3 (0.3%) |
| PI3KCA | 0 | 13 (2.6%) | 32 (3.7%) |
| RET * | 0 | 2 (0.4%) | 0 |
| TP53 | 9 (47.4%) | 251 (49.6%) | 402 (46.2%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, S.; Fung, A.S.; Perdrizet, K.A.; Chen, K.; Li, J.J.N.; Le, L.W.; Cabanero, M.; Karsaneh, O.A.A.; Tsao, M.S.; Morganstein, J.; et al. Upfront Next Generation Sequencing in Non-Small Cell Lung Cancer. Curr. Oncol. 2022, 29, 4428-4437. https://doi.org/10.3390/curroncol29070352
Kuang S, Fung AS, Perdrizet KA, Chen K, Li JJN, Le LW, Cabanero M, Karsaneh OAA, Tsao MS, Morganstein J, et al. Upfront Next Generation Sequencing in Non-Small Cell Lung Cancer. Current Oncology. 2022; 29(7):4428-4437. https://doi.org/10.3390/curroncol29070352
Chicago/Turabian StyleKuang, Shelley, Andrea S. Fung, Kirstin A. Perdrizet, Kaitlin Chen, Janice J. N. Li, Lisa W. Le, Michael Cabanero, Ola Abu Al Karsaneh, Ming S. Tsao, Josh Morganstein, and et al. 2022. "Upfront Next Generation Sequencing in Non-Small Cell Lung Cancer" Current Oncology 29, no. 7: 4428-4437. https://doi.org/10.3390/curroncol29070352
APA StyleKuang, S., Fung, A. S., Perdrizet, K. A., Chen, K., Li, J. J. N., Le, L. W., Cabanero, M., Karsaneh, O. A. A., Tsao, M. S., Morganstein, J., Ranich, L., Smith, A. C., Wei, C., Cheung, C., Shepherd, F. A., Liu, G., Bradbury, P., Pal, P., Schwock, J., ... Leighl, N. B. (2022). Upfront Next Generation Sequencing in Non-Small Cell Lung Cancer. Current Oncology, 29(7), 4428-4437. https://doi.org/10.3390/curroncol29070352





