Method for the Intraoperative Detection of IDH Mutation in Gliomas with Differential Mobility Spectrometry
Abstract
1. Introduction
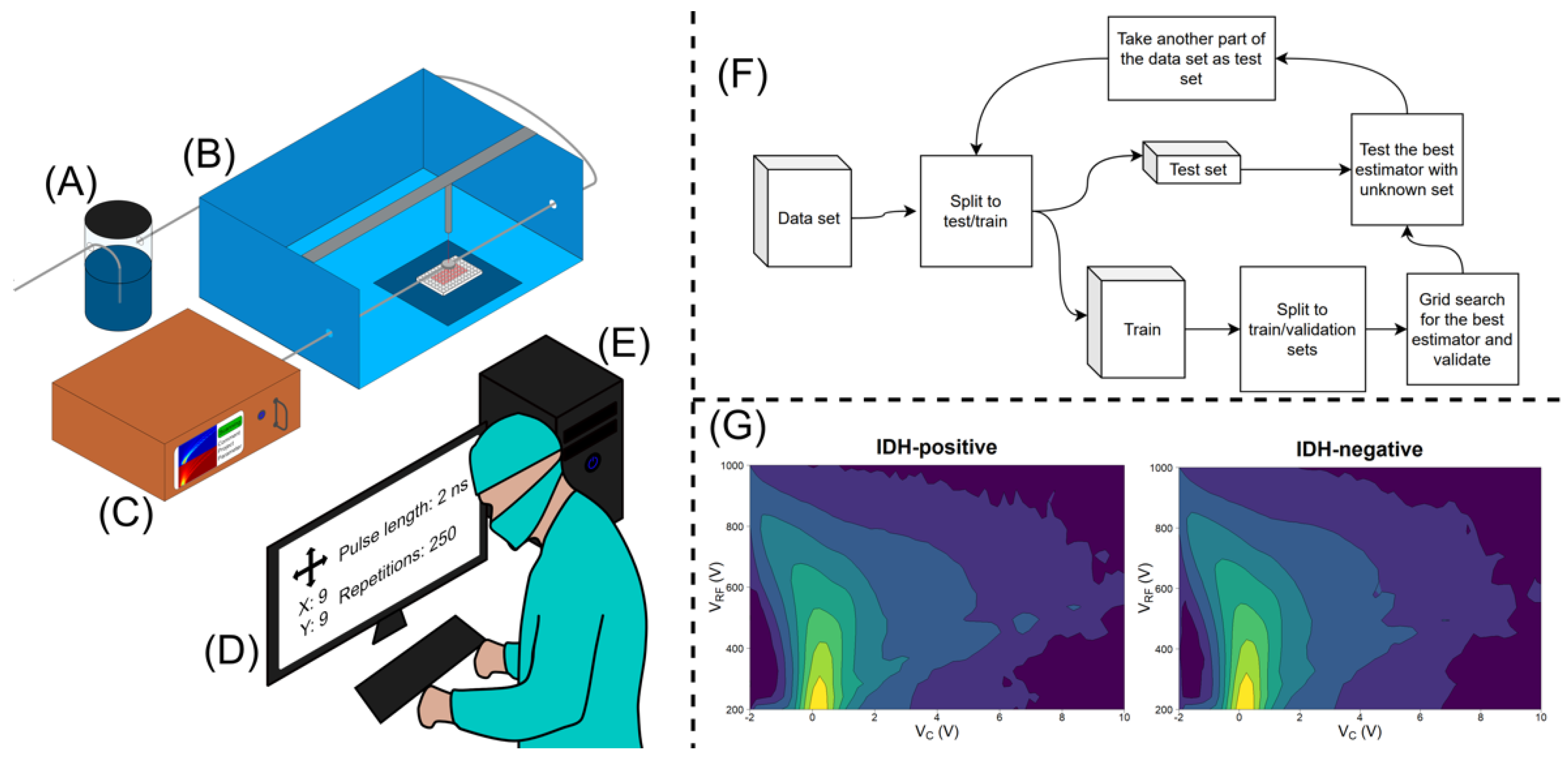
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.W.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Houillier, C.; Wang, X.; Kaloshi, G.; Mokhtari, K.; Guillevin, R.; Laffaire, J.; Paris, S.; Boisselier, B.; Idbaih, A.; Laigle-Donadey, F.; et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010, 75, 1560–1566. Available online: https://n.neurology.org/content/75/17/1560.short (accessed on 7 April 2021). [CrossRef] [PubMed]
- Metellus, P.; Coulibaly, B.; Colin, C.; de Paula, A.M.; Vasiljevic, A.; Taieb, D.; Barlier, A.; Boisselier, B.; Mokhtari, K.; Wang, X.W.; et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010, 120, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Songtao, Q.; Lei, Y.; Si, G.; Yanqing, D.; Huixia, H.; Xuelin, Z.; Lanxiao, W.; Fei, Y. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Wiley Online Libr. 2012, 103, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Juratli, T.A.; Kirsch, M.; Geiger, K.; Klink, B.; Leipnitz, E.; Pinzer, T.; Soucek, S.; Schrok, E.; Schackert, G.; Krex, D. The prognostic value of IDH mutations and MGMT promoter status in secondary high-grade gliomas. J. Neurooncol. 2012, 110, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Ricci, A.D.; Tober, N.; Nigro, M.C.; Mosca, M.; Palloni, A.; Abbati, F.; Frega, G.; De Lorenzo, S.; Tavolari, S.; et al. Second-line Treatment in Advanced Biliary Tract Cancer: Today and Tomorrow. Anticancer Res. 2020, 40, 3013–3030. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Ricci, A.D.; Brandi, G. IDH inhibitors in advanced cholangiocarcinoma: Another arrow in the quiver? Cancer Treat. Res. Commun. 2021, 27, 100356. [Google Scholar] [CrossRef]
- Rizzo, A.; Brandi, G. First-line Chemotherapy in Advanced Biliary Tract Cancer Ten Years after the ABC-02 Trial: “And Yet It Moves!”. Cancer Treat. Res. Commun. 2021, 27, 100335. [Google Scholar] [CrossRef]
- Waitkus, M.S.; Diplas, B.H.; Yan, H. Isocitrate dehydrogenase mutations in gliomas. Neuro-Oncology 2015, 18, 16–26. Available online: https://academic.oup.com/neuro-oncology/article-abstract/18/1/16/2509155 (accessed on 7 April 2021). [CrossRef]
- Fack, F.; Tardito, S.; Hochart, G.; Oudin, A.; Zheng, L.; Fritah, S.; Golebiewska, A.; Nazarov, P.; Bernard, A.; Hau, A.; et al. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol. Med. 2017, 9, 1681–1695. [Google Scholar] [CrossRef]
- Garrett, M.; Sperry, J.; Braas, D.; Yan, W.; Le, T.M.; Mottahedeh, J.; Ludwig, K.; Eskin, A.; Qin, Y.; Levy, R.; et al. Metabolic characterization of isocitrate dehydrogenase (IDH) mutant and IDH wildtype gliomaspheres uncovers cell type-specific vulnerabilities. Cancer Metab. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Beiko, J.; Suki, D.; Hess, K.R.; Fox, B.D.; Cheung, V.; Cabral, M.; Shonka, N.; Gilbert, M.R.; Sawaya, R.; Prabhu, S.S.; et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro-Oncology 2014, 16, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, T.; Sonoda, Y.; Shibahara, I.; Saito, R.; Kanamori, M.; Kumabe, T.; Tominaga, T. Impact of gross total resection in patients with WHO grade III glioma harboring the IDH 1/2 mutation without the 1p/19q co-deletion. J. Neurooncol. 2016, 129, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.; Bander, E.D.; Venn, R.A.; Powell, T.; Cederquist, G.Y.-M.; Schaefer, P.M.; Puchi, L.A.; Akhmerov, A.; Ogilvie, S.; Reiner, A.S.; et al. The role of extent of resection in IDH1 wild-type or mutant low-grade gliomas. Neurosurgery 2018, 82, 808–814. [Google Scholar] [CrossRef]
- Montemurro, N.; Fanelli, G.N.; Scatena, C.; Ortenzi, V.; Pasqualetti, F.; Mazzanti, C.M.; Morganti, R.; Paiar, F.; Naccarato, A.G.; Perrini, P. Surgical outcome and molecular pattern characterization of recurrent glioblastoma multiforme: A single-center retrospective series. Clin. Neurol Neurosurg. 2021, 207, 106735. [Google Scholar] [CrossRef]
- Van Tellingen, O.; Yetkin-Arik, B.; De Gooijer, M.C.; Wesseling, P.; Wurdinger, T.; De Vries, H.E. Overcoming the blood–brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 2015, 19, 1–12. [Google Scholar] [CrossRef]
- Longuespée, R.; Wefers, A.K.; De Vita, E.; Miller, A.K.; Reuss, D.E.; Wick, W.; Herold-Mende, C.; Kriegsmann, M.; Schirmacher, P.; Von Deimling, A.; et al. Rapid detection of 2-hydroxyglutarate in frozen sections of IDH mutant tumors by MALDI-TOF mass spectrometry. Acta Neuropathol. Commun. 2018, 6, 21. [Google Scholar] [CrossRef]
- Haapala, I.; Karjalainen, M.; Kontunen, A.S.; Vehkaoja, A.; Nordfors, K.; Haapasalo, H.; Haapasalo, J.; Oksala, N.; Roine, A. Identifying brain tumors by differential mobility spectrometry analysis of diathermy smoke. J. Neurosurg. 2020, 133, 100–106. [Google Scholar] [CrossRef]
- Schneider, B.B.; Nazarov, E.G.; Londry, F.; Vouros, P.; Covey, T.R. Differential mobility spectrometry/mass spectrometry history, theory, design optimization, simulations, and applications: Differential Mobility Spectrometry/Mass Spectrometry Non-radioactive Ion Source for Operation in Ambient Pressure View project Properties of Gas Phase Molecular Clusters View project Differential Mobility Spectrometry/Mass Spectrometry History, Theory, Design Optimization, Simulations, and Applications. Wiley Online Libr. 2016, 35, 687–737. [Google Scholar] [CrossRef]
- Sciortino, T.; Secoli, R.; D’Amico, E.; Moccia, S.; Nibali, M.C.; Gay, L.; Rossi, M.; Pecco, N.; Castellano, A.; De Momi, E.; et al. Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies. Cancers 2021, 13, 4196. [Google Scholar] [CrossRef]
- Decordova, S.; Shastri, A.; Tsolaki, A.G.; Yasmin, H.; Klein, L.; Singh, S.K.; Kishore, U. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front. Immunol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Campos, B.; Olsen, L.R.; Urup, T.; Poulsen, H.S. A comprehensive profile of recurrent glioblastoma. Oncogene 2016, 35, 5819–5825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Min, W.; Shen, L.; Zhang, Y.; Yue, Z. SURG-25. A novel bio-impedance spectroscopy system real-time intraoperatively discriminates glioblastoma from brain tissue in mice. Neuro-Oncology 2017, 19 (Suppl. 6), vi240. [Google Scholar] [CrossRef]
| IDH Mutation | − | 150 | 26 |
|---|---|---|---|
| + | 25 | 151 | |
| − | + | ||
| Classification result | |||
| Sens. 0.85 | Spec. 0.85 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haapala, I.; Rauhameri, A.; Roine, A.; Mäkelä, M.; Kontunen, A.; Karjalainen, M.; Laakso, A.; Koroknay-Pál, P.; Nordfors, K.; Haapasalo, H.; et al. Method for the Intraoperative Detection of IDH Mutation in Gliomas with Differential Mobility Spectrometry. Curr. Oncol. 2022, 29, 3252-3258. https://doi.org/10.3390/curroncol29050265
Haapala I, Rauhameri A, Roine A, Mäkelä M, Kontunen A, Karjalainen M, Laakso A, Koroknay-Pál P, Nordfors K, Haapasalo H, et al. Method for the Intraoperative Detection of IDH Mutation in Gliomas with Differential Mobility Spectrometry. Current Oncology. 2022; 29(5):3252-3258. https://doi.org/10.3390/curroncol29050265
Chicago/Turabian StyleHaapala, Ilkka, Anton Rauhameri, Antti Roine, Meri Mäkelä, Anton Kontunen, Markus Karjalainen, Aki Laakso, Päivi Koroknay-Pál, Kristiina Nordfors, Hannu Haapasalo, and et al. 2022. "Method for the Intraoperative Detection of IDH Mutation in Gliomas with Differential Mobility Spectrometry" Current Oncology 29, no. 5: 3252-3258. https://doi.org/10.3390/curroncol29050265
APA StyleHaapala, I., Rauhameri, A., Roine, A., Mäkelä, M., Kontunen, A., Karjalainen, M., Laakso, A., Koroknay-Pál, P., Nordfors, K., Haapasalo, H., Oksala, N., Vehkaoja, A., & Haapasalo, J. (2022). Method for the Intraoperative Detection of IDH Mutation in Gliomas with Differential Mobility Spectrometry. Current Oncology, 29(5), 3252-3258. https://doi.org/10.3390/curroncol29050265





